Evolutionary Insights and Flowering Regulation of SPLs in Coconut Palm
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources and Sequence Retrieval
2.2. Genome-Wide Identification of SPLs Genes
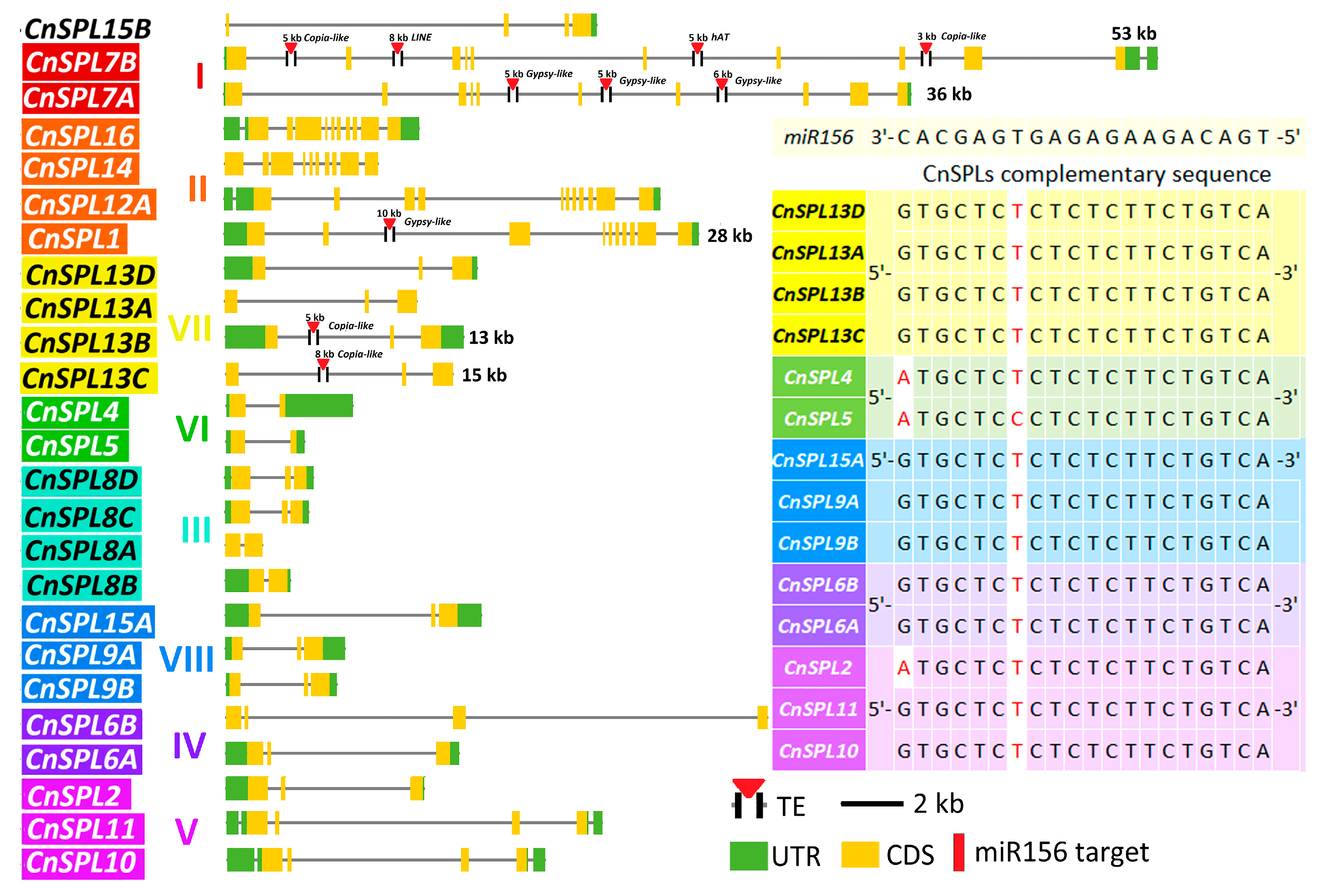
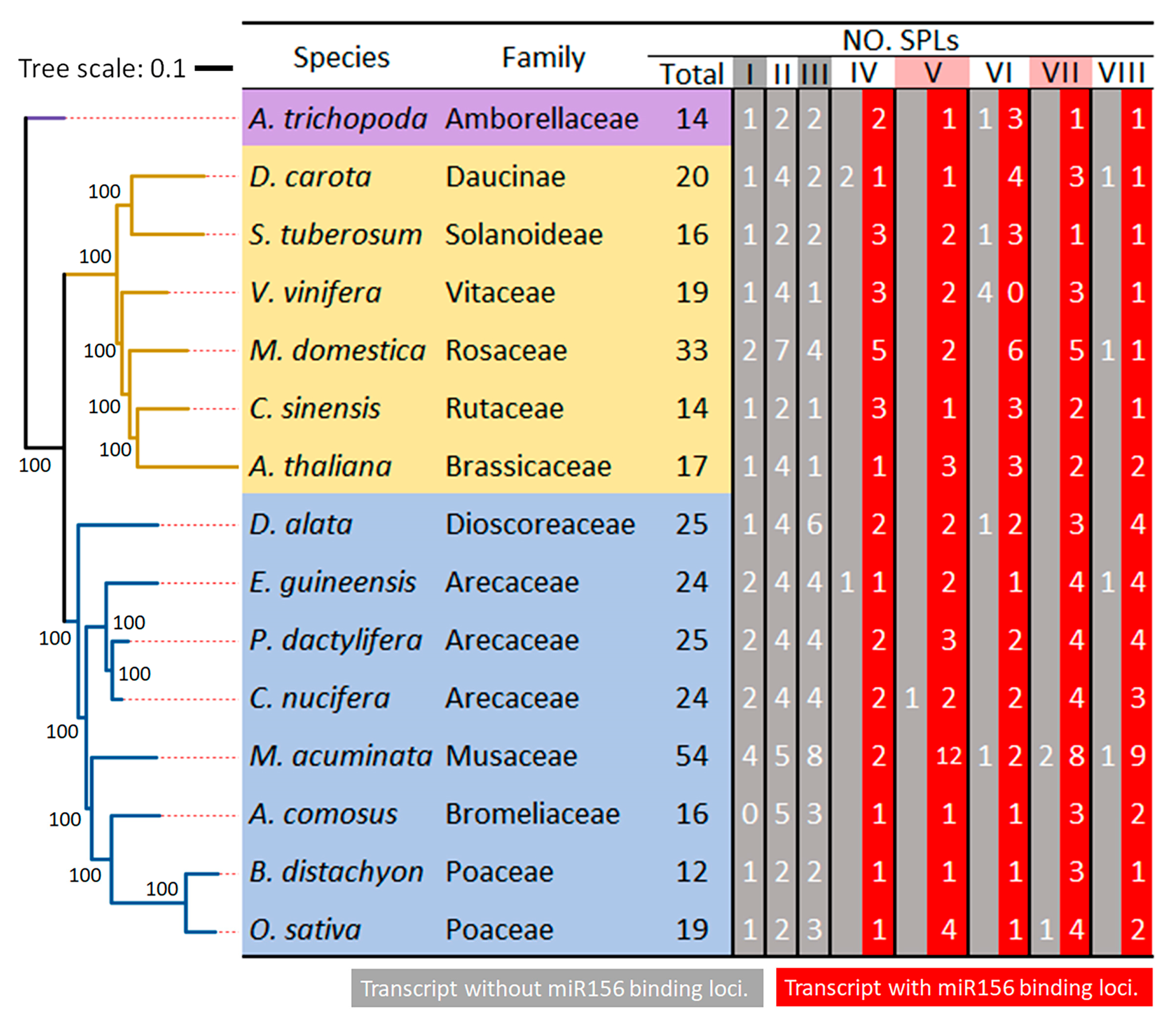
2.3. Evolutionary Analysis of CnSPL Genes
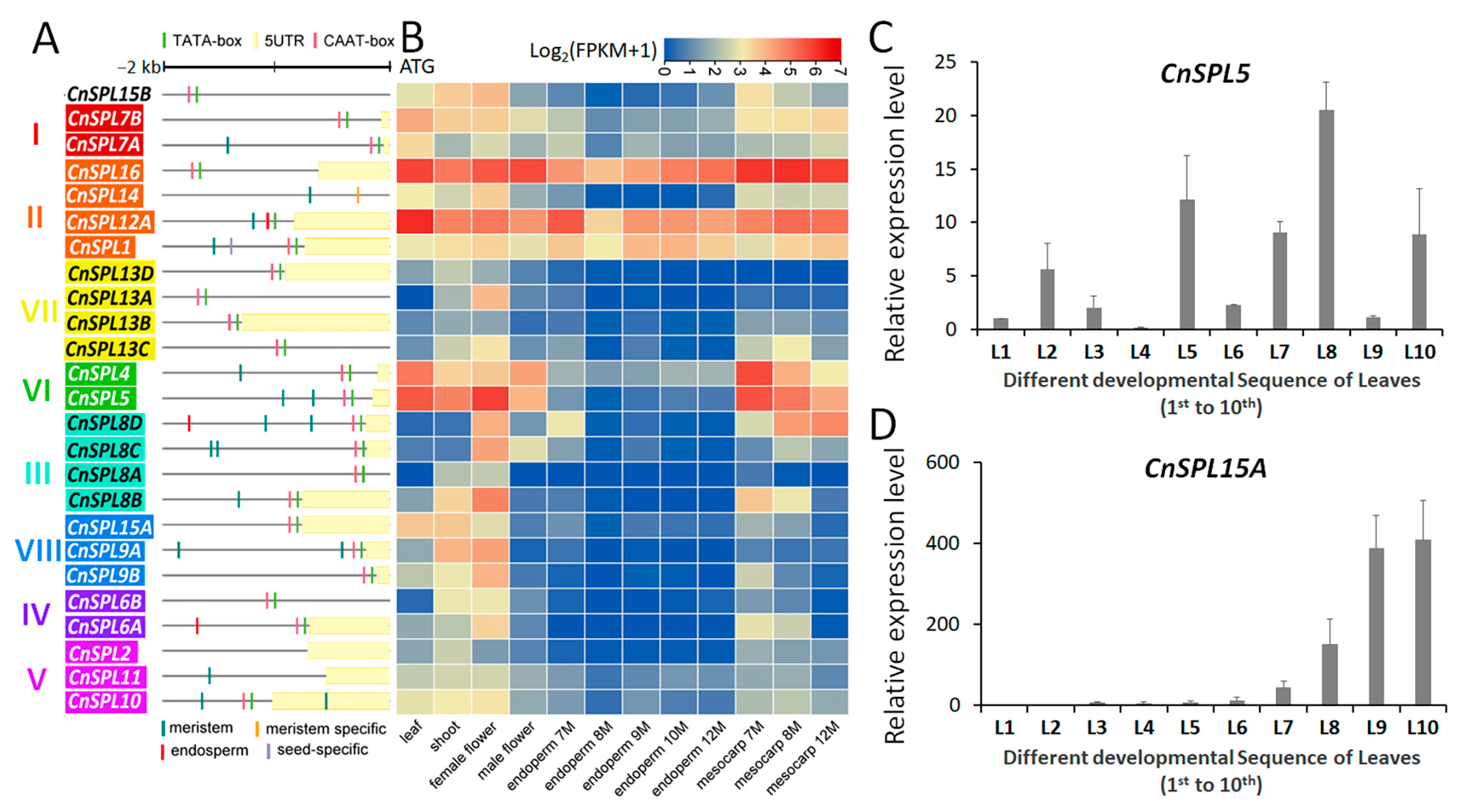
2.4. Promoter Analysis and Gene Expression Pattern Analysis Based on Transcriptome Datasets
2.5. Identification the Complementary Loci of miR156 in SPL Transcripts
2.6. Analysis of the Expression Patterns of CnSPLs Using Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR) Assays
2.7. Transient Expression of CnSPL15A in Tobacco Epidermal Cells for Subcellular Localization
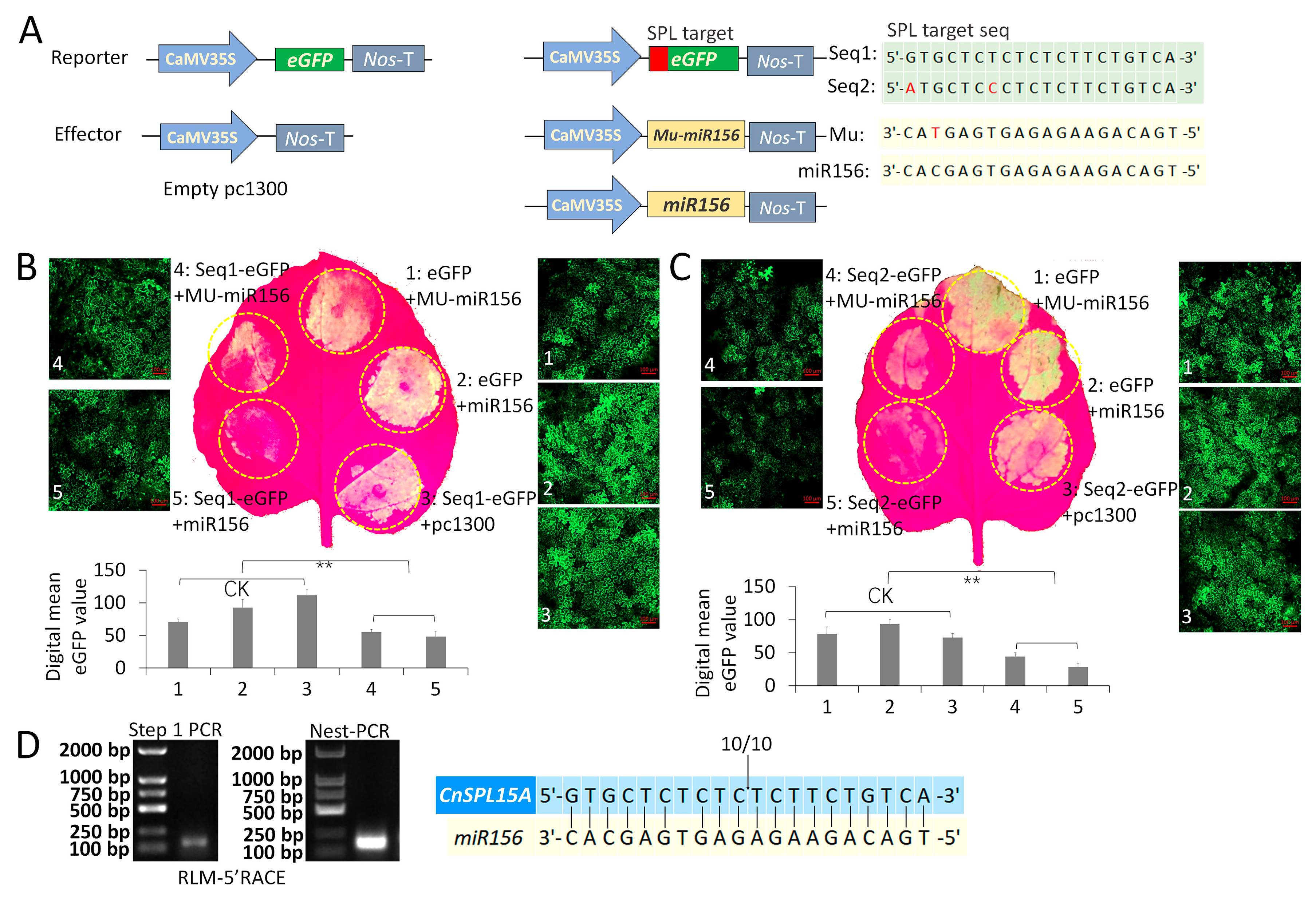
2.8. Transient Expression of miR156 and SPL-eGFP Fusion in Tobacco Epidermal Cells for Target Sequence Validation
2.9. RLM-5′ RACE for miR156-Targeted SPL Validation
2.10. Plant Transformation and Transgenic Plants Phenotype Investigation
2.11. Statistical Analysis
3. Results
3.1. Gene Characters of the SPL Gene Family (CnSPLs) in Coconut Palm
3.2. Evolutionary Character of miR156-Targeted Loci in SPLs
3.3. The Divergent Expression Patterns of CnSPLs
3.4. CnSPL15A Is Targeted by miR156 by Transcript Cleavage
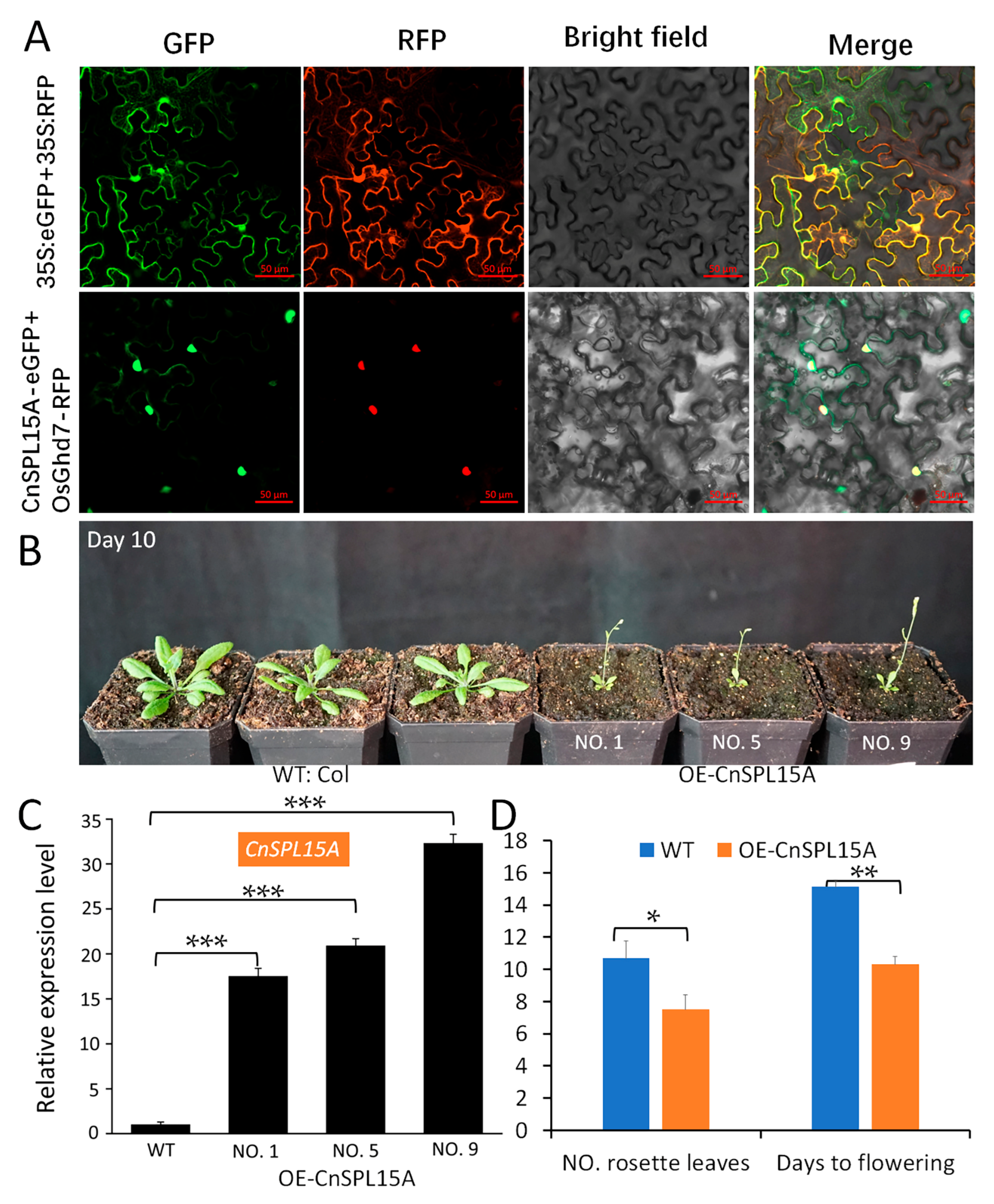
3.5. CnSPL15A Localized to the Nucleus and Affected the Vegetative Phase Change
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Tomo, Y.; Terada, T.; et al. An Arabidopsis SBP-domain fragment with a disrupted C-terminal zinc-binding site retains its tertiary structure. FEBS Lett. 2006, 580, 2109–2116. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Yu, N.; Niu, Q.W.; Ng, K.H.; Chua, N.H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015, 83, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Rojas, C.H.; Rocha, G.H.B.; Polverari, L.; Pinheiro Brito, D.A.; Batista, D.S.; Notini, M.M.; da Cruz, A.C.F.; Morea, E.G.O.; Sabatini, S.; Otoni, W.C.; et al. miR156-targeted SPL10 controls Arabidopsis root meristem activity and root-derived de novo shoot regeneration via cytokinin responses. J. Exp. Bot. 2019, 71, 934–950. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Fu, C.; Liu, S.; Tang, C.; Debnath, S.; Flanagan, A.; Ge, Y.; Tang, Y.; Jiang, Q.; Larson, P.R.; et al. The miR156-SPL4 module predominantly regulates aerial axillary bud formation and controls shoot architecture. New Phytol. 2017, 216, 829–840. [Google Scholar] [CrossRef]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Lal, S.; Pacis, L.B.; Smith, H.M. Regulation of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE genes/microRNA156 module by the homeodomain proteins PENNYWISE and POUND-FOOLISH in Arabidopsis. Mol. Plant 2011, 4, 1123–1132. [Google Scholar] [CrossRef]
- Cui, M.; Wang, C.; Zhang, W.; Pervaiz, T.; Haider, M.S.; Tang, W.; Fang, J. Characterization of Vv-miR156: Vv-SPL pairs involved in the modulation of grape berry development and ripening. Mol. Genet. Genom. 2018, 293, 1333–1354. [Google Scholar] [CrossRef]
- Miao, C.; Wang, Z.; Zhang, L.; Yao, J.; Hua, K.; Liu, X.; Shi, H.; Zhu, J.-K. The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 2019, 10, 3822. [Google Scholar] [CrossRef] [PubMed]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, K.; Cheng, Y.; Yu, S.; Shang, G.; Wang, F.-x.; Wu, L.; Xu, Z.; Mai, Y.-x.; Zhao, X.; et al. A robust mechanism for resetting juvenility during each generation in Arabidopsis. Nat. Plants 2022, 8, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.J.; Shang, G.D.; Xu, Z.G.; Yu, S.; Wu, L.Y.; Zhai, D.; Tian, S.L.; Gao, J.; Wang, L.; Wang, J.W. Cell division in the shoot apical meristem is a trigger for miR156 decline and vegetative phase transition in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, 2115667118. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hou, Y.; Xie, X.; Li, H.; Zhu, Y.; Zhai, L.; Zhang, C.; Bian, S. A blueberry MIR156a-SPL12 module coordinates the accumulation of chlorophylls and anthocyanins during fruit ripening. J. Exp. Bot. 2020, 71, 5976–5989. [Google Scholar] [CrossRef]
- Wu, J.W.; Zhao, Z.Y.; Hu, R.C.; Huang, Y.F. Genome-wide identification, stress- and hormone-responsive expression characteristics, and regulatory pattern analysis of Scutellaria baicalensis SbSPLs. Plant Mol. Biol. 2024, 114, 20. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, M.; He, X.; Zheng, Q.; Huang, Y.; Li, Y.; Ahmad, S.; Liu, D.; Lan, S.; Liu, Z. Genome-wide identification and expression analysis of the SPL gene family in three orchids. Int. J. Mol. Sci. 2023, 24, 10039. [Google Scholar] [CrossRef]
- He, F.; Long, R.; Wei, C.; Zhang, Y.; Li, M.; Kang, J.; Yang, Q.; Wang, Z.; Chen, L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family and its important role in salt stress in Medicago sativa L. BMC Plant Biol. 2022, 22, 295. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, Z.; Xu, Y.; Kong, L.; Shi, J.; Liu, Y.; Fu, C.; Wang, X.; Wang, Z.Y.; Zhou, C.; et al. Genome-wide characterization of SPL family in Medicago truncatula reveals the novel roles of miR156/SPL module in spiky pod development. BMC Genom. 2019, 20, 552. [Google Scholar] [CrossRef]
- Wei, L.; Liu, J.; Huang, J.; Wang, C.; Zhang, L.; Feng, S. Genome-wide identification of miR156 and SPL family genes and phenotypic analysis of vegetative phase change in pepper (Capsicum annuum L.). Gene 2023, 877, 4. [Google Scholar] [CrossRef]
- He, A.; Zhou, H.; Ma, C.; Bai, Q.; Yang, H.; Yao, X.; Wu, W.; Xue, G.; Ruan, J. Genome-wide identification and expression analysis of the SPL gene family and its response to abiotic stress in barley (Hordeum vulgare L.). BMC Genom. 2024, 25, 846. [Google Scholar] [CrossRef]
- Li, J.; Zeng, X.; Jin, Z.; Zhou, T.; Lang, C.; Qin, J.; Zhang, Q.; Lan, H.; Li, Y.; An, H.; et al. Genome-wide analysis of the SPL family in Zanthoxylum armatum and ZaSPL21 promotes flowering and improves salt tolerance in transgenic Nicotiana benthamiana. Plant Mol. Biol. 2025, 115, 23. [Google Scholar] [CrossRef]
- Xia, W.; Liu, R.; Zhang, J.; Mason, A.S.; Li, Z.; Gong, S.; Zhong, Y.; Dou, Y.; Sun, X.; Fan, H.; et al. Alternative splicing of flowering time gene FT is associated with halving of time to flowering in coconut. Sci. Rep. 2020, 10, 11640. [Google Scholar] [CrossRef]
- Wang, S.; Xiao, Y.; Zhou, Z.-W.; Yuan, J.; Guo, H.; Yang, Z.; Yang, J.; Sun, P.; Sun, L.; Deng, Y.; et al. High-quality reference genome sequences of two coconut cultivars provide insights into evolution of monocot chromosomes and differentiation of fiber content and plant height. Genome Biol. 2021, 22, 304. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bocs, S.; Fan, H.; Armero, A.; Baudouin, L.; Xu, P.; Xu, J.; This, D.; Hamelin, C.; Iqbal, A.; et al. Coconut genome assembly enables evolutionary analysis of palms and highlights signaling pathways involved in salt tolerance. Commu Biol. 2021, 4, 105. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, Z.; Xu, H.; Li, Y.; Huang, S.; Cao, G.; Shi, M.; Zhu, J.; Zhou, J.; Li, R.; et al. ArecaceaeMDB: A comprehensive multi-omics database for Arecaceae breeding and functional genomics studies. Plant Biotechnol. J. 2023, 21, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Lao, Z.; Mao, J.; Chen, R.; Xu, R.; Yang, Z.; Wang, Y.; Zhou, J.; Mu, Z.; Xu, H.; Li, F.; et al. Genome-wide identification and characterization of BASIC PENTACYSTEINE transcription factors and their binding motifs in coconut palm. Front. Plant Sci. 2024, 15, 1491139. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, 4. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.V.; Shahmuradov, I.A.; Salamov, A.A. Identification of promoter regions and regulatory sites. Methods Mol. Biol. 2010, 674, 57–83. [Google Scholar] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Feng, Y.; Yong, X.; Wang, Y.; Zhang, D.; Shi, P.; Xia, W.; Sun, X. Analysis of characteristics of coconut flowering related miR156 and target gene SPL. Mol. Plant Breed. 2022, 20, 3894–3901. (In Chinese) [Google Scholar]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Bo, X.; Wang, S. TargetFinder: A software for antisense oligonucleotide target site selection based on MAST and secondary structures of target mRNA. Bioinformatics 2004, 21, 1401–1402. [Google Scholar] [CrossRef]
- Xiao, Y.; Yang, Y.; Cao, H.; Fan, H.; Ma, Z.; Lei, X.; Manson, A.S.; Xia, Z.; Huang, X. Efficient isolation of high quality RNA from tropical palms for RNA-seq analysis. Plant Omics 2012, 5, 584–589. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- D’Hont, A.; Denoeud, F.; Aury, J.-M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Xia, W.; Liu, Z.; Yang, Y.; Xiao, Y.; Mason, A.S.; Zhao, S.; Ma, Z. Selection of reference genes for quantitative real-time PCR in Cocos nucifera during abiotic stress. Botany 2013, 92, 179–186. [Google Scholar] [CrossRef]
- Llave, C.; Xie, Z.; Kasschau, K.D.; Carrington, J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002, 297, 2053–2056. [Google Scholar] [CrossRef]
- Kasschau, K.D.; Xie, Z.; Allen, E.; Llave, C.; Chapman, E.J.; Krizan, K.A.; Carrington, J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 2003, 4, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Yang, J.C.; Yin, G.T.; Li, R.S.; Zou, W.T. Genome-wide characterization of the SPL gene family involved in the age development of Jatropha curcas. BMC Genom. 2020, 21, 368. [Google Scholar] [CrossRef]
- Li, L.; Shi, F.; Wang, G.; Guan, Y.; Zhang, Y.; Chen, M.; Chang, J.; Yang, G.; He, G.; Wang, Y.; et al. Conservation and divergence of SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family between wheat and rice. Int. J. Mol. Sci. 2022, 23, 2099. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, Z.; Liu, D.; Zhang, K.; Li, A.; Mao, L. SQUAMOSA promoter-binding protein-like transcription factors: Star players for plant growth and development. J. Integr. Plant Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef]
- Wang, J.-W.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Li, F.; Ma, Q.; Shen, T.; Jiang, J.; Li, H. The miR156-SPL4/SPL9 module regulates leaf and lateral branch development in Betula platyphylla. Plant Sci. 2024, 338, 111869. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Zhai, L.; Cui, Y.; Tang, G.; Huo, J.; Li, X.; Bian, S. The miR156/SPL12 module orchestrates fruit colour change through directly regulating ethylene production pathway in blueberry. Plant Biotechnol. J. 2024, 22, 386–400. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Tang, C.; Chen, N.; Wang, H.; Debnath, S.; Sun, L.; Flanagan, A.; Tang, Y.; Jiang, Q.; Allen, R.D.; et al. SPL7 and SPL8 represent a novel flowering regulation mechanism in switchgrass. New Phytol. 2019, 222, 1610–1623. [Google Scholar] [CrossRef]
- Zhou, Q.; Shi, J.; Li, Z.; Zhang, S.; Zhang, J.; Bao, M.; Liu, G. miR156/157 Targets SPLs to Regulate Flowering Transition, Plant Architecture and Flower Organ Size in Petunia. Plant Cell Physiol. 2021, 62, 839–857. [Google Scholar] [CrossRef]
- Wang, L.; Yu, P.; Lyu, J.; Hu, Y.; Han, C.; Bai, M.-Y.; Fan, M. BZR1 physically interacts with SPL9 to regulate the vegetative phase change and cell elongation in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 10415. [Google Scholar] [CrossRef]
- Zhou, B.; Luo, Q.; Shen, Y.; Wei, L.; Song, X.; Liao, H.; Ni, L.; Shen, T.; Du, X.; Han, J.; et al. Coordinated regulation of vegetative phase change by brassinosteroids and the age pathway in Arabidopsis. Nat. Commun. 2023, 14, 2608. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, R.; Chen, Y.; Cui, J.; Ge, W.; Zhang, K. Molecular identification and functional verification of SPL9 and SPL15 of Lilium. Mol. Genet. Genom. 2022, 297, 63–74. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, R.; Feng, Y.; Zhou, J.; Wang, Y.; Zhang, F.; Rehman, S.; Yang, Z.; Lao, Z.; Xu, H.; Xiao, Y.; et al. Evolutionary Insights and Flowering Regulation of SPLs in Coconut Palm. Plants 2025, 14, 2532. https://doi.org/10.3390/plants14162532
Chen R, Feng Y, Zhou J, Wang Y, Zhang F, Rehman S, Yang Z, Lao Z, Xu H, Xiao Y, et al. Evolutionary Insights and Flowering Regulation of SPLs in Coconut Palm. Plants. 2025; 14(16):2532. https://doi.org/10.3390/plants14162532
Chicago/Turabian StyleChen, Runan, Yalan Feng, Jin Zhou, Ying Wang, Fengyi Zhang, Shazia Rehman, Zhuang Yang, Zifen Lao, Hang Xu, Yong Xiao, and et al. 2025. "Evolutionary Insights and Flowering Regulation of SPLs in Coconut Palm" Plants 14, no. 16: 2532. https://doi.org/10.3390/plants14162532
APA StyleChen, R., Feng, Y., Zhou, J., Wang, Y., Zhang, F., Rehman, S., Yang, Z., Lao, Z., Xu, H., Xiao, Y., Luo, J., & Xia, W. (2025). Evolutionary Insights and Flowering Regulation of SPLs in Coconut Palm. Plants, 14(16), 2532. https://doi.org/10.3390/plants14162532





