Association Mapping Analysis of Morphological Characteristics in F2 Population of Perilla (Perilla frutescens L.) Using SSR Markers
Abstract
1. Introduction
2. Results
2.1. SSR Identification and Polymorphisms
2.2. Phenotypic Variation and Association Analysis of 13 Qualitative and Quantitative Traits
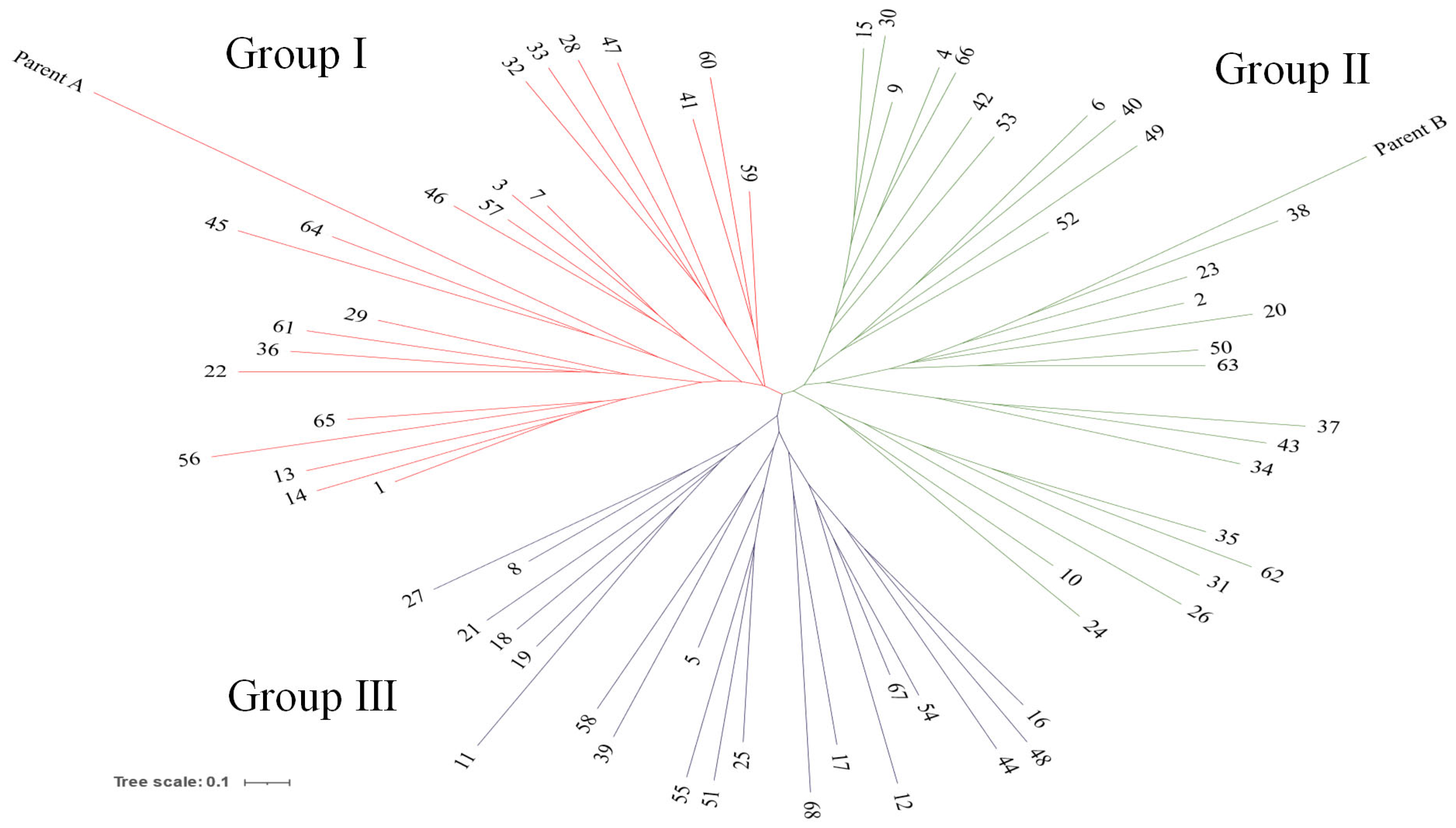
2.3. Genetic Verification of SSR Markers Among the F2 Population of Perilla
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Morphological Characteristics of F2 Population
4.2. DNA Extraction and SSR Analysis
4.3. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, J.K.; Ohnishi, O. Geographical differentiation of morphological characters among Perilla crop and their weedy types in East Asia. Breed. Sci. 2001, 51, 247–255. [Google Scholar] [CrossRef]
- Nitta, M.; Lee, J.K.; Ohnishi, O. Asian Perilla crop and their weedy forms: Their cultivation, utilization and genetic relationships. Econ. Bot. 2003, 57, 245–253. [Google Scholar] [CrossRef]
- Cho, J.; Park, H.; Heo, T.H.; Lee, J.K. Association mapping analysis (AMA) for morpho-agronomic traits and leaf aromatic compounds using SSR markers in three types of Perilla crop collected from South Korea. Genes Genom. 2024, 46, 1399–1413. [Google Scholar] [CrossRef]
- Honda, G.; Koezuka, Y.; Tabata, M. Genetic studies of fruit color and hardness in Perilla frutescens. Jpn. J. Breed. 1990, 40, 469–474. [Google Scholar] [CrossRef]
- Honda, G.; Yuba, A.; Kojima, T.; Tabata, M. Chemotaxonomic and cytogenetic studies on Perilla frutescens var. citriodora (‘Lemon Egoma’). Nat. Med. 1994, 48, 185–190. [Google Scholar]
- Lim, S.E.; Sa, K.J.; Ha, Y.J.; Lee, J.K. Bulk segregant analysis identifies SSR markers associated with leaf- and seed-related traits in Perilla crop (Perilla frutescens L.). Genes Genom. 2021, 43, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Yamane, Y. Cytogenetic studies in Perilla and Coleus. I. Chromosome numbers. Jpn. J. Genet. 1950, 25, 220. (In Japanese) [Google Scholar]
- Nitta, M.; Ohnishi, O. Genetic relationships among two Perilla crop, shiso and egoma, and the weedy type revealed by RAPD markers. Jpn. J. Genet. 1999, 74, 43–48. [Google Scholar] [CrossRef]
- Mazzucato, A.; Papa, R.; Bitocchi, E.; Mosconi, P.; Nanni, L.; Negri, V.; Picarella, M.E.; Siligato, F.; Soressi, G.P.; Tiranti, B.; et al. Genetic diversity, structure and marker-trait associations in a collection of Italian tomato (Solanum lycopersicum L.) landraces. Theor. Appl. Genet. 2008, 116, 657–669. [Google Scholar] [CrossRef]
- Mackay, T.F. The genetic architecture of quantitative traits. Annu. Rev. Genet. 2001, 35, 303–339. [Google Scholar] [CrossRef]
- Buckler, E.S.; Thornsberry, J.M. Plant molecular diversity and applications to genomics. Curr. Opin. Plant Biol. 2002, 5, 107–111. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Davasi, A.; Shifman, S. The beauty of admixture. Nat. Genet. 2005, 37, 118–119. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Skot, L.; Humphreys, M.O.; Armstead, I. An association mapping approach to identify flowering time genes in natural populations of Lolium perenne (L.). Mol. Breed. 2005, 15, 233–245. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, C.; Ren, F.; Li, Y.; Zhang, C. Association analysis of important agronomical traits of maize inbred lines with SSRs. Aust. J. Crop Sci. 2012, 6, 1131–1138. [Google Scholar]
- Tanksley, S.D.; Nelson, J.C. Advanced backcross QTL analysis: A method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 1996, 92, 191–203. [Google Scholar] [CrossRef]
- Yan, J.; Wu, Y.; Li, W.; Qin, X.; Wang, Y.; Yue, B. Genetic mapping with testcrossing associations and F2:3 populations reveals the importance of heterosis in chilling tolerance at maize seedling stage. Sci. Rep. 2017, 7, 3232. [Google Scholar] [CrossRef] [PubMed]
- Farré, A.; Sayers, L.; Leverington-Waite, M.; Goram, R.; Orford, S.; Wingen, L.; Mumford, C.; Griffiths, S. Application of a library of near isogenic lines to understand context dependent expression of QTL for grain yield and adaptive traits in bread wheat. BMC Plant Biol. 2016, 16, 161. [Google Scholar] [CrossRef]
- Choi, J.K.; Sa, K.J.; Park, D.H.; Lim, S.E.; Ryu, S.H.; Park, J.Y.; Park, K.J.; Rhee, H.I.; Lee, M.; Lee, J.K. Construction of genetic linkage map and identification of QTLs related to agronomic traits in DH population of maize (Zea mays L.) using SSR markers. Genes Genom. 2019, 41, 667–678. [Google Scholar] [CrossRef]
- Kim, J.Y.; Sa, K.J.; Ha, Y.J.; Lee, J.K. Genetic variation and association mapping in the F2 population of the Perilla crop (Perilla frutescens L.) using newly developed Perilla SSR markers. Euphytica 2021, 217, 135. [Google Scholar] [CrossRef]
- Zhao, M.; Shu, G.; Hu, Y.; Cao, G.; Wang, Y. Pattern and variation in simple sequence repeat (SSR) at different genomic regions and its implications to maize evolution and breeding. BMC Genomics. 2023, 24, 136. [Google Scholar] [CrossRef]
- Sa, K.J.; Choi, I.K.; Park, K.C.; Lee, J.K. Genetic diversity and population structure among accessions of Perilla frutescens (L.) Britton in East Asia using newly developed microsatellite markers. Genes Genom. 2018, 40, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.M.; Joel, J.A.; Raveendran, M.; Pushpam, R.; Muthuramu, S.; Pushpa, R.; Sritharan, N.; Prasanna, P.; Suresh, R. Unravelling population structure and marker trait association using SSR markers among the identified drought tolerant rice landraces (Oryza sativa L.). Czech J. Genet. Plant Breed. 2025, 61, 1–22. [Google Scholar] [CrossRef]
- Crossa, J.; Burgueño, J.; Dreisigacker, S.; Vargas, M.; Herrera-Foessel, S.A.; Lillemo, M.; Singh, R.P.; Trethowan, R.; Warburton, M.; Franco, J.; et al. Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 2007, 177, 1889–1913. [Google Scholar] [CrossRef]
- Heo, T.H.; Park, H.; Cho, J.; Lee, D.H.; Lee, J.K. Morphological variation of F2 population derived from the cross between Perilla frutescens var. crispa and var. frutescens. Plant Breed. Biotech. 2025, 13, 119–130. [Google Scholar] [CrossRef]
- Park, H.; Heo, T.H.; Cho, J.; Choi, H.Y.; Lee, D.H.; Lee, J.K. Evaluation and characteristic analysis of SSRs from the transcriptomic sequences of Perilla crop (Perilla frutescens L.). Gene 2025, 933, 148938. [Google Scholar] [CrossRef]
- Strome, S.; Bhalla, N.; Kamakaka, R.; Sharma, U.; Sullivan, W. Clarifying Mendelian vs non-Mendelian inheritance. Genetics 2024, 227, iyae078. [Google Scholar] [CrossRef]
- Najafabadi, M.Y.; Hesami, M.; Rajcan, I. Unveiling the Mysteries of Non-Mendelian Heredity in Plant Breeding. Plants 2023, 12, 1956. [Google Scholar] [CrossRef] [PubMed]
- Dittrich-Reed, D.R.; Fitzpatrick, B.M. Transgressive Hybrids as Hopeful Monsters. Evol. Biol. 2013, 40, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Scheid, O.M. Mendelian and non-Mendelian genetics in model plants. Plant Cell 2022, 34, 2455–2461. [Google Scholar] [CrossRef]
- Martínez-Castilla, L.P.; Álvarez-Buylla, E.R. Adaptive evolution in the Arabidopsis MADS-box gene family inferred from the substitution rate variation among lineages and sites. Mol. Biol. Evol. 2003, 20, 1951–1963. [Google Scholar] [CrossRef]
- Wolko, J.; Lopatynska, A.; Wolko, L.; Bocianowski, J.; Mikołajczyk, K.; Liersch, A. Identification of SSR Markers Associated with Yield-Related Traits and Heterosis Effect in Winter Oilseed Rape (Brassica Napus L.). Agronomy 2022, 12, 1544. [Google Scholar] [CrossRef]
- Sun, Z.; An, H.; Qiu, Z.; Li, J.; Li, J.; Yang, B.; Liu, J.; Chen, T.; Zhang, Y.; Lu, B.; et al. Identification of QTLs and a candidate gene affecting rice grain volume via high-density genetic mapping. Front. Plant Sci. 2025, 16, 1579589. [Google Scholar] [CrossRef]
- Jamison, D.R.; Chen, P.; Hettiarachchy, N.S.; Miller, D.M.; Shakiba, E. Identification of Quantitative Trait Loci (QTL) for Sucrose and Protein Content in Soybean Seed. Plants 2024, 13, 650. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Myles, S.; Peiffer, J.; Brown, P.J.; Ersoz, E.S.; Zhang, Z.; Costich, D.E.; Buckler, E.S. Association mapping: Critical considerations shift from genotyping to experimental design. Plant Cell 2009, 21, 2194–2202. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996, 2, 225–238. [Google Scholar] [CrossRef]
- Yazdani, R.; Scotti, I.; Jansson, G.; Plomion, C.; Mathur, G. Inheritance and diversity of simple sequence repeat (SSR) microsatellite markers in various families of Picea abies. Hereditas 2003, 138, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, Y.; Cai, Q.; Liao, C.; Mao, X.; Xie, H.; Zhu, Y.; Lian, L.; Luo, X.; Xie, H.; et al. Population structure and association analysis of yield and grain quality traits in hybrid rice primal parental lines. Euphytica 2016, 212, 261–273. [Google Scholar] [CrossRef]
- Galal, A.A.; Safhi, F.A.; Al Aboud, N.M.; Aljabri, M.; Kucher, D.E.; Kamara, M.M.; El-Mogy, M.M.; Ibrahim, O.M.; El-Moneim, D.A.; Hassanin, A.A.; et al. Molecular diversity and genetic potential of new maize inbred lines across varying sowing conditions in arid environment. Sci. Rep. 2025, 15, 2809. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, J.; Li, M.; Zhao, X.; Zhao, L. Genetic analysis and QTL mapping of growth period traits and plant height traits in soybean recombinant inbred lines from Dongnong 47 × PI 317334-B. Oil Crop Sci. 2023, 6, 66–73. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]


| Marker | GD | PIC | MAF | Separation of the F2 Population | |||
|---|---|---|---|---|---|---|---|
| AA | AB | BB | NULL | ||||
| GBPFM179 | 0.523 | 0.468 | 0.643 | 12 | 45 | 11 | 0 |
| KWPE19 | 0.625 | 0.552 | 0.486 | 21 | 34 | 13 | 0 |
| KWPE58 | 0.625 | 0.555 | 0.500 | 17 | 35 | 16 | 0 |
| KNUPF2 | 0.599 | 0.519 | 0.500 | 7 | 35 | 25 | 1 |
| KNUPF3 | 0.608 | 0.531 | 0.500 | 10 | 35 | 23 | 0 |
| KNUPF4 | 0.676 | 0.618 | 0.429 | 10 | 30 | 22 | 6 |
| KNUPF9 | 0.630 | 0.559 | 0.486 | 15 | 34 | 19 | 0 |
| KNUPF12 | 0.570 | 0.507 | 0.586 | 15 | 41 | 12 | 0 |
| KNUPF14 | 0.569 | 0.504 | 0.586 | 11 | 41 | 16 | 0 |
| KNUPF15 | 0.644 | 0.570 | 0.443 | 15 | 31 | 22 | 0 |
| KNUPF16 | 0.589 | 0.522 | 0.557 | 17 | 39 | 12 | 0 |
| KNUPF23 | 0.570 | 0.507 | 0.586 | 15 | 41 | 12 | 0 |
| KNUPF29 | 0.597 | 0.528 | 0.543 | 18 | 38 | 12 | 0 |
| KNUPF30 | 0.622 | 0.551 | 0.500 | 14 | 35 | 19 | 0 |
| KNUPF31 | 0.606 | 0.525 | 0.486 | 9 | 34 | 25 | 0 |
| KNUPF36 | 0.650 | 0.575 | 0.400 | 25 | 28 | 15 | 0 |
| KNUPF37 | 0.601 | 0.534 | 0.543 | 15 | 38 | 15 | 0 |
| KNUPF39 | 0.601 | 0.534 | 0.543 | 15 | 38 | 15 | 0 |
| KNUPF40 | 0.620 | 0.548 | 0.500 | 13 | 35 | 20 | 0 |
| KNUPF42 | 0.643 | 0.567 | 0.429 | 24 | 30 | 14 | 0 |
| KNUPF50 | 0.616 | 0.546 | 0.514 | 14 | 36 | 18 | 0 |
| KNUPF59 | 0.640 | 0.565 | 0.443 | 14 | 31 | 23 | 0 |
| KNUPF61 | 0.630 | 0.555 | 0.471 | 22 | 33 | 13 | 0 |
| KNUPF81 | 0.653 | 0.580 | 0.429 | 19 | 30 | 19 | 0 |
| KNUPF82 | 0.563 | 0.505 | 0.600 | 15 | 42 | 10 | 1 |
| KNUPF83 | 0.558 | 0.497 | 0.600 | 11 | 42 | 15 | 0 |
| KNUPF93 | 0.639 | 0.569 | 0.471 | 13 | 33 | 21 | 1 |
| KNUPF112 | 0.495 | 0.430 | 0.657 | 5 | 46 | 17 | 0 |
| KNUPF127 | 0.626 | 0.550 | 0.471 | 12 | 33 | 23 | 0 |
| KNUPF130 | 0.591 | 0.524 | 0.557 | 13 | 39 | 16 | 0 |
| KNUPF156 | 0.592 | 0.516 | 0.529 | 9 | 37 | 22 | 0 |
| KNUPF162 | 0.625 | 0.552 | 0.486 | 13 | 34 | 21 | 0 |
| KNUPF163 | 0.661 | 0.591 | 0.414 | 22 | 29 | 16 | 1 |
| KNUPF167 | 0.464 | 0.418 | 0.700 | 8 | 49 | 11 | 0 |
| KNUPF168 | 0.640 | 0.565 | 0.443 | 14 | 31 | 23 | 0 |
| KNUPF169 | 0.645 | 0.578 | 0.471 | 17 | 33 | 17 | 1 |
| KNUPF170 | 0.608 | 0.531 | 0.500 | 10 | 35 | 23 | 0 |
| KNUPF176 | 0.570 | 0.507 | 0.586 | 15 | 41 | 12 | 0 |
| KNUPF182 | 0.630 | 0.555 | 0.471 | 13 | 33 | 22 | 0 |
| KNUPF191 | 0.656 | 0.582 | 0.400 | 17 | 28 | 23 | 0 |
| Max | 0.676 | 0.618 | 0.700 | ||||
| Min | 0.464 | 0.418 | 0.400 | ||||
| Mean | 0.607 | 0.537 | 0.511 | ||||
| QL1 | QL2 | QL3 | QL4 | QN1 | QN2 | QN3 | QN4 | QN5 | QN6 | QN7 | QN8 | QN9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parent A | G/P | Purple | Purple | Purple | 124 | 134 | 168 | 151.4 | 8.6 | 33.3 | 12.6 | 8.3 | 59.8 |
| Parent B | Green | Green | Green | White | 119 | 126 | 157 | 105.4 | 8.5 | 44 | 12.6 | 11.2 | 88.8 |
| 1 | Green | G/P | G/P | Pink | 119 | 131 | 164 | 149.8 | 12.7 | 48 | 11.1 | 7.1 | 50.6 |
| 2 | Green | Green | Green | White | 116 | 130 | 160 | 139.4 | 12.2 | 52 | 11.6 | 8.6 | 62.4 |
| 3 | Green | G/P | G/P | Pink | 121 | 131 | 161 | 143.2 | 14.5 | 56 | 10.8 | 7.2 | 46.5 |
| 4 | Green | Green | G/P | White | 122 | 132 | 166 | 154.8 | 14.7 | 52 | 10.9 | 7.5 | 49.2 |
| 5 | Green | Green | Green | White | 122 | 136 | 165 | 174.3 | 13.8 | 52 | 12.4 | 9.1 | 71.5 |
| 6 | Green | Green | G/P | Pink | 121 | 133 | 171 | 148.7 | 9.1 | 32 | 11.6 | 8.9 | 64.9 |
| 7 | Green | Purple | G/P | Purple | 121 | 130 | 152 | 130.8 | 13.3 | 44 | 11.5 | 9.2 | 64.9 |
| 8 | Green | Purple | Purple | Purple | 119 | 129 | 158 | 159.8 | 6 | 32 | 12.2 | 8.3 | 58.6 |
| 9 | Green | G/P | G/P | White | 120 | 128 | 159 | 120.2 | 9.2 | 40 | 10.2 | 7.8 | 48.0 |
| 10 | Green | G/P | G/P | White | 116 | 125 | 157 | 150.6 | 13.8 | 44 | 11.8 | 8.9 | 67.7 |
| 11 | Green | Green | G/P | Pink | 123 | 133 | 164 | 163.9 | 10.5 | 36 | 11.9 | 8.1 | 56.9 |
| 12 | Green | Green | G/P | Pink | 124 | 128 | 166 | 158.7 | 11.7 | 44 | 13.2 | 9.2 | 75.7 |
| 13 | Green | Green | G/P | Pink | 121 | 132 | 160 | 165.4 | 15.4 | 48 | 11.6 | 9.2 | 64.3 |
| 14 | Green | Green | G/P | Pink | 125 | 133 | 161 | 133.4 | 11.5 | 48 | 11.5 | 8.7 | 64.3 |
| 15 | Green | Green | G/P | White | 116 | 126 | 158 | 149.4 | 8.6 | 32 | 11.4 | 9.0 | 63.2 |
| 16 | Green | Purple | G/P | White | 116 | 128 | 157 | 106.4 | 12.3 | 52 | 11.5 | 10.5 | 75.5 |
| 17 | Green | Green | Green | White | 123 | 131 | 159 | 171.4 | 13.3 | 44 | 11.5 | 8.9 | 64.6 |
| 18 | Green | G/P | G/P | Pink | 121 | 132 | 161 | 134.7 | 11.6 | 48 | 10.5 | 7.4 | 47.0 |
| 19 | Green | G/P | G/P | White | 117 | 129 | 159 | 156.3 | 13.4 | 48 | 10.8 | 8.1 | 55.8 |
| 20 | Green | G/P | G/P | Pink | 123 | 135 | 164 | 138.7 | 11.6 | 52 | 12.0 | 8.3 | 63.9 |
| 21 | Green | G/P | G/P | Purple | 121 | 131 | 164 | 149.2 | 12.8 | 44 | 10.0 | 6.2 | 39.5 |
| 22 | Green | Purple | G/P | Purple | 125 | 131 | 163 | 140.6 | 9.9 | 36 | 11.0 | 7.0 | 46.3 |
| 23 | Green | G/P | G/P | Pink | 121 | 131 | 166 | 160.8 | 11.8 | 40 | 11.8 | 7.7 | 53.0 |
| 24 | Green | Purple | G/P | Purple | 120 | 128 | 159 | 174.2 | 8.2 | 28 | 11.4 | 7.9 | 53.3 |
| 25 | Green | Purple | G/P | Purple | 122 | 131 | 167 | 174.1 | 10.5 | 40 | 13.0 | 8.3 | 65.3 |
| 26 | Green | Green | G/P | Pink | 125 | 135 | 166 | 164.1 | 10.6 | 40 | 12.1 | 7.8 | 56.3 |
| 27 | Green | G/P | G/P | Pink | 121 | 133 | 165 | 158.4 | 10.6 | 44 | 11.4 | 6.9 | 46.1 |
| 28 | Green | G/P | G/P | Pink | 119 | 130 | 160 | 144.4 | 7.4 | 28 | 11.4 | 7.0 | 37.7 |
| 29 | Green | G/P | G/P | Pink | 120 | 130 | 161 | 134.7 | 9.7 | 44 | 12.1 | 8.8 | 41.3 |
| 30 | Green | G/P | G/P | Purple | 118 | 128 | 162 | 141.1 | 14.3 | 48 | 12.4 | 9.3 | 46.1 |
| 31 | Green | G/P | G/P | Pink | 124 | 135 | 165 | 163.2 | 12.2 | 44 | 12.5 | 9.1 | 69.2 |
| 32 | Green | G/P | G/P | White | 117 | 129 | 161 | 134.8 | 18.6 | 60 | 11.7 | 7.7 | 53.7 |
| 33 | Green | G/P | G/P | Purple | 121 | 136 | 171 | 138.7 | 8.8 | 36 | 12.4 | 8.3 | 60.0 |
| 34 | Green | Purple | G/P | Pink | 121 | 131 | 166 | 161.9 | 13.5 | 44 | 12.0 | 7.9 | 55.4 |
| 35 | Green | G/P | G/P | Pink | 120 | 129 | 160 | 154.3 | 9.2 | 28 | 12.1 | 7.8 | 57.3 |
| 36 | Green | Green | G/P | Purple | 124 | 135 | 167 | 143.1 | 9.6 | 44 | 10.8 | 7.8 | 51.1 |
| 37 | Green | G/P | G/P | Purple | 124 | 132 | 160 | 154.9 | 11.4 | 44 | 11.5 | 7.3 | 49.6 |
| 38 | Green | Green | Green | White | 120 | 130 | 164 | 164.9 | 16.2 | 56 | 12.1 | 8.9 | 63.9 |
| 39 | Green | Green | G/P | White | 120 | 131 | 171 | 156.3 | 14 | 44 | 11.7 | 6.9 | 46.7 |
| 40 | Green | G/P | G/P | Pink | 121 | 131 | 163 | 165.8 | 15.6 | 52 | 11.9 | 8.9 | 63.2 |
| 41 | Green | Green | Green | White | 125 | 134 | 170 | 164.3 | 10.7 | 52 | 13.5 | 10.4 | 86.5 |
| 42 | Green | G/P | G/P | White | 123 | 133 | 162 | 172.7 | 17.9 | 52 | 12.0 | 9.3 | 69.3 |
| 43 | Green | Purple | G/P | Purple | 121 | 132 | 163 | 170.9 | 19.2 | 64 | 15.0 | 10.2 | 87.6 |
| 44 | Green | G/P | G/P | Pink | 125 | 135 | 163 | 166.2 | 13.8 | 44 | 12.7 | 8.9 | 69.4 |
| 45 | Green | G/P | G/P | Pink | 125 | 132 | 167 | 157.3 | 15 | 44 | 13.2 | 8.8 | 68.4 |
| 46 | Green | G/P | Green | White | 121 | 132 | 166 | 146.3 | 13.1 | 40 | 14.7 | 10.9 | 95.6 |
| 47 | Green | G/P | G/P | Pink | 127 | 137 | 161 | 145.8 | 5.7 | 28 | 14.6 | 9.9 | 88.1 |
| 48 | Green | G/P | G/P | Pink | 122 | 135 | 166 | 168.3 | 11.9 | 44 | 12.7 | 9.0 | 72.9 |
| 49 | Green | G/P | G/P | White | 121 | 130 | 161 | 159.6 | 9.8 | 28 | 13.0 | 9.9 | 80.8 |
| 50 | Green | Purple | G/P | Purple | 124 | 132 | 168 | 167.7 | 11.5 | 48 | 14.9 | 10.6 | 98.9 |
| 51 | Green | G/P | G/P | Pink | 124 | 133 | 169 | 154.3 | 13.8 | 56 | 13.6 | 10.2 | 86.5 |
| 52 | Green | Green | G/P | White | 121 | 131 | 167 | 150.3 | 8.5 | 28 | 13.5 | 10.2 | 86.2 |
| 53 | Green | G/P | G/P | White | 126 | 132 | 172 | 154.2 | 15.7 | 48 | 11.5 | 8.1 | 57.7 |
| 54 | Green | Green | G/P | White | 124 | 131 | 166 | 150.3 | 15.4 | 48 | 11.5 | 8.2 | 55.1 |
| 55 | Green | Purple | G/P | Purple | 126 | 134 | 173 | 170.3 | 10.2 | 36 | 11.0 | 7.5 | 49.6 |
| 56 | Green | Purple | G/P | Purple | 125 | 131 | 172 | 153.8 | 14.3 | 48 | 9.6 | 6.6 | 38.3 |
| 57 | Green | Green | G/P | White | 124 | 135 | 167 | 154.5 | 15 | 56 | 13.2 | 9.9 | 79.0 |
| 58 | Green | G/P | G/P | White | 120 | 131 | 161 | 139.6 | 19.3 | 64 | 12.7 | 9.6 | 75.6 |
| 59 | Green | G/P | G/P | Pink | 128 | 137 | 170 | 137.2 | 12.1 | 44 | 12.4 | 9.1 | 68.8 |
| 60 | Green | G/P | G/P | Pink | 121 | 133 | 168 | 149.1 | 11.6 | 48 | 12.1 | 10.2 | 76.0 |
| 61 | Green | G/P | G/P | Pink | 128 | 137 | 173 | 153.2 | 12.6 | 44 | 12.6 | 9.5 | 73.3 |
| 62 | Green | G/P | G/P | Purple | 121 | 131 | 166 | 154.3 | 7.9 | 28 | 14.0 | 8.8 | 75.9 |
| 63 | Green | G/P | G/P | Purple | 125 | 134 | 166 | 154.2 | 14 | 44 | 10.7 | 6.5 | 41.5 |
| 64 | Green | G/P | G/P | Pink | 126 | 135 | 172 | 140.1 | 13.3 | 40 | 11.1 | 7.8 | 53.2 |
| 65 | Green | G/P | G/P | Pink | 131 | 141 | 173 | 147.3 | 7.9 | 32 | 11.1 | 7.8 | 54.1 |
| 66 | Green | Purple | G/P | Pink | 125 | 131 | 166 | 164.1 | 12.8 | 48 | 11.1 | 7.6 | 53.5 |
| 67 | G/P | Purple | Purple | Purple | 123 | 131 | 164 | 165.8 | 10.9 | 48 | 12.1 | 7.9 | 56.8 |
| 68 | Green | Purple | G/P | Pink | 120 | 130 | 161 | 169.3 | 17.3 | 48 | 11.6 | 8.2 | 57.0 |
| Max | 131 | 141 | 173 | 174.3 | 19.3 | 64 | 15.0 | 10.9 | 98.9 | ||||
| Min | 116 | 125 | 152 | 106.4 | 5.7 | 28 | 9.6 | 6.2 | 37.7 | ||||
| Mean | 122 | 132 | 164 | 153.1 | 12.3 | 44.1 | 12.0 | 8.5 | 62.2 |
| Trait | Marker | p Value | Marker R2 | Trait | Marker | p Value | Marker R2 |
|---|---|---|---|---|---|---|---|
| QL1 | GBPFM179 | 0.01 | 0.31 | QN3 | KNUPF14 | 0.04 | 0.21 |
| KNUPF4 | 0.04 | 0.22 | KNUPF16 | 0.02 | 0.26 | ||
| KNUPF14 | 0.04 | 0.22 | KNUPF23 | 0.03 | 0.25 | ||
| KNUPF23 | 0.04 | 0.22 | KNUPF182 | 0.00 | 0.38 | ||
| KNUPF31 | 0.04 | 0.22 | QN5 | GBPFM179 | 0.02 | 0.27 | |
| KNUPF59 | 0.04 | 0.22 | KNUPF83 | 0.04 | 0.22 | ||
| KNUPF156 | 0.04 | 0.22 | KNUPF167 | 0.01 | 0.28 | ||
| KNUPF167 | 0.04 | 0.22 | QN6 | GBPFM179 | 0.04 | 0.21 | |
| QL2 | KNUPF23 | 0.02 | 0.25 | KNUPF37 | 0.04 | 0.21 | |
| KNUPF30 | 0.04 | 0.22 | KNUPF83 | 0.02 | 0.27 | ||
| QL3 | GBPFM179 | 0.00 | 0.38 | KNUPF167 | 0.01 | 0.29 | |
| KNUPF59 | 0.01 | 0.29 | QN7 | KNUPF170 | 0.04 | 0.21 | |
| KNUPF112 | 0.02 | 0.26 | QN8 | KNUPF31 | 0.04 | 0.22 | |
| QN1 | KNUPF16 | 0.01 | 0.28 | KNUPF93 | 0.03 | 0.24 | |
| KNUPF30 | 0.01 | 0.29 | KNUPF162 | 0.03 | 0.24 | ||
| KNUPF40 | 0.03 | 0.23 | KNUPF167 | 0.03 | 0.24 | ||
| KNUPF59 | 0.01 | 0.32 | QN9 | KNUPF93 | 0.04 | 0.22 | |
| QN2 | KNUPF30 | 0.02 | 0.27 | KNUPF162 | 0.04 | 0.22 | |
| KNUPF59 | 0.02 | 0.25 | KNUPF167 | 0.03 | 0.23 | ||
| KNUPF182 | 0.01 | 0.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heo, T.H.; Park, H.; Cho, J.; Lee, D.H.; Lee, J.K. Association Mapping Analysis of Morphological Characteristics in F2 Population of Perilla (Perilla frutescens L.) Using SSR Markers. Plants 2025, 14, 2799. https://doi.org/10.3390/plants14172799
Heo TH, Park H, Cho J, Lee DH, Lee JK. Association Mapping Analysis of Morphological Characteristics in F2 Population of Perilla (Perilla frutescens L.) Using SSR Markers. Plants. 2025; 14(17):2799. https://doi.org/10.3390/plants14172799
Chicago/Turabian StyleHeo, Tae Hyeon, Hyeon Park, Jungeun Cho, Da Hyeon Lee, and Ju Kyong Lee. 2025. "Association Mapping Analysis of Morphological Characteristics in F2 Population of Perilla (Perilla frutescens L.) Using SSR Markers" Plants 14, no. 17: 2799. https://doi.org/10.3390/plants14172799
APA StyleHeo, T. H., Park, H., Cho, J., Lee, D. H., & Lee, J. K. (2025). Association Mapping Analysis of Morphological Characteristics in F2 Population of Perilla (Perilla frutescens L.) Using SSR Markers. Plants, 14(17), 2799. https://doi.org/10.3390/plants14172799






