The Transcription Factor CaNAC81 Is Involved in the Carotenoid Accumulation in Chili Pepper Fruits
Abstract
1. Introduction
2. Results
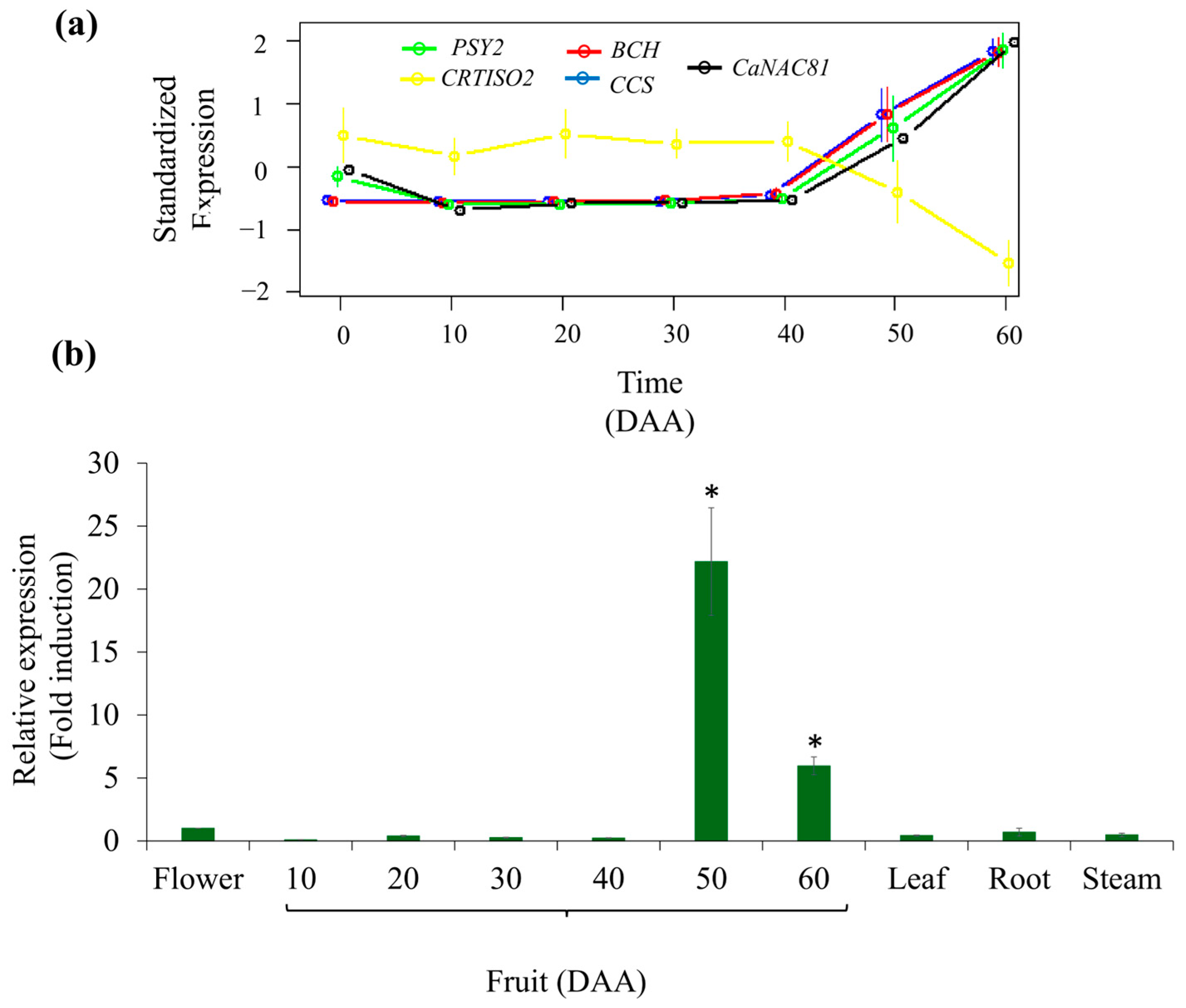
2.1. Selection of the CaNAC81 TF Gene
2.2. Characterization of the CaNAC81 TF Gene
2.3. Virus-Induced Silencing of CaNAC81 in Chili Pepper
3. Discussion
3.1. Selection and Characterization of CaNAC81 TF Gene
3.2. Virus-Induced Silencing Assays of CaNAC81 in Chili Pepper
4. Materials and Methods
4.1. Selection and Characterization of the CaNAC81 TF Gene
4.2. Plant Growth Conditions
4.3. Virus-Induced Silencing of the CaNAC81 Gene
4.4. CaNAC81 Expression Analysis by q-PCR
4.5. Separation and Quantification of Carotenoids by HPLC
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tripodi, P.; Kumar, S. The Capsicum Crop: An Introduction. In The Capsicum Genome. Compendium of Plant Genomes; Ramchiary, N., Kole, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 1–8. [Google Scholar] [CrossRef]
- Idrees, S.; Hanif, M.A.; Ayub, M.A.; Hanif, A.; Ansari, T.M. Chapter 9—Chili Pepper. In Medicinal Plants of South Asia; Hanif, M.A., Nawaz, H., Khan, M.M., Byrne, H.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 113–124. [Google Scholar] [CrossRef]
- Yuca, H. Capsicum annuum L. In Novel Drug Targets with Traditional Herbal Medicines: Scientific and Clinical Evidence; Gürağaç Dereli, F.T., Ilhan, M., Belwal, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2022; pp. 95–108. [Google Scholar] [CrossRef]
- Batiha, G.E.; Alqahtani, A.; Ojo, O.A.; Shaheen, H.M.; Wasef, L.; Elzeiny, M.; Ismail, M.; Shalaby, M.; Murata, T.; Zaragoza-Bastida, A.; et al. Biological Properties, Bioactive Constituents, and Pharmacokinetics of Some Capsicum spp. and Capsaicinoids. Int. J. Mol. Sci. 2020, 21, 5179. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Villegas, R.; Gonzalez-Amaro, R.M.; Figueroa-Hernandez, C.Y.; Rodriguez-Buenfil, I.M. The Genus Capsicum: A Review of Bioactive Properties of Its Polyphenolic and Capsaicinoid Composition. Molecules 2023, 28, 4239. [Google Scholar] [CrossRef]
- Meléndez-Martínez, A.J.; Mandic, A.I.; Bantis, F.; Bohm, V.; Borge, G.I.A.; Brncic, M.; Bysted, A.; Cano, M.P.; Dias, M.G.; Elgersma, A.; et al. A comprehensive review on carotenoids in foods and feeds: Status quo, applications, patents, and research needs. Crit. Rev. Food Sci. Nutr. 2022, 62, 1999–2049. [Google Scholar] [CrossRef]
- Meléndez-Martínez, A.J.; Mapelli-Brahm, P.; Hornero-Méndez, D.; Vicario, I.M. CHAPTER 1. Structures, Nomenclature and General Chemistry of Carotenoids and Their Esters. In Carotenoid Esters in Foods; Mercadante, A.Z., Ed.; The Royal Society of Chemistry: London, UK, 2019; pp. 1–50. [Google Scholar] [CrossRef]
- Mohd Hassan, N.; Yusof, N.A.; Yahaya, A.F.; Mohd Rozali, N.N.; Othman, R. Carotenoids of Capsicum fruits: Pigment and health-promoting functional attributes. Antioxidants 2019, 8, 469. [Google Scholar] [CrossRef]
- Ro, N.; Oh, H.; Ko, H.C.; Yi, J.; Na, Y.W.; Haile, M. Genome-Wide Analysis of Fruit Color and Carotenoid Content in Capsicum Core Collection. Plants 2024, 13, 2562. [Google Scholar] [CrossRef]
- Berry, H.M.; Rickett, D.V.; Baxter, C.J.; Enfissi, E.M.A.; Fraser, P.D. Carotenoid biosynthesis and sequestration in red chilli pepper fruit and its impact on colour intensity traits. J. Exp. Bot. 2019, 70, 2637–2650. [Google Scholar] [CrossRef]
- Sun, T.; Tadmor, Y.; Li, L. Pathways for Carotenoid Biosynthesis, Degradation, and Storage. In Plant and Food Carotenoids: Methods and Protocols; Rodríguez-Concepción, M., Welsch, R., Eds.; Springer: New York, NY, USA, 2020; pp. 3–23. [Google Scholar] [CrossRef]
- Gómez-García, M.d.R.; Ochoa-Alejo, N. Biochemistry and molecular biology of carotenoid biosynthesis in chili peppers (Capsicum spp.). Int. J. Mol. Sci. 2013, 14, 19025–19053. [Google Scholar] [CrossRef]
- Llorente, B.; Martinez-Garcia, J.F.; Stange, C.; Rodriguez-Concepcion, M. Illuminating colors: Regulation of carotenoid biosynthesis and accumulation by light. Curr. Opin. Plant Biol. 2017, 37, 49–55. [Google Scholar] [CrossRef]
- Quian-Ulloa, R.; Stange, C. Carotenoid Biosynthesis and Plastid Development in Plants: The Role of Light. Int. J. Mol. Sci. 2021, 22, 1184. [Google Scholar] [CrossRef]
- Shen, L.F.; Zhang, C.F.; Xia, Y.H.; Yang, S.S.; Chang, T.; Ullah, S.; Ji, X.H. Transcript Analysis Reveals Positive Regulation of CA12g04950 on Carotenoids of Pigment Pepper Fruit under Nitrogen Reduction. Agriculture 2024, 14, 521. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Chen, C.; Zhang, S.; Wang, J.; Huang, Z.; Chen, M.; Cao, B.; Zhu, Z.; Lei, J. Systematic analysis of the Capsicum ERF transcription factor family: Identification of regulatory factors involved in the regulation of species-specific metabolites. BMC Genom. 2020, 21, 573. [Google Scholar] [CrossRef] [PubMed]
- Arce-Rodriguez, M.L.; Martinez, O.; Ochoa-Alejo, N. Genome-Wide Identification and Analysis of the MYB Transcription Factor Gene Family in Chili Pepper (Capsicum spp.). Int. J. Mol. Sci. 2021, 22, 2229. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.H.; Yang, S.P.; Yu, Y.N.; Khan, A.; Feng, P.L.; Ali, M.; Shao, D.K.; Wang, Y.Y.; Zhang, R.X.; Gai, W.X.; et al. Comprehensive transcriptome-based characterization of differentially expressed genes involved in carotenoid biosynthesis of different ripening stages of different ripening stages of Capsicum. Sci. Hortic. 2021, 288, 110311. [Google Scholar] [CrossRef]
- Liu, R.; Song, J.; Liu, S.; Chen, C.; Zhang, S.; Wang, J.; Xiao, Y.; Cao, B.; Lei, J.; Zhu, Z. Genome-wide identification of the Capsicum bHLH transcription factor family: Discovery of a candidate regulator involved in the regulation of species-specific bioactive metabolites. BMC Plant Biol. 2021, 21, 262. [Google Scholar] [CrossRef]
- Villa-Rivera, M.G.; Martinez, O.; Ochoa-Alejo, N. Putative Transcription Factor Genes Associated with Regulation of Carotenoid Biosynthesis in Chili Pepper Fruits Revealed by RNA-Seq Coexpression Analysis. Int. J. Mol. Sci. 2022, 23, 11774. [Google Scholar] [CrossRef]
- Villa-Rivera, M.G.; Martínez, O.; Ochoa-Alejo, N. Transcriptomics of Chili Pepper Fruit with Emphasis on the Carotenoid Biosynthetic Pathway. In Peppers, 1st ed.; Prasad, S., Variyar, I.P.S., Vanshika, A., Penna, S., Eds.; CRC Press: Boca Raton, FL, USA, 2024; pp. 80–97. [Google Scholar] [CrossRef]
- Ma, X.; Yu, Y.-N.; Jia, J.-H.; Li, Q.-H.; Gong, Z.-H. The pepper MYB transcription factor CaMYB306 accelerates fruit coloration and negatively regulates cold resistance. Sci. Hortic. 2022, 295, 110892. [Google Scholar] [CrossRef]
- Ma, J.; Dai, J.-x.; Liu, X.-w.; Lin, D. The transcription factor CaBBX20 regulates capsanthin accumulation in pepper (Capsicum annuum L.). Sci. Hortic. 2023, 314, 111907. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Oh, S.-K.; Lee, S.; Yu, S.H.; Choi, D. Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta 2005, 222, 876–887. [Google Scholar] [CrossRef]
- Guo, W.L.; Wang, S.B.; Chen, R.G.; Chen, B.H.; Du, X.H.; Yin, Y.X.; Gong, Z.H.; Zhang, Y.Y. Characterization and expression profile of CaNAC2 pepper gene. Front. Plant Sci. 2015, 6, 755. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.M.; Zhou, Q.; Zhou, X.; Wei, B.D.; Ji, S.J. Transcription factor CaNAC1 regulates low-temperature-induced phospholipid degradation in green bell pepper. J. Exp. Bot. 2020, 71, 1078–1091. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.M.; Zhang, H.F.; Liu, S.Y.; Wang, X.K.; Zhang, Y.M.; Meng, Y.C.; Luo, D.; Chen, R.G. The NAC transcription factor CaNAC064 is a regulator of cold stress tolerance in peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ma, F.; Wang, X.; Liu, S.; Saeed, U.H.; Hou, X.; Zhang, Y.; Luo, D.; Meng, Y.; Zhang, W.; et al. Molecular and Functional Characterization of CaNAC035, an NAC Transcription Factor From Pepper (Capsicum annuum L.). Front. Plant Sci. 2020, 11, 14. [Google Scholar] [CrossRef]
- Borras, D.; Plazas, M.; Moglia, A.; Lanteri, S. The influence of acute water stresses on the biochemical composition of bell pepper (Capsicum annuum L.) berries. J. Sci. Food Agric. 2021, 101, 4724–4734. [Google Scholar] [CrossRef]
- Jia, G.; Shwe Zin Thinn, K.; Kim, S.H.; Min, J.; Oh, S.-K. Capsicum annuum NAC4 (CaNAC4) Is a Transcription Factor with Roles in Biotic and Abiotic Stresses. Plant Pathol. J. 2024, 40, 512–524. [Google Scholar] [CrossRef]
- Xiao, J.; Sui, X.; Xu, Z.; Liang, L.; Tang, W.; Tang, Y.; Sun, B.; Lai, Y.; Huang, Z.; Zheng, Y.; et al. CaNAC76 enhances lignin content and cold resistance in pepper by regulating CaCAD1. Int. J. Biol. Macromol. 2025, 285, 138271. [Google Scholar] [CrossRef]
- Liu, G.-S.; Li, H.-L.; Grierson, D.; Fu, D.-Q. NAC Transcription Factor Family Regulation of Fruit Ripening and Quality: A Review. Cells 2022, 11, 525. [Google Scholar] [CrossRef]
- Song, S.; Song, S.Y.; Nian, P.; Lv, D.; Jing, Y.; Lu, S.; Wang, Q.; Zhou, F. Transcriptomic analysis suggests a coordinated regulation of carotenoid metabolism in ripening chili pepper (Capsicum annuum var. conoides) fruits. Antioxidants 2022, 11, 2245. [Google Scholar] [CrossRef]
- Diao, W.; Snyder, J.C.; Wang, S.; Liu, J.; Pan, B.; Guo, G.; Ge, W.; Dawood, M. Genome-Wide Analyses of the NAC Transcription Factor Gene Family in Pepper (Capsicum annuum L.): Chromosome Location, Phylogeny, Structure, Expression Patterns, Cis-Elements in the Promoter, and Interaction Network. Int. J. Mol. Sci. 2018, 19, 1028. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, J.; Sun, H.; Xiong, C.; Sun, X.; Wang, X.; Wang, Z.; Jarret, R.; Wang, J.; Tang, B.; et al. Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation. Nat. Commun. 2023, 14, 5487. [Google Scholar] [CrossRef] [PubMed]
- Escoto-Sandoval, C.; Flores-Díaz, A.; Reyes-Valdés, M.H.; Ochoa-Alejo, N.; Martínez, O. A method to analyze time expression profiles demonstrated in a database of chili pepper fruit development. Sci. Rep. 2021, 11, 13181. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Liu, C.; Han, L.; Wang, S.; Xue, Z. NAC transcription factors play an important role in ethylene biosynthesis, reception and signaling of tomato fruit ripening. Mol. Genet. Genom. 2016, 291, 1205–1217. [Google Scholar] [CrossRef]
- Gao, Y.; Wei, W.; Zhao, X.; Tan, X.; Fan, Z.; Zhang, Y.; Jing, Y.; Meng, L.; Zhu, B.; Zhu, H.; et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 2018, 5, 75. [Google Scholar] [CrossRef]
- Bhatt, T.; Patel, K. Carotenoids: Potent to Prevent Diseases Review. Nat. Prod. Bioprospect. 2020, 10, 109–117. [Google Scholar] [CrossRef]
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 2014, 26, 3167–3184. [Google Scholar] [CrossRef]
- Weryszko-Chmielewska, E.; Michałojć, Z. Anatomical traits of sweet pepper (Capsicum annuum L.) fruit. Acta Agrobot. 2012, 64, 181–188. [Google Scholar] [CrossRef]
- Ernst, H.A.; Olsen, A.N.; Larsen, S.; Lo Leggio, L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Qiu, H.; Chen, R.; Xiong, A.; Xu, Z.; Miao, W.; Chen, R.; Wang, P.; Hou, X.; et al. The MADS-RIPENING INHIBITOR-DIVARICATA1 module regulates carotenoid biosynthesis in nonclimacteric Capsicum fruits. Plant Physiol. 2025, 197. [Google Scholar] [CrossRef]
- Wei, X.; Meng, C.; Yuan, Y.; Nath, U.K.; Zhao, Y.; Wang, Z.; Yang, S.; Li, L.; Niu, L.; Yao, Q.; et al. CaPSY1 gene plays likely the key role in carotenoid metabolism of pepper (Capsicum annuum) at ripening. Funct. Plant Biol. 2021, 48, 141–155. [Google Scholar] [CrossRef]
- Tian, S.L.; Li, L.; Shah, S.N.M.; Gong, Z.H. The relationship between red fruit colour formation and key genes of capsanthin biosynthesis pathway in Capsicum annuum. Biol. Plant. 2015, 59, 507–513. [Google Scholar] [CrossRef]
- Chow, C.-N.; Yang, C.-W.; Wu, N.-Y.; Wang, H.-T.; Tseng, K.-C.; Chiu, Y.-H.; Lee, T.-Y.; Chang, W.-C. PlantPAN 4.0: Updated database for identifying conserved non-coding sequences and exploring dynamic transcriptional regulation in plant promoters. Nucleic Acids Res. 2023, 52, D1569–D1578. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The protein sequence classification resource in 2025. Nucleic Acids Res. 2024, 53, D444–D456. [Google Scholar] [CrossRef]
- Bailey, T.L.; Gribskov, M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 1998, 14, 48–54. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Steinegger, M.; Meier, M.; Mirdita, M.; Vöhringer, H.; Haunsberger, S.J.; Söding, J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinform. 2019, 20, 473. [Google Scholar] [CrossRef]
- Studer, G.; Tauriello, G.; Bienert, S.; Biasini, M.; Johner, N.; Schwede, T. ProMod3-A versatile homology modelling toolbox. PLoS Comput. Biol. 2021, 17, e1008667. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Prestridge, D.S. SIGNAL SCAN: A computer program that scans DNA sequences for eukaryotic transcriptional elements. Bioinformatics 1991, 7, 203–206. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Arce-Rodriguez, M.L.; Ochoa-Alejo, N. Virus-Induced Gene Silencing (VIGS) in Chili Pepper (Capsicum spp.). Methods Mol. Biol. 2020, 2172, 27–38. [Google Scholar] [CrossRef]
- Ahmed, F.; Senthil-Kumar, M.; Dai, X.; Ramu, V.S.; Lee, S.; Mysore, K.S.; Zhao, P.X. pssRNAit: A Web Server for Designing Effective and Specific Plant siRNAs with Genome-Wide Off-Target Assessment. Plant Physiol. 2020, 184, 65–81. [Google Scholar] [CrossRef]
- Arce-Rodriguez, M.L.; Ochoa-Alejo, N. An R2R3-MYB Transcription Factor Regulates Capsaicinoid Biosynthesis. Plant Physiol. 2017, 174, 1359–1370. [Google Scholar] [CrossRef]
- Martinez-Lopez, L.A.; Ochoa-Alejo, N.; Martinez, O. Dynamics of the chili pepper transcriptome during fruit development. BMC Genom. 2014, 15, 143. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Richins, R.D.; Kilcrease, J.; Rodgriguez-Uribe, L.; O’Connell, M.A. Carotenoid Extraction and Quantification from Capsicum annuum. Bio Protoc. 2014, 4, e1256. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villa-Rivera, M.G.; Castañeda-Marín, A.; Martínez, O.; Ochoa-Alejo, N. The Transcription Factor CaNAC81 Is Involved in the Carotenoid Accumulation in Chili Pepper Fruits. Plants 2025, 14, 2099. https://doi.org/10.3390/plants14142099
Villa-Rivera MG, Castañeda-Marín A, Martínez O, Ochoa-Alejo N. The Transcription Factor CaNAC81 Is Involved in the Carotenoid Accumulation in Chili Pepper Fruits. Plants. 2025; 14(14):2099. https://doi.org/10.3390/plants14142099
Chicago/Turabian StyleVilla-Rivera, Maria Guadalupe, Alejandra Castañeda-Marín, Octavio Martínez, and Neftalí Ochoa-Alejo. 2025. "The Transcription Factor CaNAC81 Is Involved in the Carotenoid Accumulation in Chili Pepper Fruits" Plants 14, no. 14: 2099. https://doi.org/10.3390/plants14142099
APA StyleVilla-Rivera, M. G., Castañeda-Marín, A., Martínez, O., & Ochoa-Alejo, N. (2025). The Transcription Factor CaNAC81 Is Involved in the Carotenoid Accumulation in Chili Pepper Fruits. Plants, 14(14), 2099. https://doi.org/10.3390/plants14142099








