Functional Analysis of the Autophagy-Related Gene OsATG4b in Rice Grain Chalkiness Regulation
Abstract
1. Introduction
2. Results
2.1. Identification of Rice OsATG4b Protein
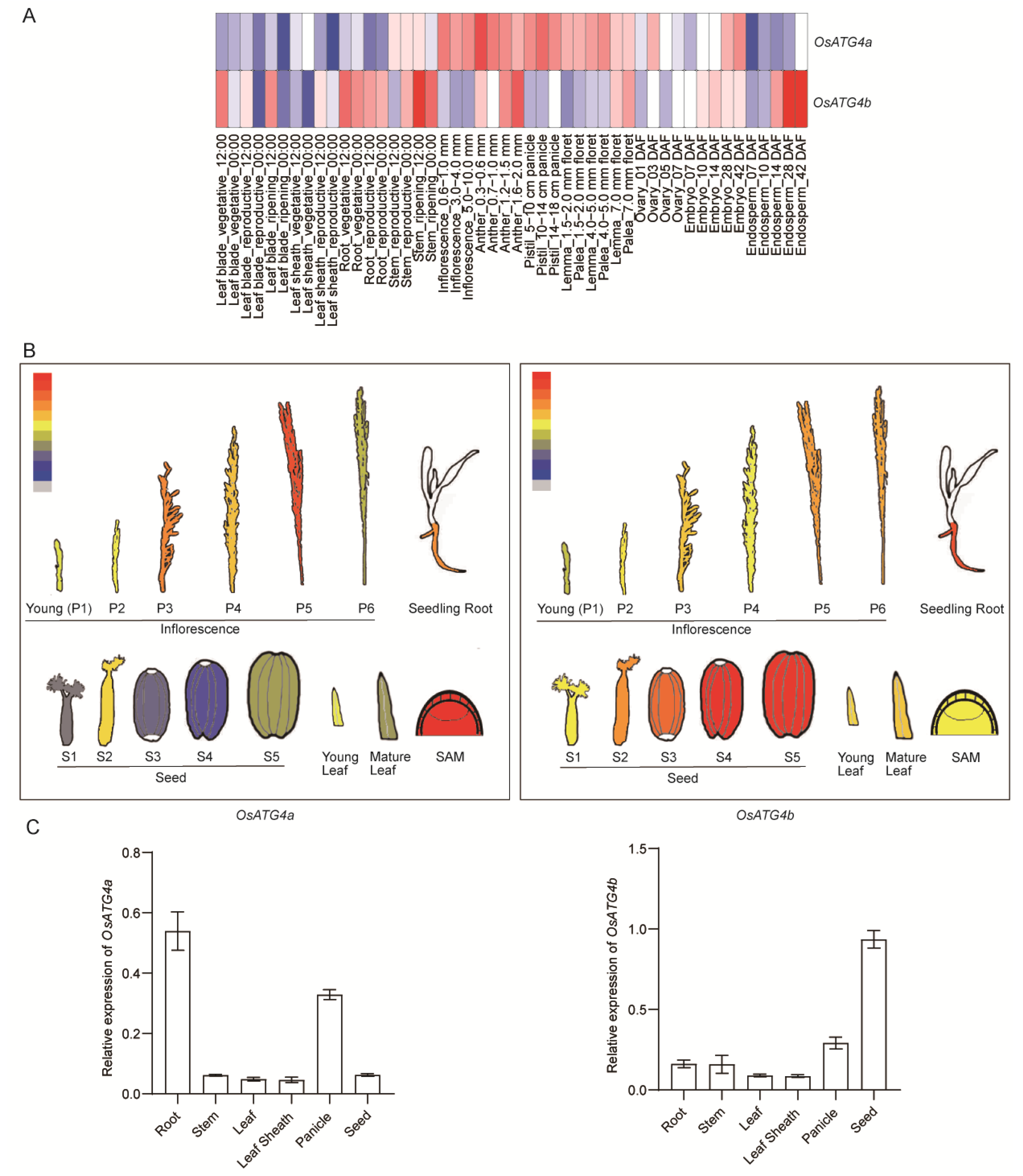
2.2. Expression Patterns of OsATG4a and OsATG4b
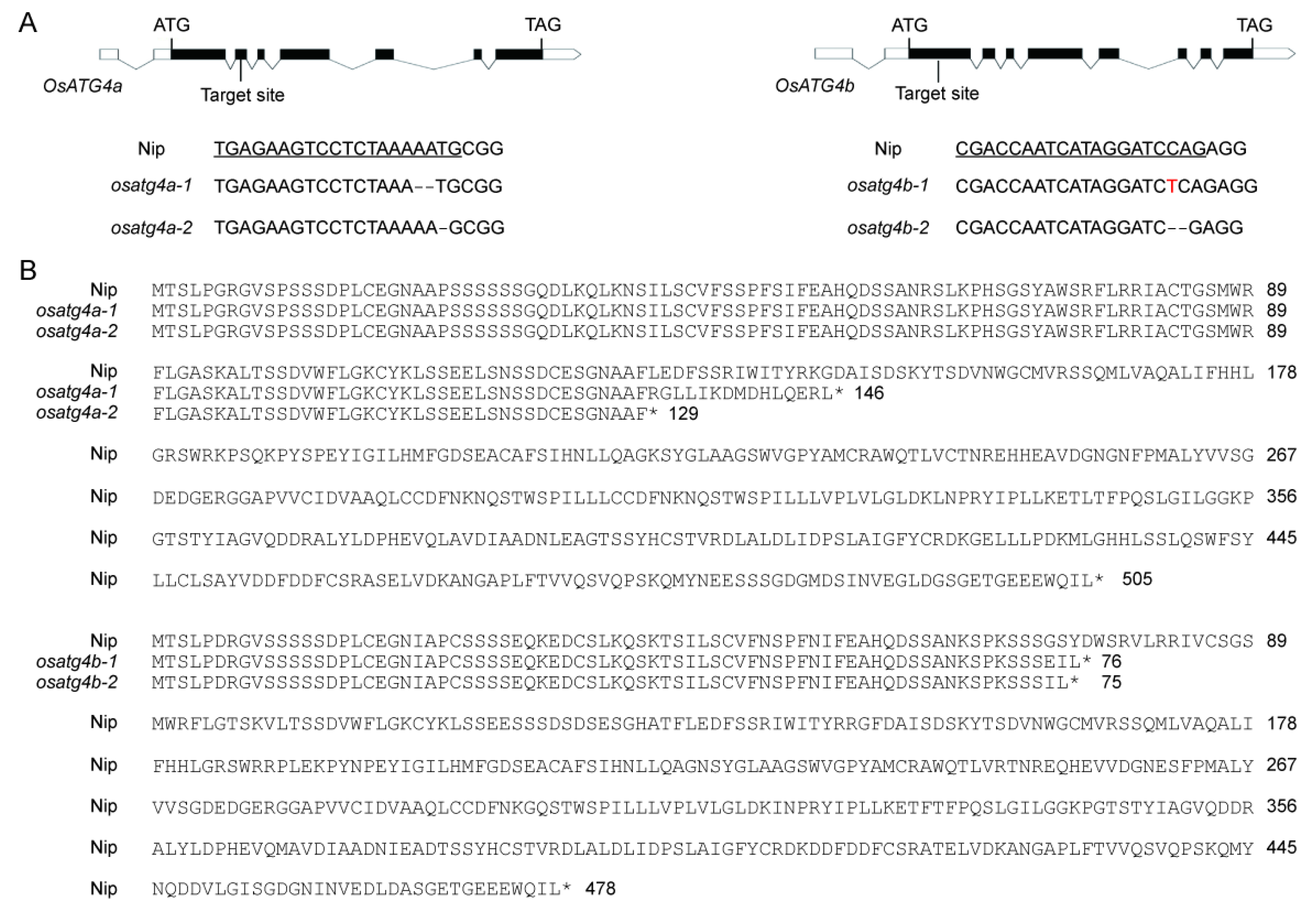
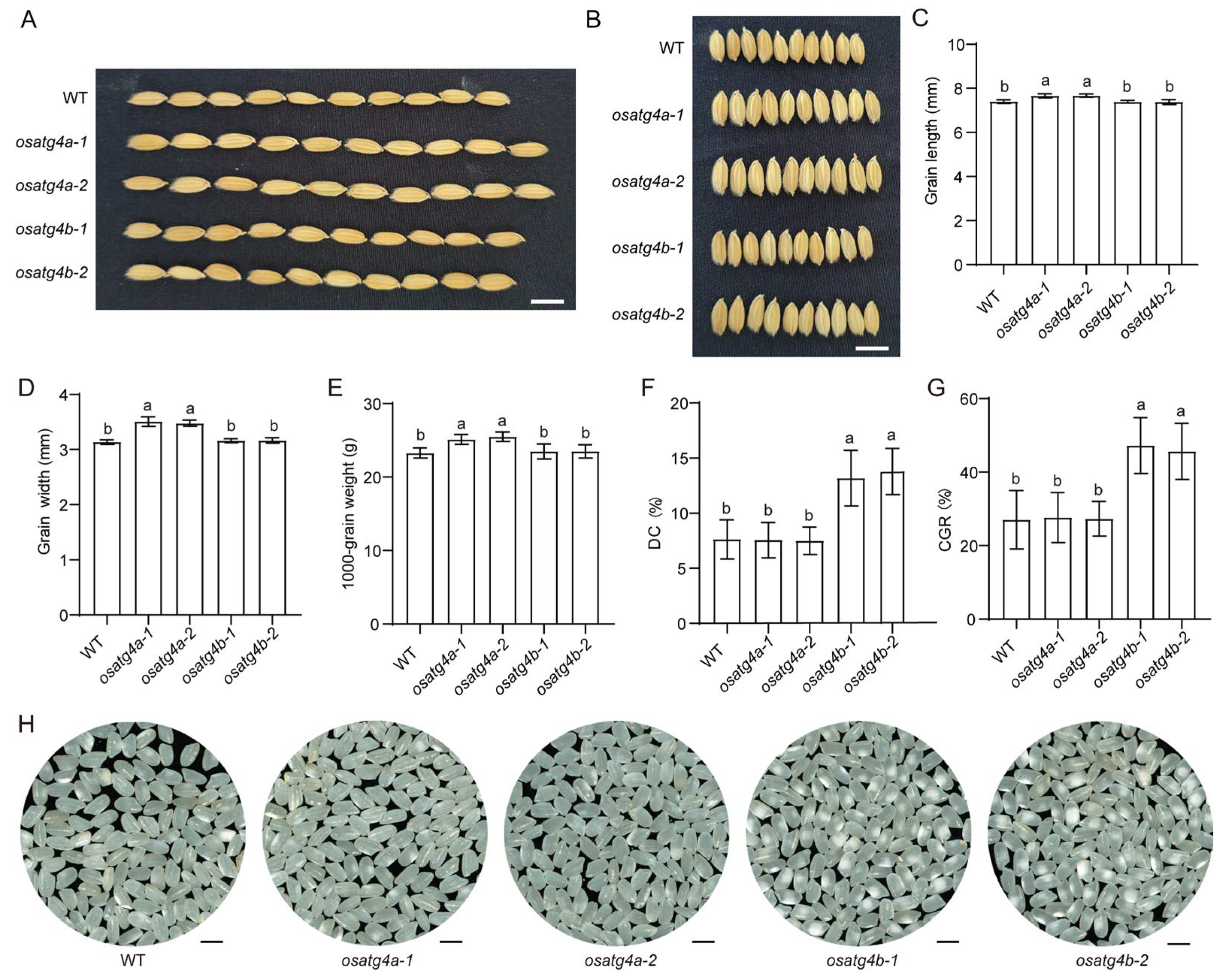
2.3. Generation and Phenotypic Characterization of OsATG4a and OsATG4b Knockout Mutants
2.4. Overexpression of OsATG4b Decreases Grain Chalkiness
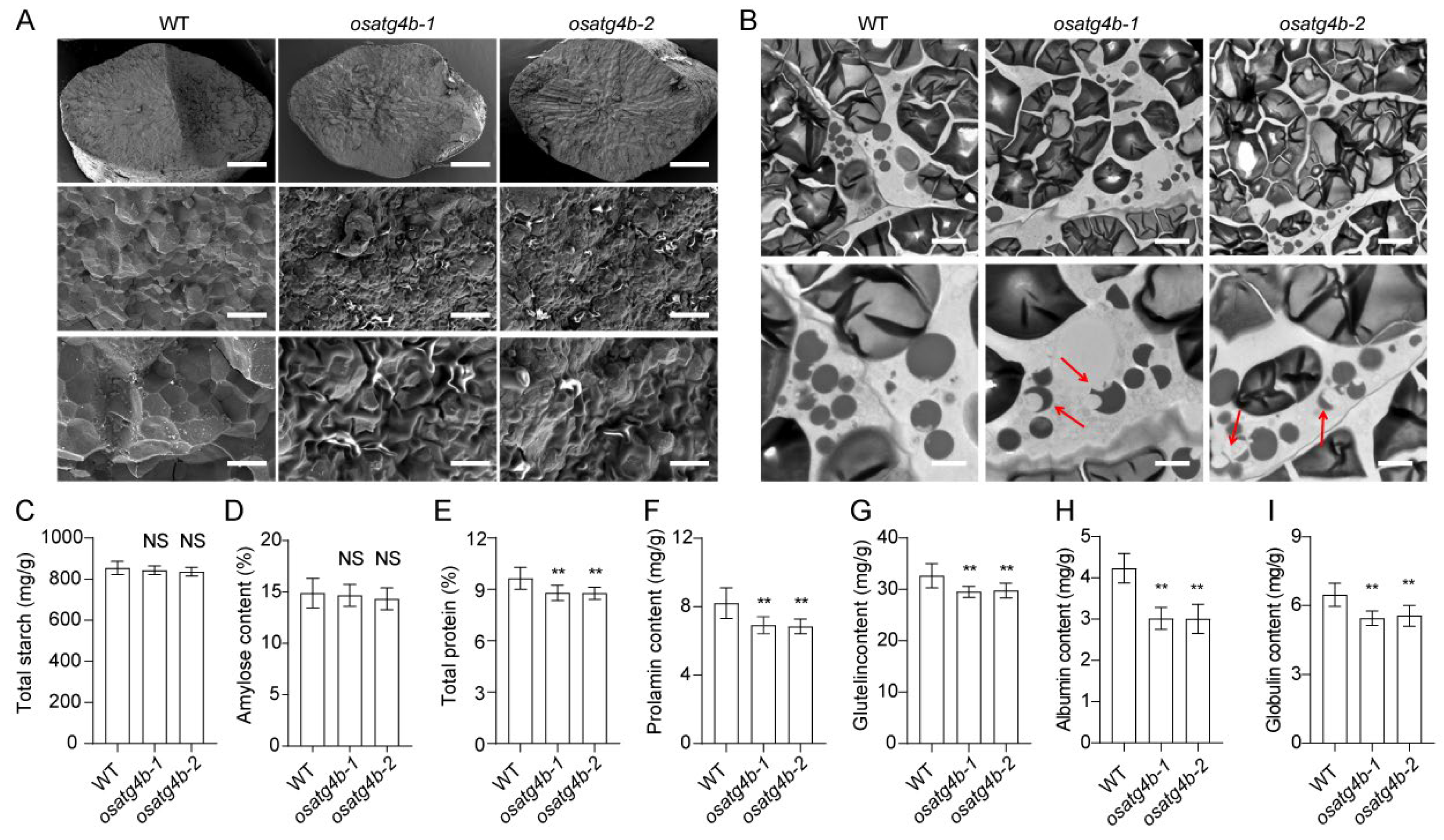
2.5. Altered Structure and Composition of Seed Storage Substances in OsATG4b Endosperm
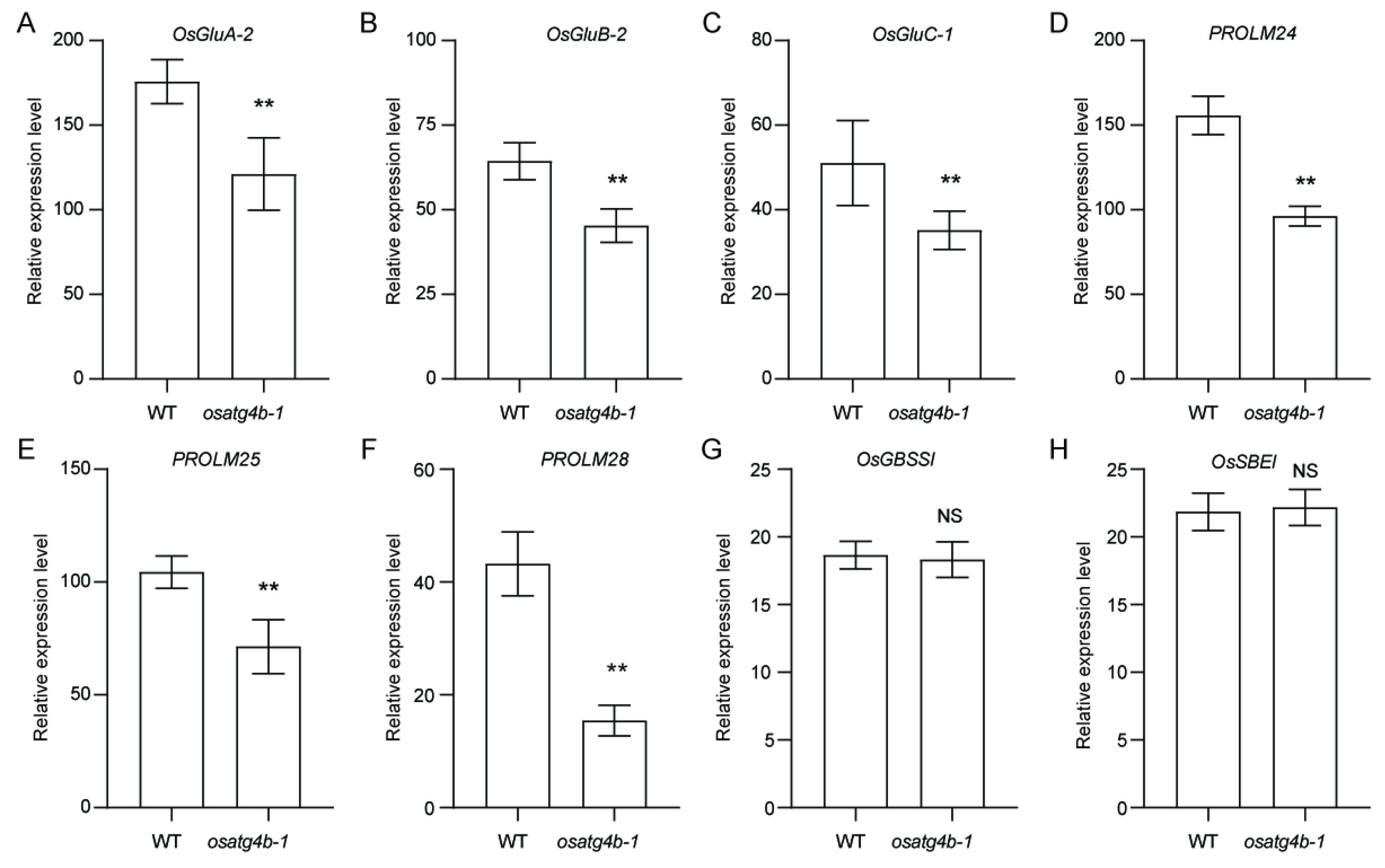
2.6. OsATG4b Mutation Alters Expression of Storage Protein Genes Without Affecting Starch Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification of ATG4 Homologs and Phylogenetic Analysis
4.3. CRISPR/Cas9-Mediated Knockout of OsATG4a and OsATG4b
4.4. Construction of OsATG4b Overexpression Lines
4.5. Expression Analysis by Quantitative RT-PCR
4.6. Measurement of Phenotype Data
4.7. Transmission Electron Microscopy
4.8. Scanning Electron Microscopy
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nutan, K.K.; Rathore, R.S.; Tripathi, A.K.; Mishra, M.; Pareek, A.; Singla-Pareek, S.L. Integrating the dynamics of yield traits in rice in response to environmental changes. J. Exp. Bot. 2020, 71, 490–506. [Google Scholar] [CrossRef]
- Zeng, D.; Tian, Z.; Rao, Y.; Dong, G.; Yang, Y.; Huang, L.; Leng, Y.; Xu, J.; Sun, C.; Zhang, G.; et al. Rational design of high-yield and superior-quality rice. Nat. Plants 2007, 3, 17031. [Google Scholar] [CrossRef]
- Custodio, M.C.; Cuevas, R.P.; Ynion, J.; Laborte, A.G.; Velasco, M.L.; Demont, M. Rice quality: How is it defined by consumers, industry, food scientists, and geneticists? Trends Food Sci. Tech. 2019, 92, 122–137. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, Z.; Li, Y.; Xu, C.; Xia, H.; Zhuang, H.; Sun, S.; Guo, M.; Yan, C. Rapid improvement of rice eating and cooking quality through gene editing toward glutelin as target. J. Integr. Plant Biol. 2022, 64, 1860–1865. [Google Scholar] [CrossRef]
- Chen, L.; Li, X.; Zheng, M.; Hu, R.; Dong, J.; Zhou, L.; Liu, W.; Liu, D.; Yang, W. Genes controlling grain chalkiness in rice. Crop J. 2024, 12, 979–991. [Google Scholar] [CrossRef]
- Zhao, D.; Zhang, C.; Li, Q.; Liu, Q. Genetic control of grain appearance quality in rice. Biotechnol. Adv. 2022, 60, 108014. [Google Scholar] [CrossRef] [PubMed]
- Siebenmorgen, T.J.; Grigg, B.C.; Lanning, S.B. Impacts of preharvest factors during kernel development on rice quality and functionality. Annu. Rev. Food Sci. Technol. 2013, 4, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Chen, S.; Han, Y.; Liu, G.; Liu, J.; Yang, Q.; Zhang, T.; Shen, J.; Fan, X.; Zhang, C.; et al. A CRISPR/Cas9-mediated mutant library of seed-preferred genes in rice. Plant Biotechnol. J. 2024, 11, 3012–3014. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Liu, B.; Wang, D.; Rasheed, A.; Xie, L.; Xia, X.; He, Z. Orchestrating seed storage protein and starch accumulation toward overcoming yield–quality trade-off in cereal crops. J. Integr. Plant Biol. 2024, 66, 468–483. [Google Scholar] [CrossRef]
- Yang, W.; Liang, J.; Hao, Q.; Luan, X.; Tan, Q.; Lin, S.; Zhu, H.; Liu, G.; Liu, Z.; Bu, S.; et al. Fine mapping of two grain chalkiness QTLs sensitive to high temperature in rice. Rice 2021, 14, 33. [Google Scholar] [CrossRef]
- Kaneko, K.; Sasaki, M.; Kuribayashi, N.; Suzuki, H.; Sasuga, Y.; Shiraya, T.; Inomata, T.; Itoh, K.; Baslam, M.; Mitsui, T. Proteomic and Glycomic Characterization of Rice Chalky Grains Produced Under Moderate and High-temperature Conditions in Field System. Rice 2016, 9, 26. [Google Scholar] [CrossRef]
- Cao, R.; Zhao, S.; Jiao, G.; Duan, Y.; Ma, L.; Dong, N.; Lu, F.; Zhu, M.; Shao, G.; Hu, S.; et al. OPAQUE3, encoding a transmembrane bZIP transcription factor, regulates endosperm storage protein and starch biosynthesis in rice. Plant Commun. 2022, 3, 100463. [Google Scholar] [CrossRef]
- Wang, G.; Wang, F.; Wang, G.; Wang, F.; Zhang, X.; Zhong, M.; Zhang, J.; Lin, D.; Tang, Y.; Xu, Z.; et al. Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 2012, 24, 3447–3462. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Wang, Y.; Liu, F.; Zhou, K.; Ding, Y.; Zhou, F.; Wang, Y.; Liu, K.; Gan, L.; Ma, W.; et al. GLUTELIN PRECURSOR ACCUMULATION3 encodes a regulator of post-Golgi vesicular traffic essential for vacuolar protein sorting in rice endosperm. Plant Cell 2014, 26, 410–425. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zheng, X.; Yang, J.; Messing, J.; Wu, Y. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 10842–10847. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The Master of Bulk and Selective Recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 2012, 63, 215–237. [Google Scholar] [CrossRef]
- Ustun, S.; Hafren, A.; Hofius, D. Autophagy as a mediator of life and death in plants. Curr. Opin. Plant Biol. 2017, 40, 122–130. [Google Scholar] [CrossRef]
- Su, W.; Bao, Y.; Yu, X.; Xia, X.; Liu, C.; Yin, W. Autophagy and Its Regulators in Response to Stress in Plants. Int. J. Mol. Sci. 2020, 21, 8889. [Google Scholar] [CrossRef]
- Yang, M.; Ismayil, A.; Liu, Y. Autophagy in Plant-Virus Interactions. Annu. Rev. Virol. 2020, 7, 403–419. [Google Scholar] [CrossRef]
- Yoshimoto, K. Beginning to Understand Autophagy, an Intracellular Self-Degradation System in Plants. Plant Cell Physiol. 2012, 53, 1355–1365. [Google Scholar] [CrossRef]
- Doelling, J.H.; Walker, J.M.; Friedman, E.M.; Thompson, A.R.; Vierstra, R.D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 33105–33114. [Google Scholar] [CrossRef]
- Chung, T.; Phillips, A.R.; Vierstra, R.D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 2010, 62, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Noda, N.N.; Ohsumi, Y.; Inagaki, F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010, 584, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Shpilka, T.; Weidberg, H.; Pietrokovski, S.; Elazar, Z. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol. 2011, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Woo, J.; Dinesh-Kumar, S.P. Arabidopsis ATG4 cysteine proteases specificity toward ATG8 substrates. Autophagy 2014, 10, 926–927. [Google Scholar] [CrossRef]
- Zhen, X.; Li, X.; Yu, J.; Xu, F. OsATG8c-Mediated Increased Autophagy Regulates the Yield and Nitrogen Use Efficiency in Rice. Int. J. Mol. Sci. 2019, 20, 4956. [Google Scholar] [CrossRef]
- Yoshimoto, K.; Hanaoka, H.; Sato, S.; Kato, T.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004, 16, 2967–2983. [Google Scholar] [CrossRef]
- Woo, J.; Park, E.; Dinesh-Kumar, S.P.; Dinesh-Kumar, S.P. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc. Natl. Acad. Sci. USA 2014, 14, 863–868. [Google Scholar] [CrossRef]
- Laureano-Marín, A.M.; Aroca, Á.; Pérez-Pérez, M.E.; Yruela, I.; Jurado-Flores, A.; Moreno, I.; Crespo, J.L.; Romero, L.C.; Gotor, C. Abscisic Acid-Triggered Persulfidation of the Cys Protease ATG4 Mediates Regulation of Autophagy by Sulfide. Plant Cell 2020, 32, 3902–3920. [Google Scholar] [CrossRef]
- Huang, S.; Wang, P.; Li, C.; Zhou, Q.; Huang, T.; Cai, Y.; Cheng, Q.; Wang, H.; Zhong, Q.; Chen, Z.; et al. Natural variation of an autophagy-family gene among rice subspecies affects grain size and weight. Crop J. 2024, 12, 121–132. [Google Scholar] [CrossRef]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro Version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef]
- Qi, H.; Xia, F.N.; Xiao, S. Autophagy in plants: Physiological roles and post-translational regulation. J. Integr. Plant Biol. 2021, 63, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, Q.; Wu, P.; Li, Y.; Lin, Y.; Liu, W.; Guo, S.; Liu, Y.; Huang, Y.; Xu, P.; et al. Dynamic monitoring of TGW6 by selective autophagy during grain development in rice. New Phytol. 2023, 240, 2419–2435. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Zheng, N.; Yu, J.; Bi, C.; Xu, F. Autophagy mediates grain yield and nitrogen stress resistance by modulating nitrogen remobilization in rice. PLoS ONE 2021, 16, e0244996. [Google Scholar] [CrossRef] [PubMed]
- Kurusu, T.; Koyano, T.; Hanamata, S.; Kubo, T.; Noguchi, Y.; Yagi, C.; Nagata, N.; Yamamoto, T.; Ohnishi, T.; Okazaki, Y.; et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 2014, 10, 878–888. [Google Scholar] [CrossRef]
- Izumi, M.; Hidema, J.; Wada, S.; Kondo, E.; Kurusu, T.; Kuchitsu, K.; Makino, A.; Ishida, H. Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol. 2015, 167, 1307–1320. [Google Scholar] [CrossRef]
- Hu, Z.; Yang, Z.; Zhang, Y.; Zhang, A.; Lu, Q.; Fang, Y.; Lu, C. Autophagy targets Hd1 for vacuolar degradation to regulate rice flowering. Mol. Plant 2022, 15, 1137–1156. [Google Scholar] [CrossRef]
- Ibl, V.; Kapusi, E.; Arcalis, E.; Kawagoe, Y.; Stoger, E. Fusion, rupture, and degeneration: The fate of in vivo-labelled PSVs in developing barley endosperm. J. Exp. Bot. 2014, 65, 3249–3261. [Google Scholar] [CrossRef]
- Di Berardino, J.; Marmagne, A.; Berger, A.; Yoshimoto, K.; Cueff, G.; Chardon, F.; Masclaux-Daubresse, C.; Reisdorf-Cren, M. Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J. Exp. Bot. 2018, 69, 1403–1414. [Google Scholar] [CrossRef]
- Zhang, X.; Ding, X.; Marshall, R.S.; Paez-Valencia, J.; Lacey, P.; Vierstra, R.D.; Otegui, M.S. Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. eLife 2020, 9, e51918. [Google Scholar] [CrossRef]
- Li, Y.; Fan, C.; Xing, Y.; Yun, P.; Luo, L.; Yan, B.; Peng, B.; Xie, W.; Wang, G.; Li, X.; et al. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 2014, 46, 398–404. [Google Scholar] [CrossRef]
- Wu, B.; Yun, P.; Zhou, H.; Xia, D.; Gu, Y.; Li, P.; Yao, J.; Zhou, Z.; Chen, J.; Liu, R.; et al. Natural variation in WHITE-CORE RATE 1 regulates redox homeostasis in rice endosperm to affect grain quality. Plant Cell 2022, 34, 1912–1932. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Liu, H.; Guo, M.; Han, X.; Li, Y.; Chen, R.; Guo, Y.; Yang, Y.; Sun, S.; Zhou, Y.; et al. Natural variation of an E3 ubiquitin ligase encoding gene Chalk9 regulates grain chalkiness in rice. Nat. Commun. 2025, 16, 6653. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Yun, P.; Zhu, Y.; Wang, L.; Wang, Y.; Li, P.; Zhou, H.; Cheng, S.; Liu, R.; Gao, G.; et al. Natural variation of WBR7 confers rice high yield and quality by modulating sucrose supply in sink organs. Plant Biotechnol. J. 2024, 22, 2985–2999. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.P.; Onodera, Y.; Touno, S.M.; Takaiwa, F. Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol. 2006, 141, 1694–1707. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Yamamoto, M.P.; Touno, S.M.; Yasuda, H.; Takaiwa, F. Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 2009, 59, 908–920. [Google Scholar] [CrossRef]
- Wu, M.-W.; Liu, J.; Bai, X.; Chen, W.-Q.; Ren, Y.; Liu, J.-L.; Chen, M.-M.; Zhao, H.; Yao, X.; Zhang, J.-D.; et al. Transcription factors NAC20 and NAC26 interact with RPBF to activate albumin accumulations in rice endosperm. Plant Biotechnol. J. 2023, 21, 890–892. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Naito, Y.; Hino, K.; Bono, H.; Ui-Tei, K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 2015, 31, 1120–1123. [Google Scholar] [CrossRef]
- Liu, W.; Xie, X.; Ma, X.; Li, J.; Chen, J.; Liu, Y.-G. DSDecode: A Web-Based Tool for Decoding of Sequencing Chromatograms for Genotyping of Targeted Mutations. Mol. Plant 2015, 8, 1431–1433. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, M.; Sun, S.; Zou, Y.; Yin, S.; Liu, Y.; Tang, S.; Gu, M.; Yang, Z.; Yan, C. Natural variation of OsGluA2 is involved in grain protein content regulation in rice. Nat. Commun. 2019, 10, 1949. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Z.; Han, X.; Yuan, Y.; Xing, R.; Liu, H.; Li, C.; Shen, H.; Guo, Y.; Sun, S.; Yang, Y.; et al. Functional Analysis of the Autophagy-Related Gene OsATG4b in Rice Grain Chalkiness Regulation. Plants 2025, 14, 2530. https://doi.org/10.3390/plants14162530
Hu Z, Han X, Yuan Y, Xing R, Liu H, Li C, Shen H, Guo Y, Sun S, Yang Y, et al. Functional Analysis of the Autophagy-Related Gene OsATG4b in Rice Grain Chalkiness Regulation. Plants. 2025; 14(16):2530. https://doi.org/10.3390/plants14162530
Chicago/Turabian StyleHu, Zhi, Xiang Han, Yumeng Yuan, Ruishan Xing, Hongchun Liu, Chenming Li, Hongli Shen, Yifan Guo, Shengyuan Sun, Yihao Yang, and et al. 2025. "Functional Analysis of the Autophagy-Related Gene OsATG4b in Rice Grain Chalkiness Regulation" Plants 14, no. 16: 2530. https://doi.org/10.3390/plants14162530
APA StyleHu, Z., Han, X., Yuan, Y., Xing, R., Liu, H., Li, C., Shen, H., Guo, Y., Sun, S., Yang, Y., Guo, M., & Yan, C. (2025). Functional Analysis of the Autophagy-Related Gene OsATG4b in Rice Grain Chalkiness Regulation. Plants, 14(16), 2530. https://doi.org/10.3390/plants14162530





