Identification of the Glyceraldehyde-3-Phosphate Dehydrogenase (GeGAPDH) Gene Family in Gastrodia elata Revealing Its Response Characteristics to Low-Temperature and Pathogen Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Characterization of the GeGAPDH Family
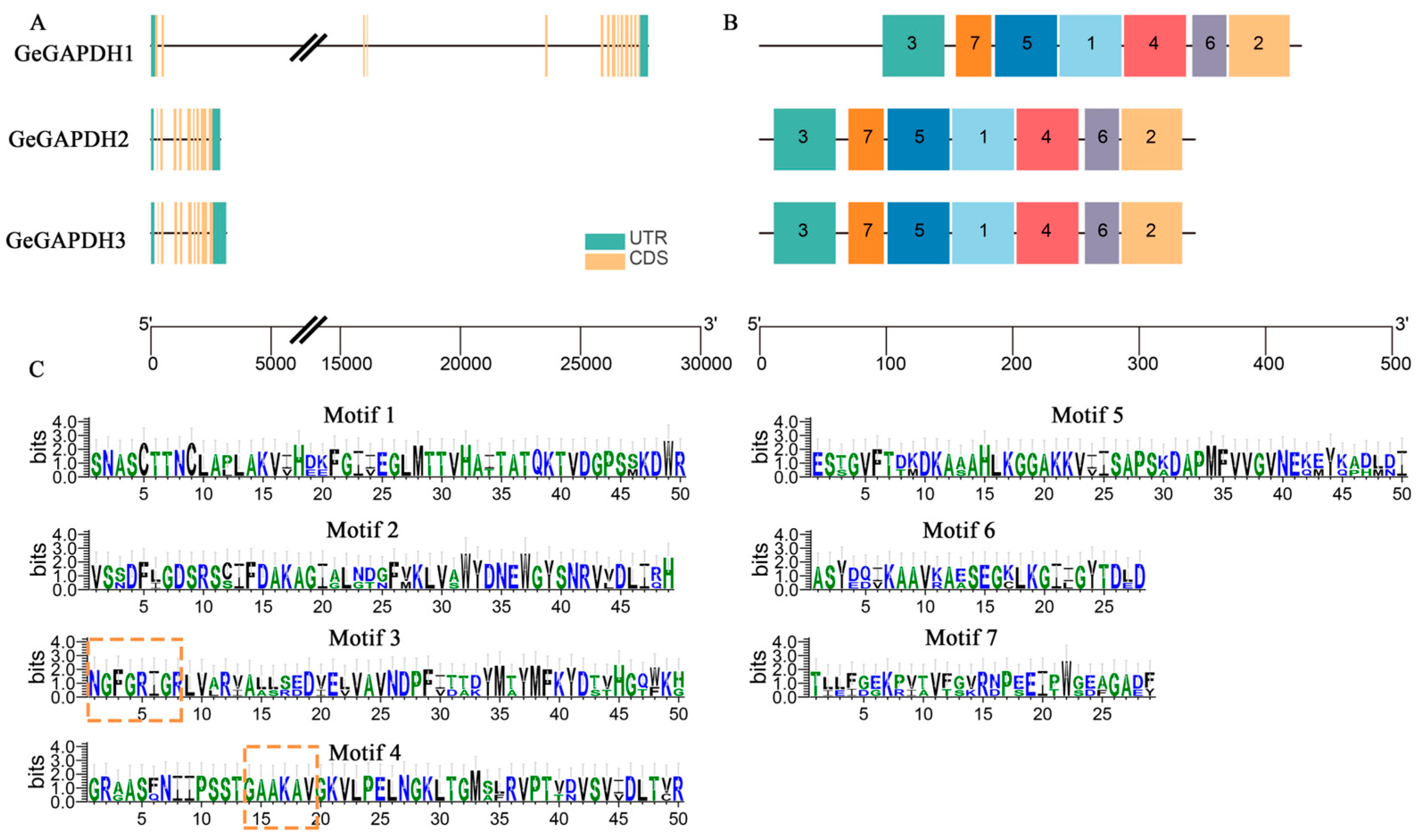
2.2. Gene Structure and Conserved Domain Analysis of the GeGAPDH Family
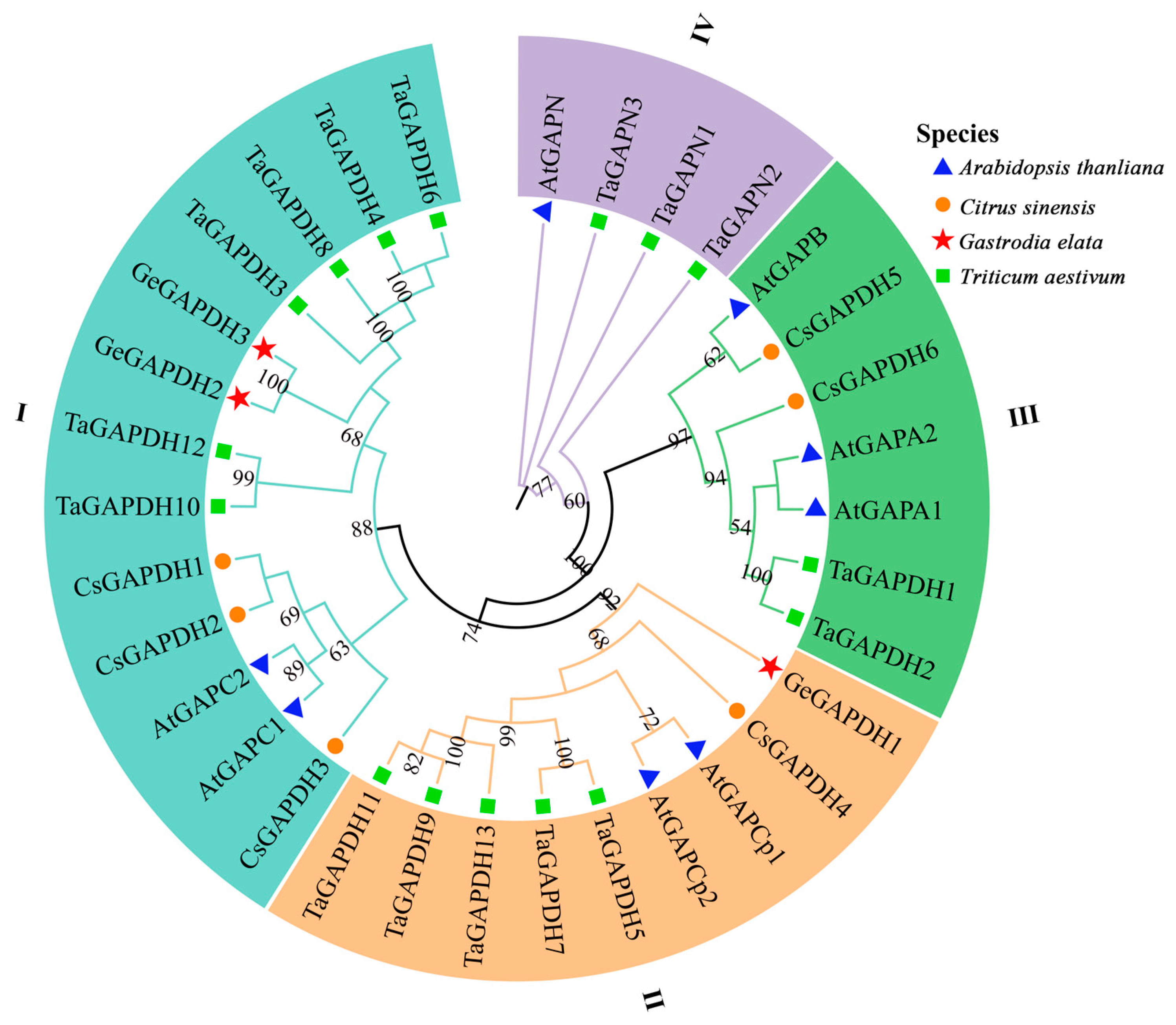
2.3. Phylogenetic Analysis of the GAPDH Family
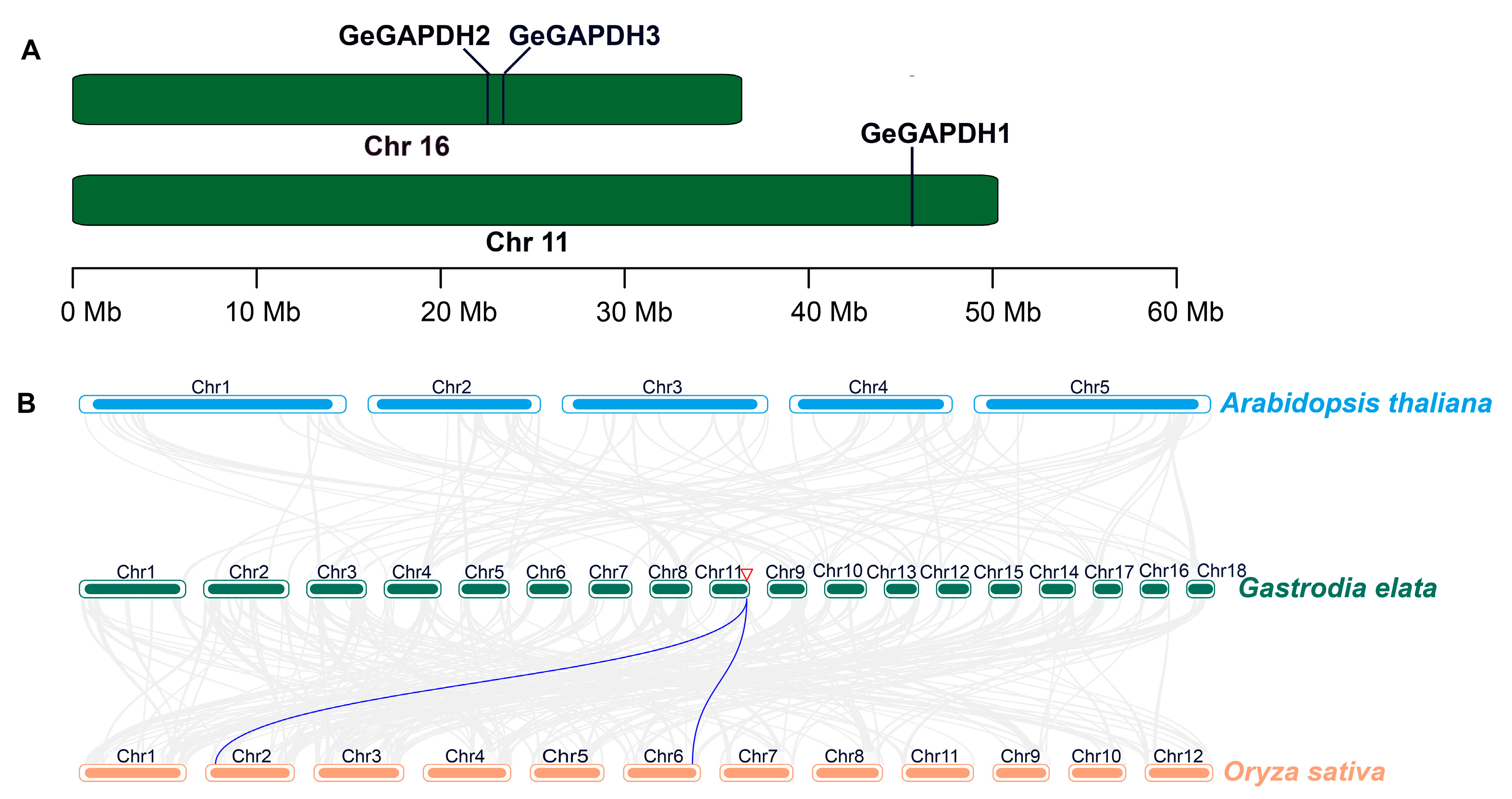
2.4. Chromosomal Localization and Collinearity Analysis of the GeGAPDH Family
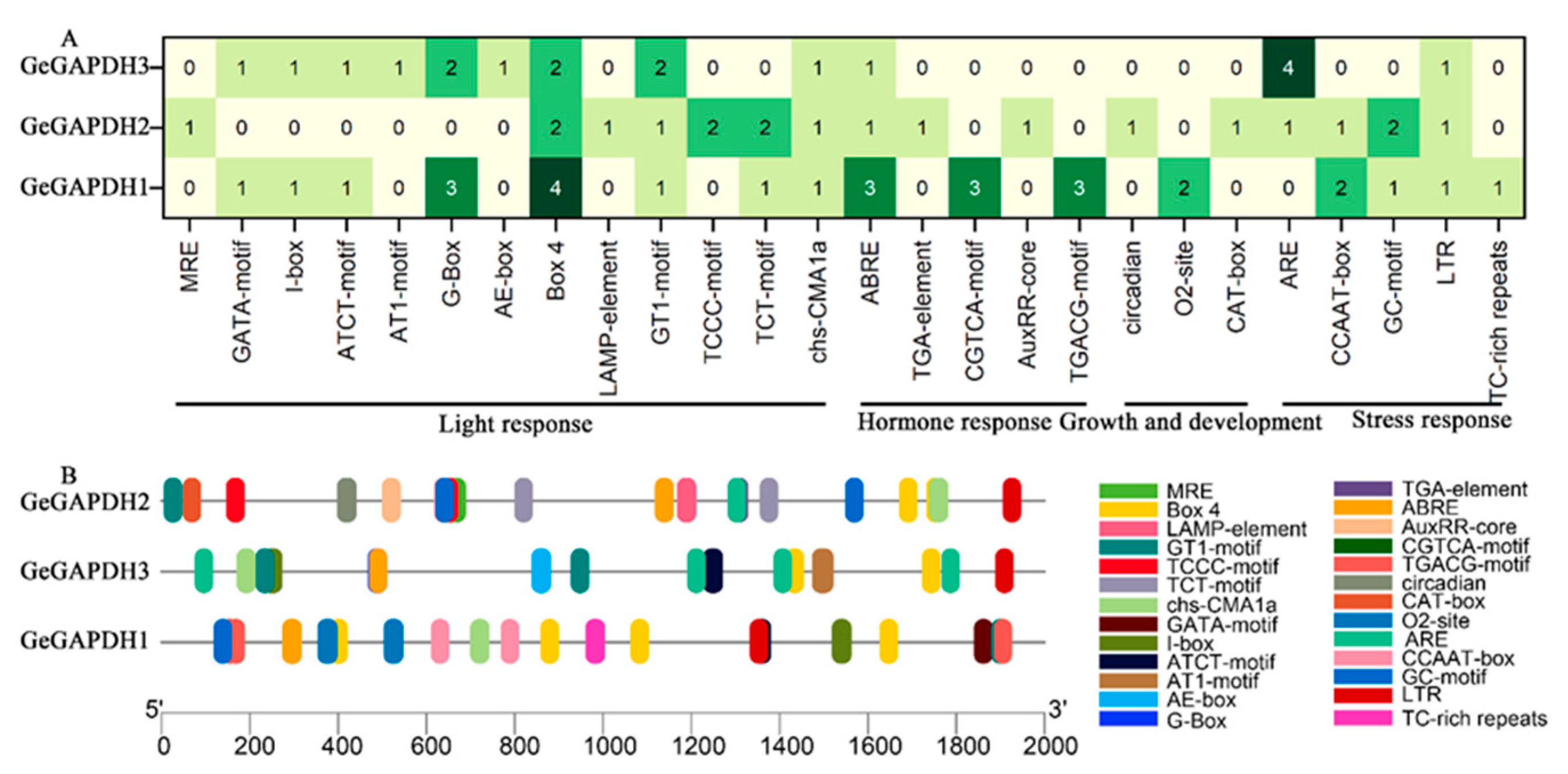
2.5. Cis-Acting Element Analysis of the GeGAPDH Family
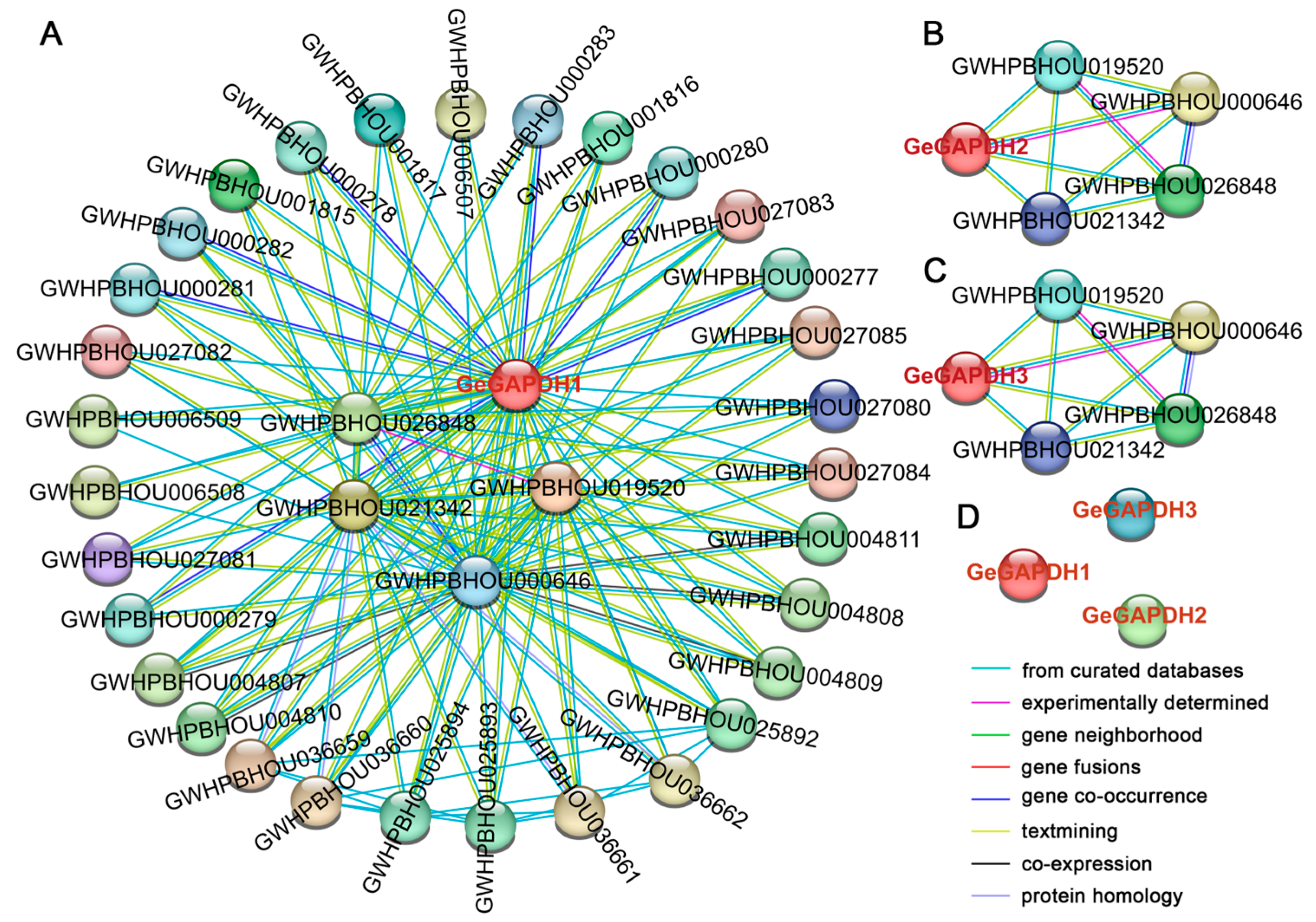
2.6. Interaction Analysis of GeGAPDH Proteins
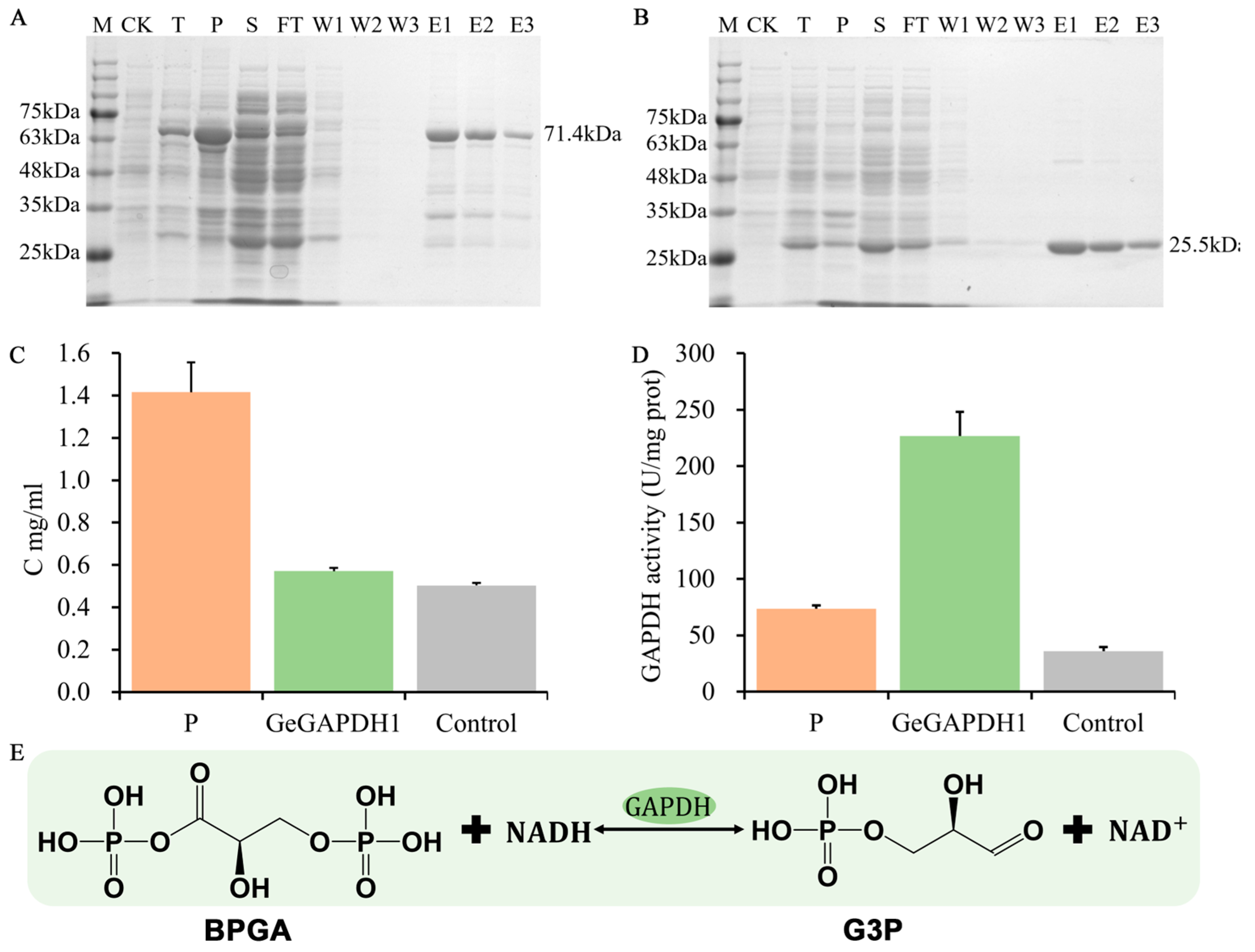
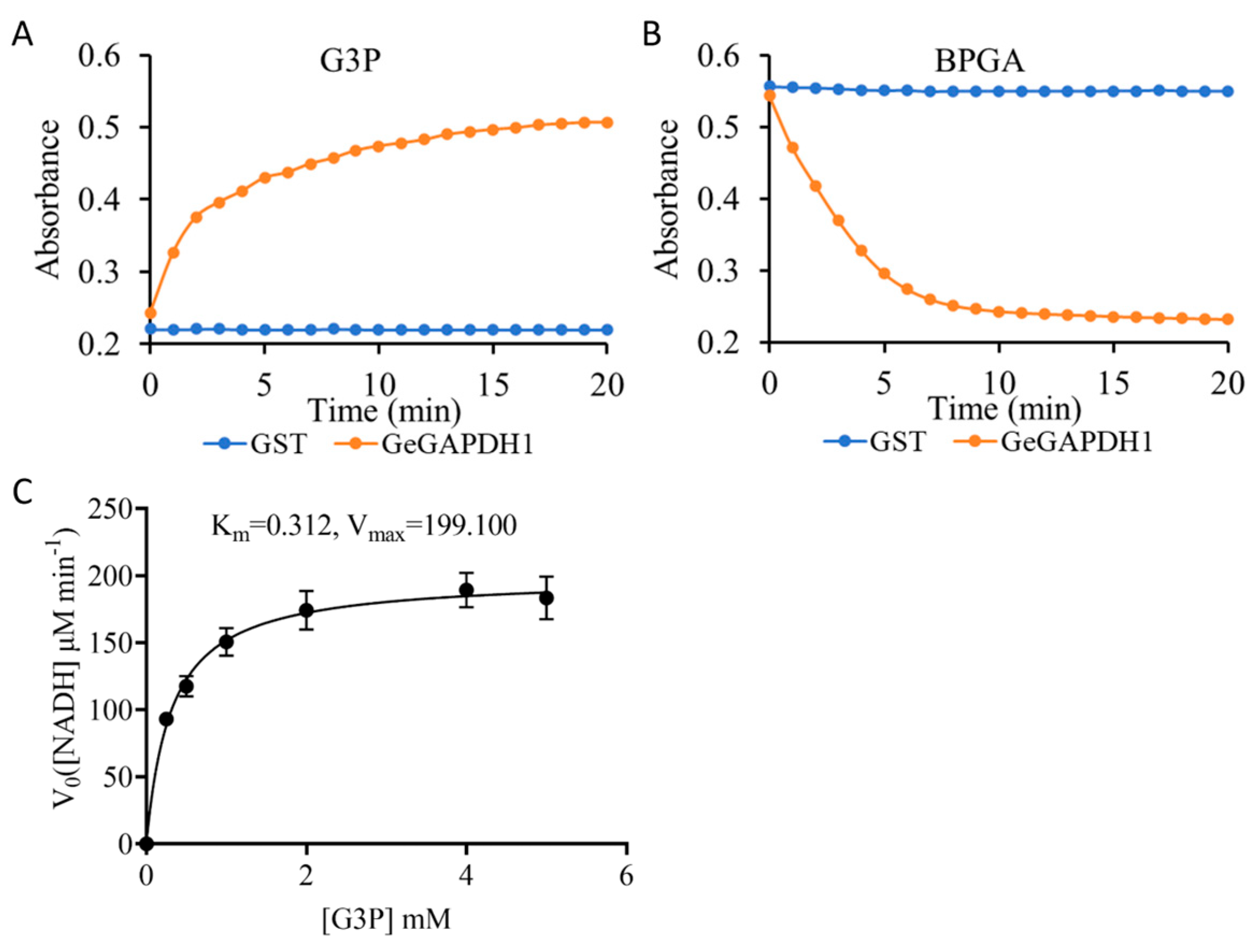
2.7. Enzyme Activity and Kinetics Detection of the GeGAPDH1
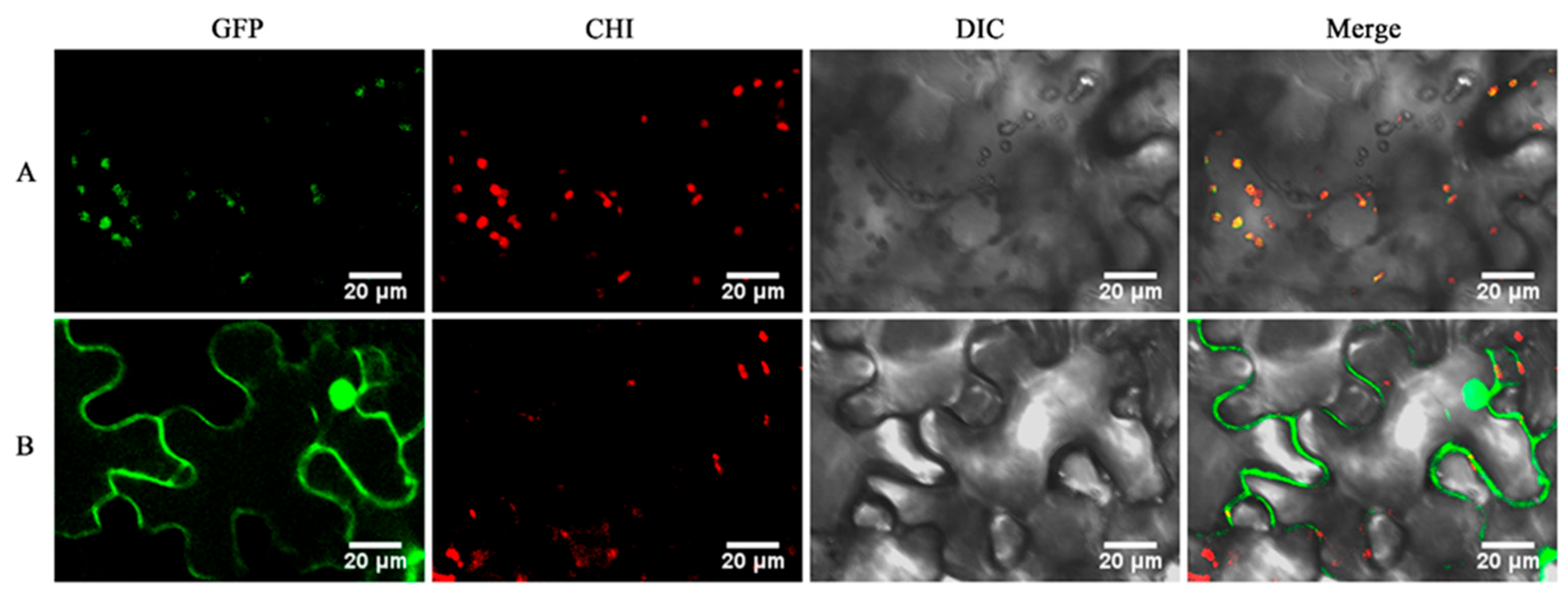
2.8. Subcellular Localization Analysis of GeGAPDH1
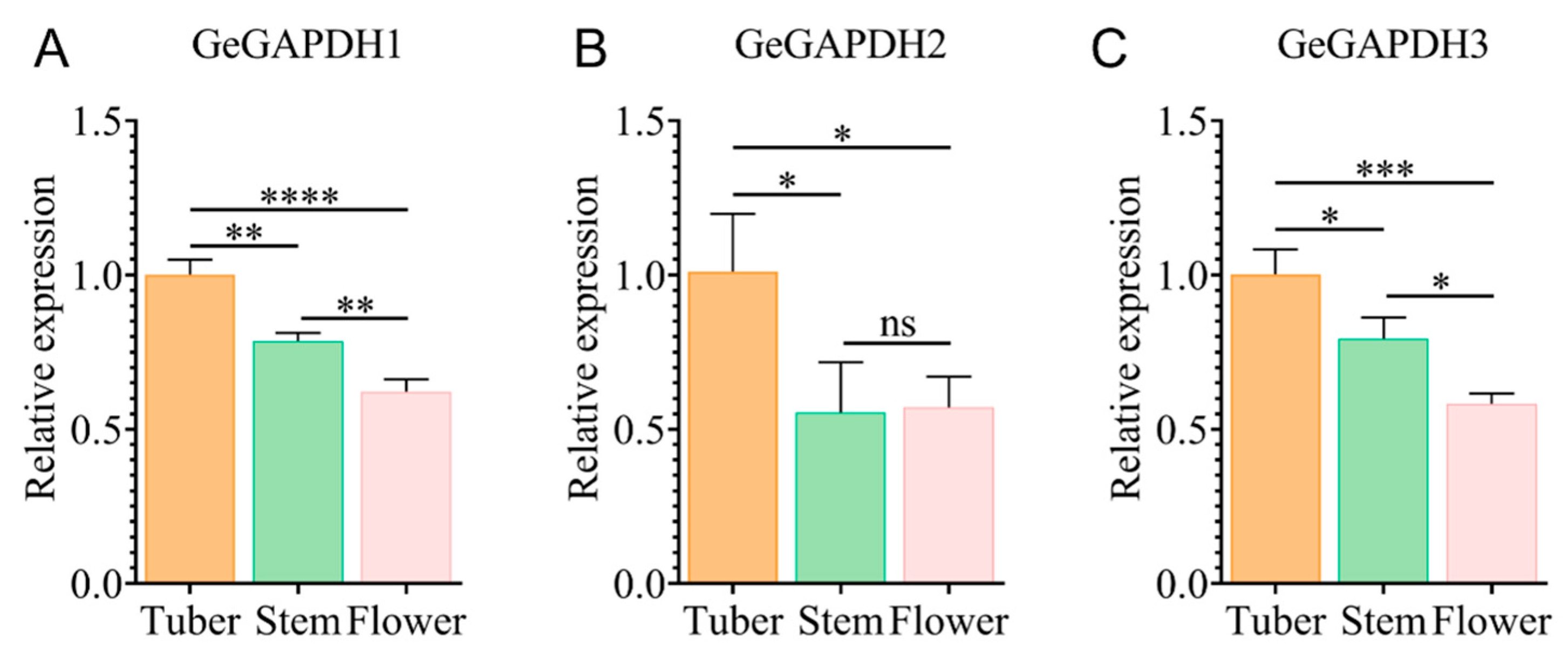
2.9. Expression Characteristics of the GeGAPDH Family Among Tissues
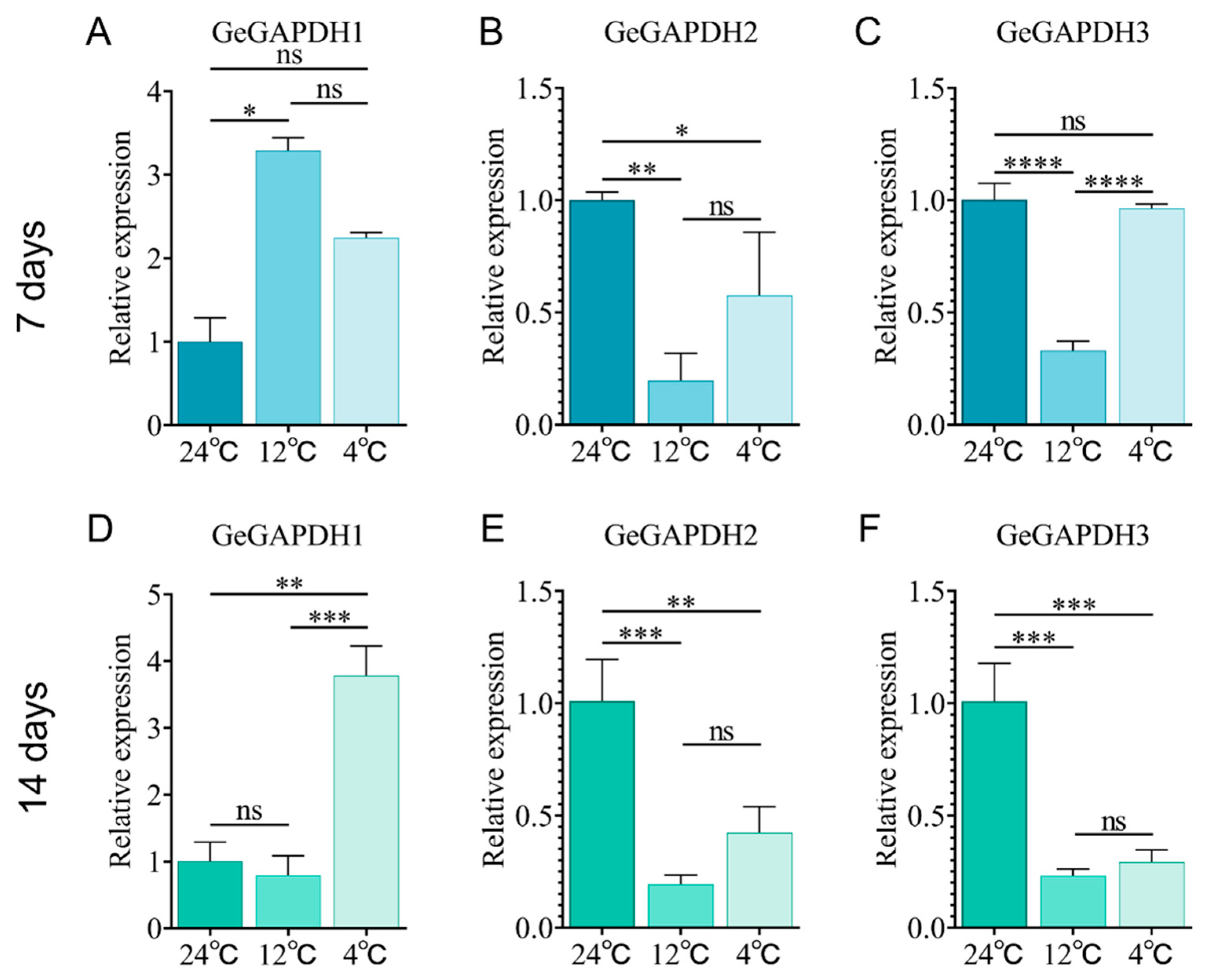
2.10. Expression Characteristics of the GeGAPDH Family Under Low-Temperature Stress
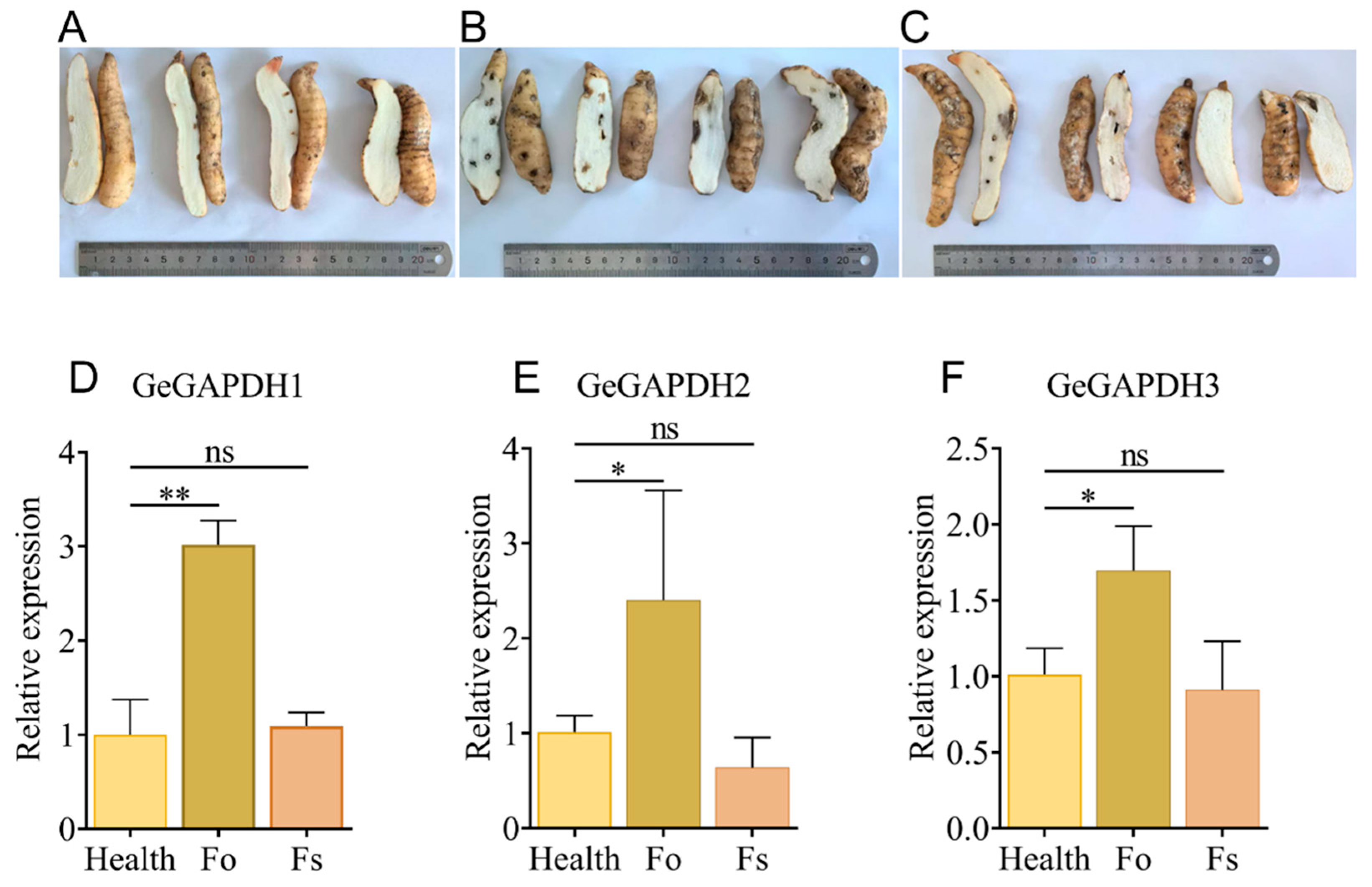
2.11. Expression Characteristics of the GeGAPDH Family Under Pathogen Stress
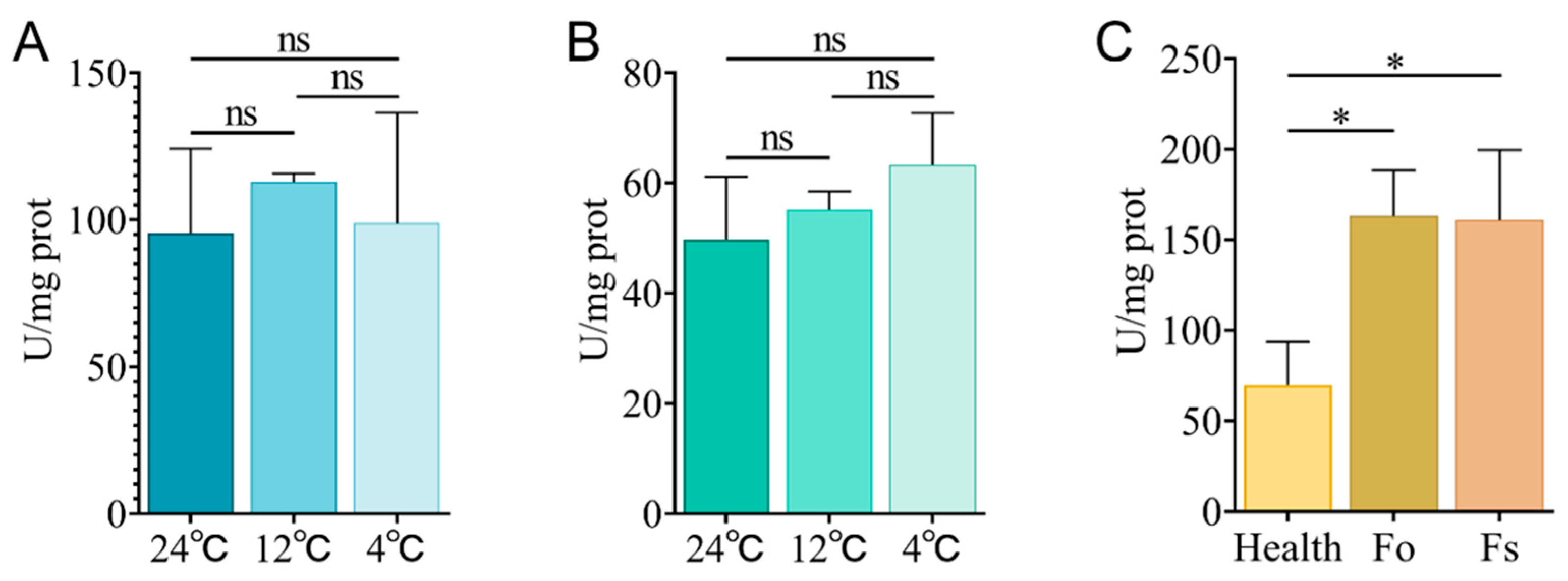
2.12. Detection of GAPDH Enzyme Activity Under Low Temperature and Fusarium Stress
3. Discussion
4. Materials and Methods
4.1. Identification of the GeGAPDH Family in G. elata
4.2. Characterization of the GeGAPDH Family
4.3. Collinearity, Phylogenetic and Protein–Protein Interaction Network Analysis of GAPDH Family
4.4. Detection of GAPDH Activity in Total Protein Extracted from G. elata
4.5. Cloning, Expression, and Subcellular Localization of the GeGAPDH Genes
4.5.1. RNA Extraction and cDNA Synthesis
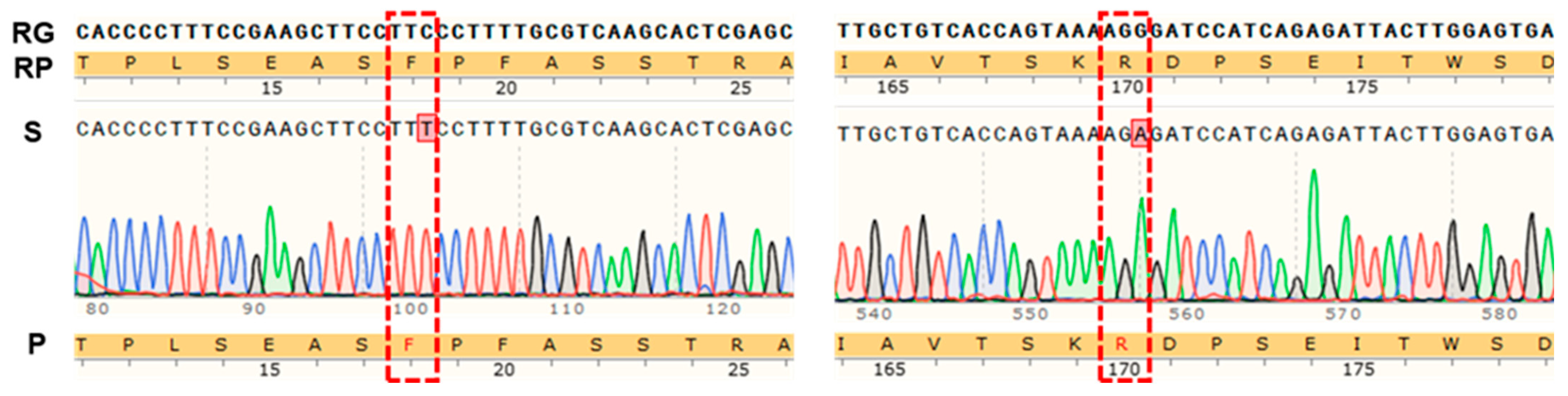
4.5.2. Cloning of the GeGAPDH Genes
4.5.3. Expression, Purification, Enzyme Activity and Kinetics Detection of the GeGAPDH Genes
4.5.4. Subcellular Localization of the GeGAPDH1 Gene
4.6. Expression Profile Analysis of the GeGAPDH Family
4.6.1. Effect of Low-Temperature Stress on GeGAPDH Expression in G. elata in Symbiotic with A. mellea
4.6.2. Effect of F. oxysporum and F. solani Stress on GeGAPDH Expression in G. elata in Symbiotic with A. mellea
4.6.3. Detection of GAPDH Enzyme Activity Under Low Temperature and Pathogen Stress Conditions
4.6.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, P.J.; Sheen, L.Y. Gastrodiae Rhizoma (tiān má): A review of biological activity and antidepressant mechanisms. J. Tradit. Complement. Med. 2011, 1, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.H.; Huang, N.K.; Shiao, Y.J.; Lu, C.K.; Chao, Y.M.; Huang, Y.J.; Yeh, C.H.; Lin, Y.L. Gastrodiae rhizoma attenuates brain aging via promoting neuritogenesis and neurodifferentiation. Phytomed. Int. J. Phytother. Phytopharm. 2021, 87, 153576. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, J.; Peng, M.; Meng, H.; Ma, H.; Cai, P.; Xu, Y.; Zhao, Q.; Si, G. A review on central nervous system effects of Gastrodin. Front. Pharmacol. 2018, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.W.; Chen, B.; Zhu, H.F.; Liang, Y.L.; Mao, L.S. Gastrodin alleviates cisplatin nephrotoxicity by inhibiting ferroptosis via the SIRT1/FOXO3A/GPX4 signaling pathway. J. Ethnopharmacol. 2024, 319, 117282. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, S.; Liu, Y.; Gong, C.; Xiang, L.; Li, S.; Wang, P.; Wang, Y.; Sun, L.; Zhang, Q.; et al. Parishin alleviates vascular ageing in mice by upregulation of Klotho. J. Cell. Mol. Med. 2023, 27, 1398–1409. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhou, S.; Yan, R.; Gong, C.; Gui, Q.; Zhang, Q.; Xiang, L.; Chen, L.; Wang, P.; Li, S.; et al. Parishin From Gastrodia elata Ameliorates Aging Phenotype in Mice in a Gut Microbiota-Related Manner. Front. Microbiol. 2022, 13, 877099. [Google Scholar] [CrossRef]
- Liao, C.C.; Yu, H.P.; Chou, A.H.; Lee, H.C.; Hu, L.M.; Liu, F.C. Gastrodin Alleviates Acetaminophen—Induced Liver Injury in a Mouse Model Through Inhibiting MAPK and Enhancing Nrf2 Pathways. Inflammation 2022, 45, 1450–1462. [Google Scholar] [CrossRef]
- Wu, G.Z.; Jia, C.Q.; Wang, X.; Chen, L.; Liu, F.; Zhang, Y.Y.; Dong, H.J.; Li, J. Research progress on extraction, purification and pharmacological activities of Gastrodiae R hizoma polysaccharides. Chin. Arch. Tradit. Chin. Med. 2022, 40, 135–139. [Google Scholar] [CrossRef]
- Zhou, C.Y.; Di, Y.G.; Qiu, Q.L.; Cai, J.L.; Liu, Y.X.; Xu, H.N.; Li, K.Z. Effects of Low Temperature Stress on Growth and Development of G. elata were Analyzed by Transcriptome and Metabolome. Mol. Plant Breed. 2023, 21, 110–122. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, J.Q.; Jiang, W.K.; Yuan, Q.S.; Wang, Y.H.; Guo, L.P.; Yang, Y.; Yang, Y.; Zhou, T. Isolation, identification, and pathogenicity research of brown rot pathogens from Gastrodia elata. China J. Chin. Mater. 2022, 47, 2288–2295. [Google Scholar] [CrossRef]
- Zhou, M.C.; Li, H.Q.; Liu, M.; Xu, Q.Z.; Cheng, Z.J.; Wang, C.Y. Research progress of Gastrodia elata disease and insect pests and their control. South Cent. Agric. Sci. Technol. 2024, 45, 230–235. [Google Scholar]
- Li, J.; He, K.; Zhang, Q.; Wu, X.; Li, Z.; Pan, X.; Wang, Y.; Li, C.; Zhang, M. Draft genome and biological characteristics of Fusarium solani and Fusarium oxysporum causing black rot in Gastrodia elata. Int. J. Mol. Sci. 2023, 24, 4545. [Google Scholar] [CrossRef] [PubMed]
- Henry, E.; Fung, N.; Liu, J.; Drakakaki, G.; Coaker, G. Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 2015, 11, e1005199. [Google Scholar] [CrossRef] [PubMed]
- Wegener, M.; Persicke, M.; Dietz, K.J. Reprogramming the translatome during daily light transitions as affected by cytosolic glyceraldehyde-3-phosphate dehydrogenases GAPC1/C2. J. Exp. Bot. 2024, 75, 2494–2509. [Google Scholar] [CrossRef]
- Marri, L.; Sparla, F.; Pupillo, P.; Trost, P. Co-ordinated gene expression of photosynthetic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase, and CP12 in Arabidopsis thaliana. J. Exp. Bot. 2005, 56, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Anoman, A.D.; Muñoz-Bertomeu, J.; Rosa-Téllez, S.; Flores-Tornero, M.; Serrano, R.; Bueso, E.; Fernie, A.R.; Segura, J.; Ros, R. Plastidial Glycolytic Glyceraldehyde-3-Phosphate Dehydrogenase Is an Important Determinant in the Carbon and Nitrogen Metabolism of Heterotrophic Cells in Arabidopsis. Plant Physiol. 2015, 169, 1619–1637. [Google Scholar] [CrossRef]
- Piattoni, C.V.; Rius, S.P.; Gomez-Casati, D.F.; Guerrero, S.A.; Iglesias, A.A. Heterologous expression of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Triticum aestivum and Arabidopsis thaliana. Biochimie 2010, 92, 909–913. [Google Scholar] [CrossRef]
- Guo, L.; Ma, F.; Wei, F.; Fanella, B.; Allen, D.K.; Wang, X. Cytosolic phosphorylating glyceraldehyde-3-phosphate dehydrogenases affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell 2014, 26, 3023–3035. [Google Scholar] [CrossRef]
- Trost, P.; Fermani, S.; Marri, L.; Zaffagnini, M.; Falini, G.; Scagliarini, S.; Pupillo, P.; Sparla, F. Thioredoxin-dependent regulation of photosynthetic glyceraldehyde-3-phosphate dehydrogenase: Autonomous vs. CP12-dependent mechanisms. Photosynth. Res. 2006, 89, 263–275. [Google Scholar] [CrossRef]
- Zeng, L.; Deng, R.; Guo, Z.; Yang, S.; Deng, X. Genome-wide identification and characterization of Glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genom. 2016, 17, 240. [Google Scholar] [CrossRef]
- Meng, X.; Lv, Y.; Mujahid, H.; Edelmann, M.J.; Zhao, H.; Peng, X.; Peng, Z. Proteome-wide lysine acetylation identification in developing rice (Oryza sativa) seeds and protein co-modification by acetylation, succinylation, ubiquitination, and phosphorylation. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Su, Y.M.; Deng, L.; Zhang, F.C. Expressoin pattern analysis of cyosol glyceraldehyde-3-phos-phatedehydrogenase genes in Arabidopsis. Biotechnology 2016, 26, 469–472. [Google Scholar] [CrossRef]
- Li, Y.M.; OuYang, M.Z.; Sun, S.R.; Zhu, L. Cloning and Expression Analysis of Glyceraldehyde-3-phosphate Dehydro genase (GAPDH) Gene CaGAPB in Pepper. Mol. Plant Breed. 2018, 16, 1382–1389. [Google Scholar] [CrossRef]
- Zhu, P.S.; Liu, H.Y.; Cao, Z.; Cong, Y.D. Identification of GAPDH Gene Family in Tomato and Its Response to GSNO Regulating NaCl Stress. Mol. Plant Breed. 2023, 21, 7682–7688. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, Z.H.; Mei, F.J.; Wang, T.C.; Li, Y.H. Cloning and Analysis of Expression Characteristics of Glyceraldehyde-3-Phosphate Dehydrogenase 2(ZmGAPDH2) Gene in Maize. J. Maize Sci. 2022, 30, 42–50. [Google Scholar] [CrossRef]
- Wang, X.L.; Ji, J.J.; Gao, J.P. Clone and expression of CpGAPDH gene in Codonopsis pilosula. China J. Chin. Mater. Medica 2018, 43, 712–720. [Google Scholar] [CrossRef]
- Zhu, H.; Li, K.; Zhao, R.Q.; Wu, Y.S. Cloning and sequencing of Panax notoginseng GAPDH. Acta Bot. Boreali Occident. Sin. 2006, 26, 1316–1319. [Google Scholar] [CrossRef]
- Zhu, L.F.; Yang, C.Q.; You, Y.H.; Liang, W.; Wang, N.N.; Ma, F.W.; Li, C.Y. Validation of reference genes for qRT-PCR analysis in peel and flesh of six apple cultivars (Malus domestica) at diverse stages of fruit development. Sci. Hortic. 2019, 244, 165–171. [Google Scholar] [CrossRef]
- Li, Y.M.; Sun, S.R.; Wang, Y.; Cai, X.X.; Yao, J.X.; Zhu, L. Identification of the GAPDH gene family in Citrullus lanatus and functional characteristics of ClGAPC2 in Arabidopsis thaliana. Plant Biol. 2023, 25, 334–342. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, J.; Xia, N.; Qu, Y.; Zhan, Y.; Teng, W.; Li, H.; Li, W.; Li, Y.; Zhao, X.; et al. Genome—wide identification and analysis of glyceraldehyde-3-phosphate dehydrogenase family reveals the role of GmGAPDH14 to improve salt tolerance in soybean (Glycine max L.). Front. Plant Sci. 2023, 14, 1193044. [Google Scholar] [CrossRef]
- Li, X.; Wei, W.; Li, F.; Zhang, L.; Deng, X.; Liu, Y.; Yang, S. The Plastidial Glyceraldehyde-3-Phosphate Dehydrogenase Is Critical for Abiotic Stress Response in Wheat. Int. J. Mol. Sci. 2019, 20, 1104. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Bertomeu, J.; Bermúdez, M.A.; Segura, J.; Ros, R. Arabidopsis plants deficient in plastidial glyceraldehyde-3-phosphate dehydrogenase show alterations in abscisic acid (ABA) signal transduction: Interaction between ABA and primary metabolism. J. Exp. Bot. 2011, 62, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Xie, Y.; Liu, G.; Lin, D.; He, C.; Shi, H. Molecular identification of GAPDHs in cassava highlights the antagonism of MeGAPCs and MeATG8s in plant disease resistance against cassava bacterial blight. Plant Mol. Biol. 2018, 97, 201–214. [Google Scholar] [CrossRef]
- Xu, J.; Wang, R.; Zhang, X.; Zhuang, W.; Zhang, Y.; Lin, J.; Zhan, P.; Chen, S.; Lu, H.; Wang, A.; et al. Identification and expression profiling of GAPDH family genes involved in response to Sclerotinia sclerotiorum infection and phytohormones in Brassica napus. Front. Plant Sci. 2024, 15, 1360024. [Google Scholar] [CrossRef]
- Liu, J.J.; Yang, X.Q.; Li, Z.Y.; Miao, J.Y.; Li, S.B.; Zhang, W.P.; Lin, Y.C.; Lin, L.B. The role of symbiotic fungi in the life cycle of Gastrodia elata Blume (Orchidaceae): A comprehensive review. Front. Plant Sci. 2023, 14, 1309038. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Chen, C.; Yao, L.; Tran, J.; Zhang, H. Genome-wide identification, characterization, interaction network and expression profile of GAPDH gene family in sweet orange (Citrus sinensis). PeerJ. 2019, 7, e7934. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Fermani, S.; Costa, A.; Lemaire, S.D.; Trost, P. Plant cytoplasmic GAPDH: Redox post-translational modifications and moonlighting properties. Front. Plant Sci. 2013, 4, 450. [Google Scholar] [CrossRef]
- Schormann, N.; Campos, J.; Motamed, R.; Hayden, K.L.; Gould, J.R.; Green, T.J.; Senkovich, O.; Banerjee, S.; Ulett, G.C.; Chattopadhyay, D. Chlamydia trachomatis glyceraldehyde 3-phosphate dehydrogenase: Enzyme kinetics, high-resolution crystal structure, and plasminogen binding. Protein Sci. A Publ. Protein Soc. 2020, 29, 2446–2458. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, H.; Yang, S. Cytosolic TaGAPC2 Enhances Tolerance to Drought Stress in Transgenic Arabidopsis Plants. Int. J. Mol. Sci. 2020, 21, 7499. [Google Scholar] [CrossRef]
- Kim, S.C.; Guo, L.; Wang, X. Nuclear moonlighting of cytosolic glyceraldehyde-3-phosphate dehydrogenase regulates Arabidopsis response to heat stress. Nat. Commun. 2020, 11, 3439. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Long, F.; Li, Y.; Xu, Y.; Hu, L.; Yao, T.; Huang, Y.; Hu, D.; Yang, Y.; Fei, Y. Comparative Transcriptome Analysis Revealing the Potential Mechanism of Low-Temperature Stress in Machilus microcarpa. Front. Plant Sci. 2022, 13, 900870. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lu, L.; Zeng, M.; Wang, D.; Zhang, T.Z.; Xie, Y.; Gao, S.B.; Fu, S.; Zhou, X.P.; Wu, J.X. Rice black-streaked dwarf virus P10 promotes phosphorylation of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) to induce autophagy in Laodelphax striatellus. Autophagy 2022, 18, 745–764. [Google Scholar] [CrossRef]
- Petersen, J.; Brinkmann, H.; Cerff, R. Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J. Mol. Evol. 2003, 57, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Bertomeu, J.; Cascales-Miñana, B.; Mulet, J.M.; Baroja-Fernández, E.; Pozueta-Romero, J.; Kuhn, J.M.; Segura, J.; Ros, R. Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 2009, 151, 541–558. [Google Scholar] [CrossRef]
- Russell, D.A.; Sachs, M.M. The maize cytosolic glyceraldehyde-3-phosphate dehydrogenase gene family: Organ-specific expression and genetic analysis. Mol. Gen. Genet. MGG 1991, 229, 219–228. [Google Scholar] [CrossRef]
- Chen, G.H.; Li, C.; Feng, X.D. Research progress of UDP—Sugar biosynthesis. J. Biol. 2023, 40, 95–100. [Google Scholar] [CrossRef]
- Chen, C.C.; Mi, H.L. Analysis of genes related to biosynthesis of gastrodin. Plant Physiol. J. 2021, 57, 1819–1828. [Google Scholar] [CrossRef]
- Rius, S.P.; Casati, P.; Iglesias, A.A.; Gomez-Casati, D.F. Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol. 2008, 148, 1655–1667. [Google Scholar] [CrossRef]
- Zhao, D.; Cheng, Z.; Qian, Y.; Hu, Z.; Tang, Y.; Huang, X.; Tao, J. PlWRKY47 Coordinates With Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenase 2 Gene to Improve Thermotolerance Through Inhibiting Reactive Oxygen Species Generation in Herbaceous Peony. Plant Cell Environ. 2025, 48, 226–243. [Google Scholar] [CrossRef]
- Selvarajan, D.; Mohan, C.; Dhandapani, V.; Nerkar, G.; Jayanarayanan, A.N.; Vadakkancherry Mohanan, M.; Murugan, N.; Kaur, L.; Chennappa, M.; Kumar, R.; et al. Differential gene expression profiling through transcriptome approach of Saccharum spontaneum L. under low temperature stress reveals genes potentially involved in cold acclimation. 3 Biotech. 2018, 8, 195. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Jin, X.; Liu, J.; Zhao, X.; Zhou, J.; Wang, X.; Wang, D.; Lai, C.; Xu, W.; Huang, J.; et al. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 2018, 9, 1615. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the Expasy Server. In The Proteomics Protocols Handbook; John, M.W., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis–acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Kruger, N.J. The Bradford method for protein quantitation. Methods Mol. Biol. 1994, 32, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.Y.; Tian, Y.; Yu, D.Q.; Wang, R.; Pei, J.J.; Chen, C.; Li, X.S. Effects of different treatments on the expression of enzymes related to storage softening of fresh-cut Gastrodia elata. Jiangsu Agric. Sci. 2022, 50, 198–205. [Google Scholar] [CrossRef]
- Yuan, M. Characteristics of Specific Microbial Community in the Rhizosphere of Continuous Cropping Salvia miltiorrhiza Bge. Based on 13C Stable Isotope Nucleic Acid Probe Technology. Master’s Thesis, Shandong University of Traditional Chinese Medicine, Jinan, China, 2023. [Google Scholar]
| Gene Name | Gene ID | Number of Amino Acids | Molecular Mass | Theoretical pI | Instability Index 1 | Grand Average of Hydropathicity 2 | Subcellular Prediction |
|---|---|---|---|---|---|---|---|
| GeGAPDH1 | GWHGBHOU003488 | 428 | 45,867.01 | 8.51 | 37.51 | −0.106 | plastid |
| GeGAPDH2 | GWHGBHOU007564 | 344 | 37,088.64 | 7.68 | 26.75 | −0.01 | cytoplasm |
| GeGAPDH3 | GWHGBHOU007572 | 344 | 37,088.64 | 7.68 | 26.75 | −0.01 | cytoplasm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Jiang, M.; Han, P.; Lin, X.; Wang, X. Identification of the Glyceraldehyde-3-Phosphate Dehydrogenase (GeGAPDH) Gene Family in Gastrodia elata Revealing Its Response Characteristics to Low-Temperature and Pathogen Stress. Plants 2025, 14, 1866. https://doi.org/10.3390/plants14121866
Yan Y, Jiang M, Han P, Lin X, Wang X. Identification of the Glyceraldehyde-3-Phosphate Dehydrogenase (GeGAPDH) Gene Family in Gastrodia elata Revealing Its Response Characteristics to Low-Temperature and Pathogen Stress. Plants. 2025; 14(12):1866. https://doi.org/10.3390/plants14121866
Chicago/Turabian StyleYan, Yaxing, Mei Jiang, Pengjie Han, Xiaohu Lin, and Xiao Wang. 2025. "Identification of the Glyceraldehyde-3-Phosphate Dehydrogenase (GeGAPDH) Gene Family in Gastrodia elata Revealing Its Response Characteristics to Low-Temperature and Pathogen Stress" Plants 14, no. 12: 1866. https://doi.org/10.3390/plants14121866
APA StyleYan, Y., Jiang, M., Han, P., Lin, X., & Wang, X. (2025). Identification of the Glyceraldehyde-3-Phosphate Dehydrogenase (GeGAPDH) Gene Family in Gastrodia elata Revealing Its Response Characteristics to Low-Temperature and Pathogen Stress. Plants, 14(12), 1866. https://doi.org/10.3390/plants14121866







