Biosynthesis and Regulatory Mechanisms of Plant Flavonoids: A Review
Abstract
1. Introduction
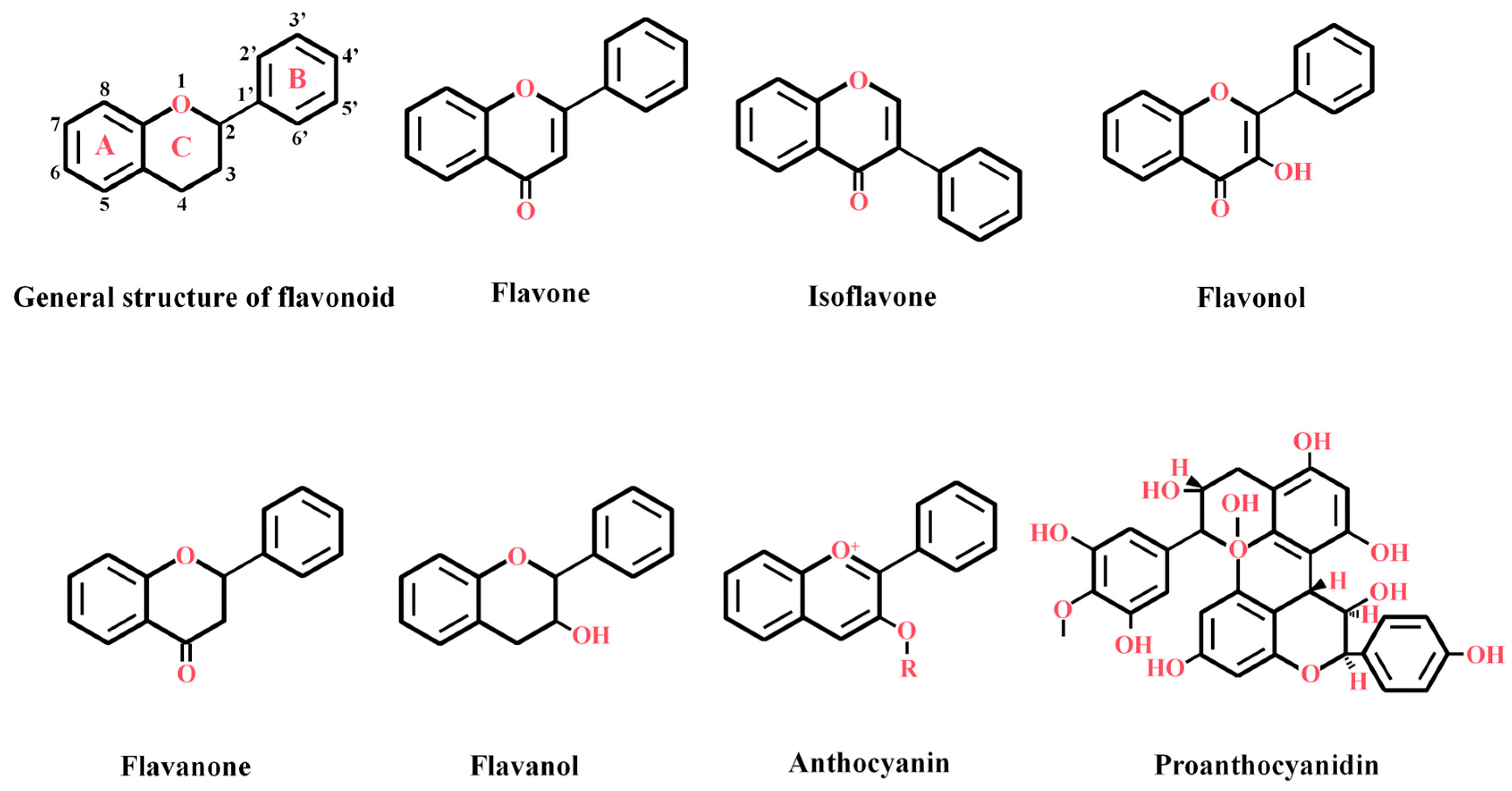
2. Structural Characteristics and Classification of Flavonoids
3. Biosynthesis of Plant Flavonoids
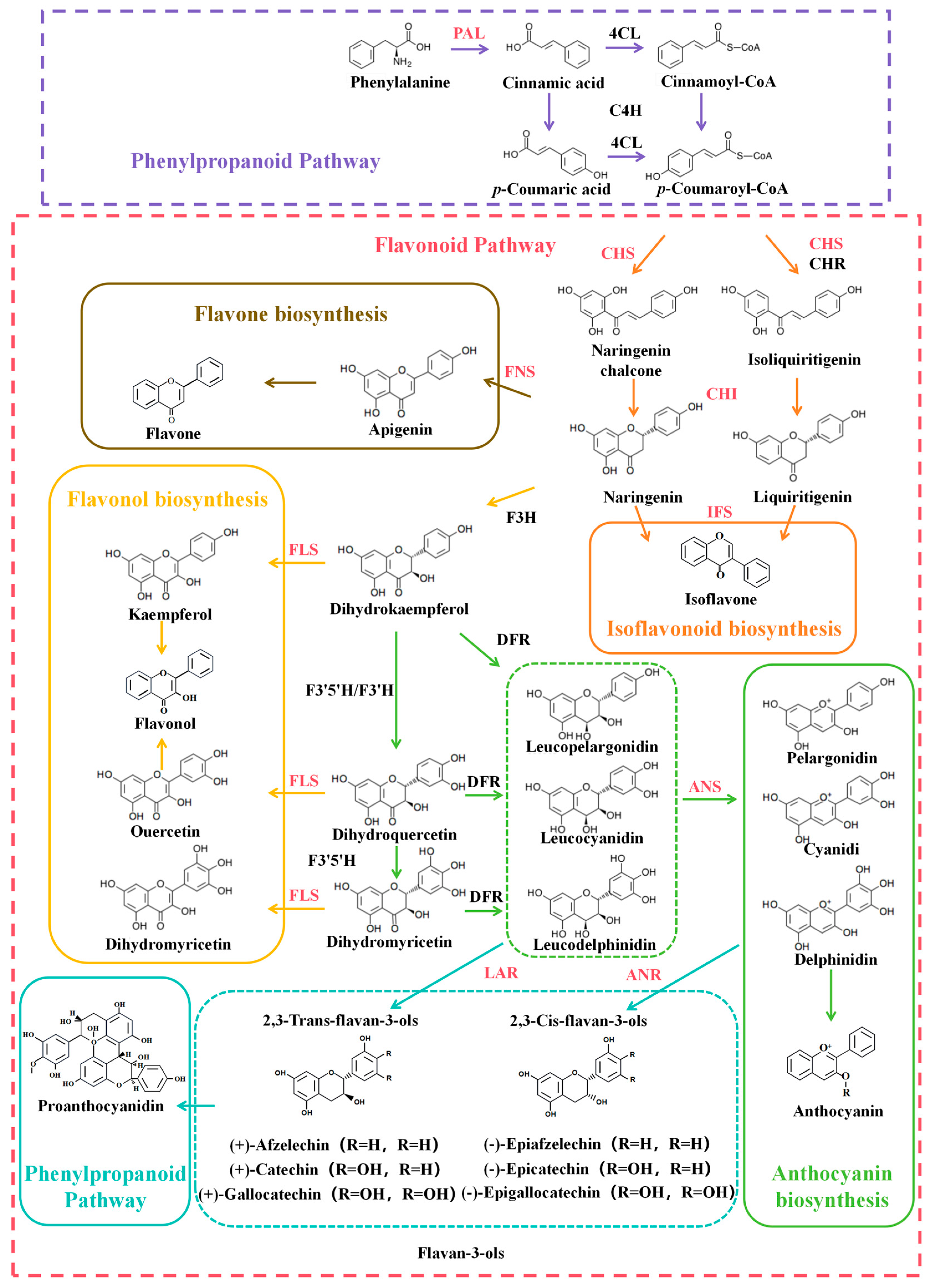
3.1. Flavonoid Biosynthetic Pathway
3.2. Key Enzymes in Flavonoid Biosynthesis
4. Regulation of Flavonoid Biosynthesis
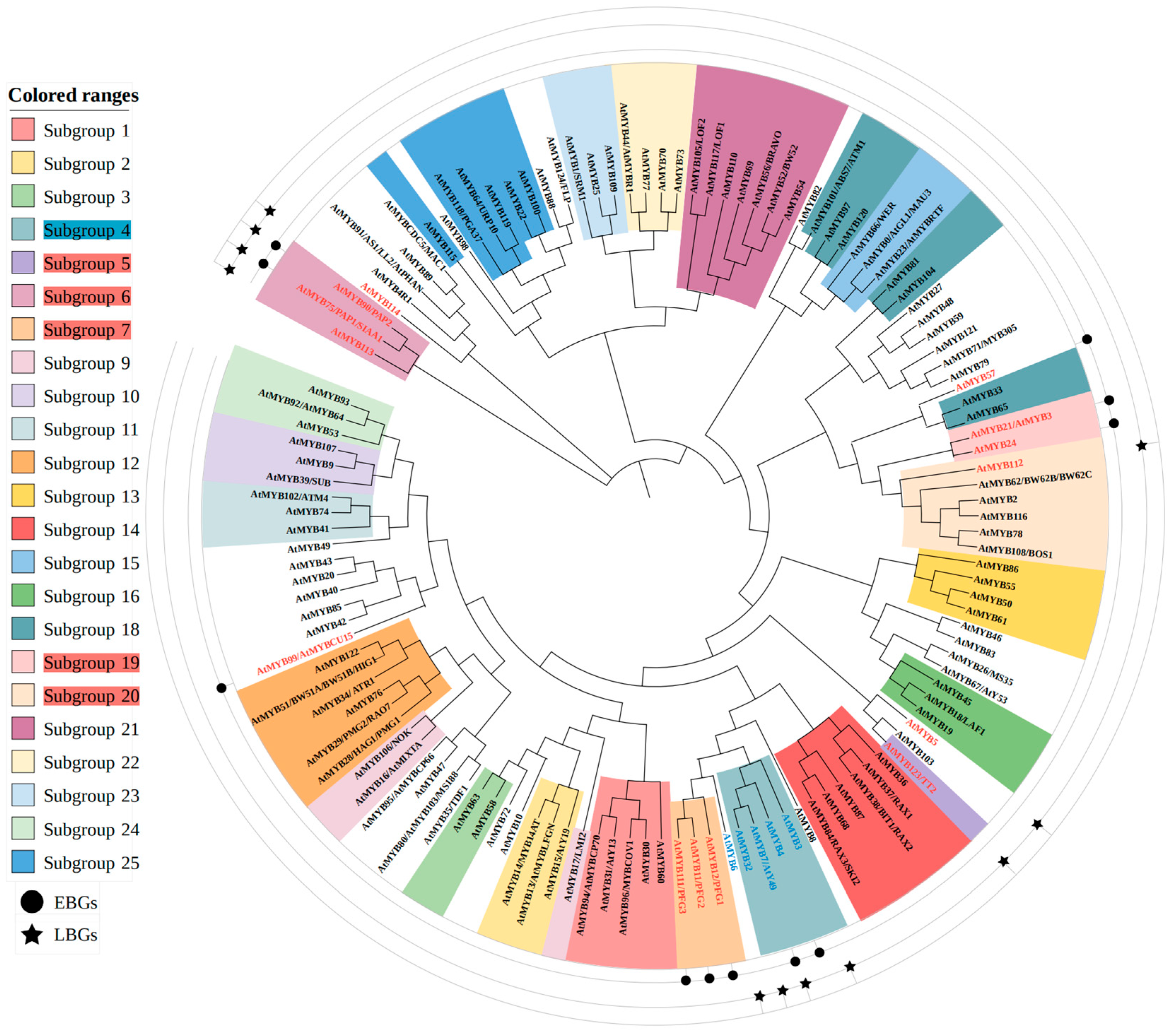
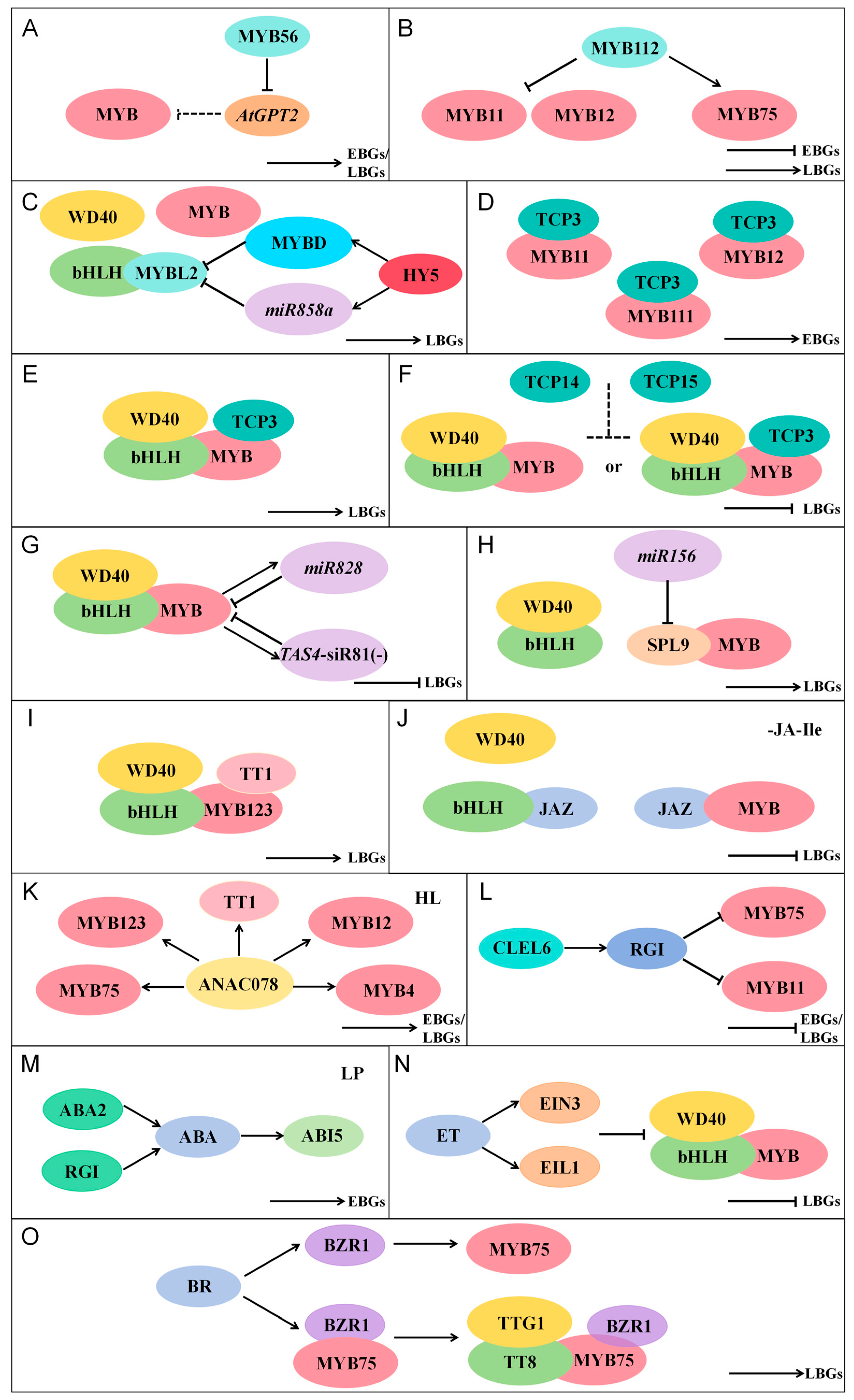
4.1. MYB Transcription Factors Regulating Flavonoid Biosynthesis
4.1.1. MYB Family Members Involved in Flavonoid Regulation in A. thaliana
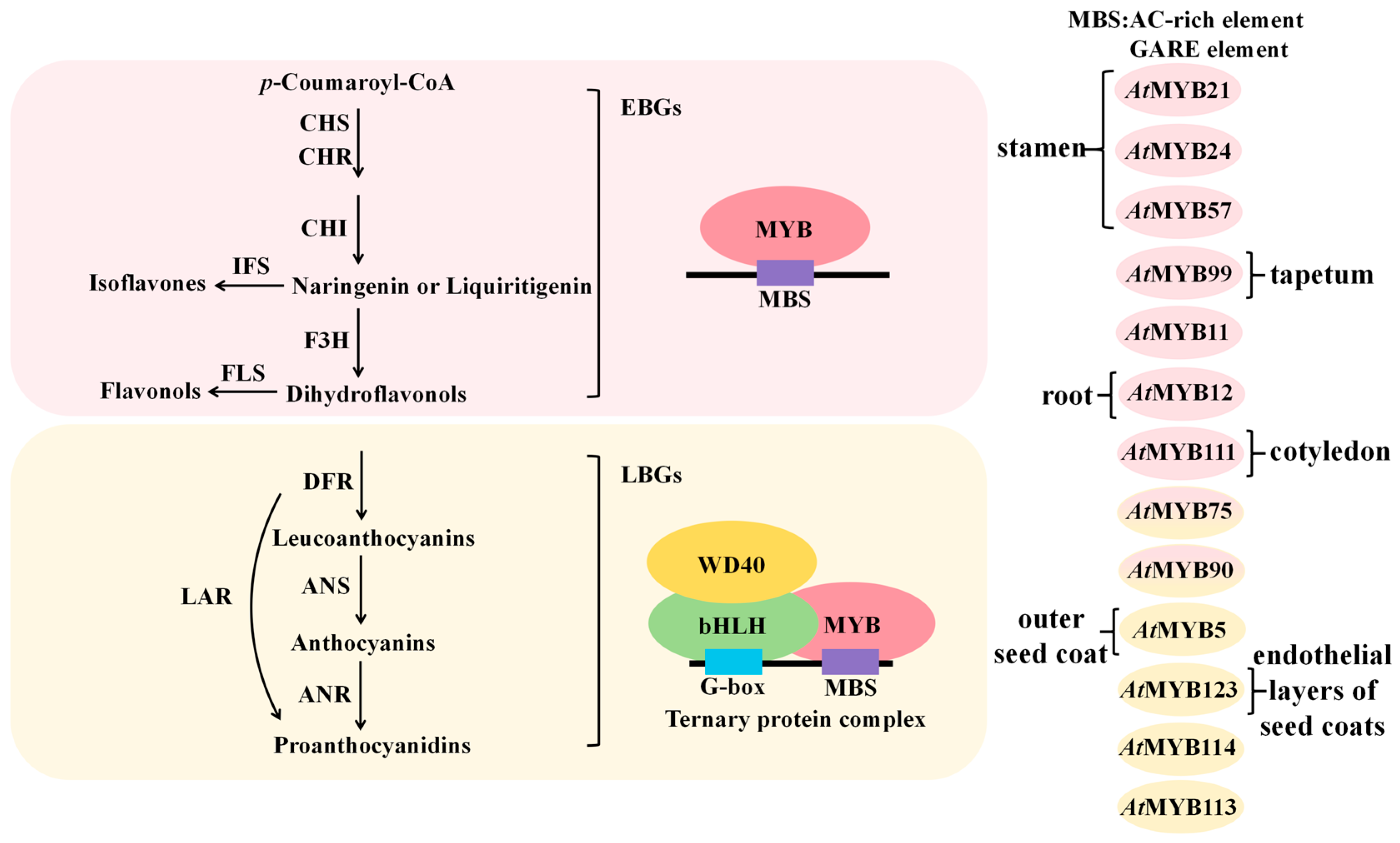
4.1.2. Positive Regulation of Flavonoid Biosynthetic Genes by MYB Transcription Factors
4.1.3. Negative Regulation of Flavonoid Biosynthetic Genes by MYB Transcription Factors
4.2. Additional Factors Regulating Flavonoid Biosynthesis
4.2.1. Regulation of Flavonoid Biosynthesis by Other TFs
4.2.2. Regulation of Flavonoid Biosynthesis by miRNAs
4.2.3. Regulation of Flavonoid Biosynthesis by Plant Hormones
4.2.4. Regulation of Flavonoid Biosynthesis by Sulfopeptide
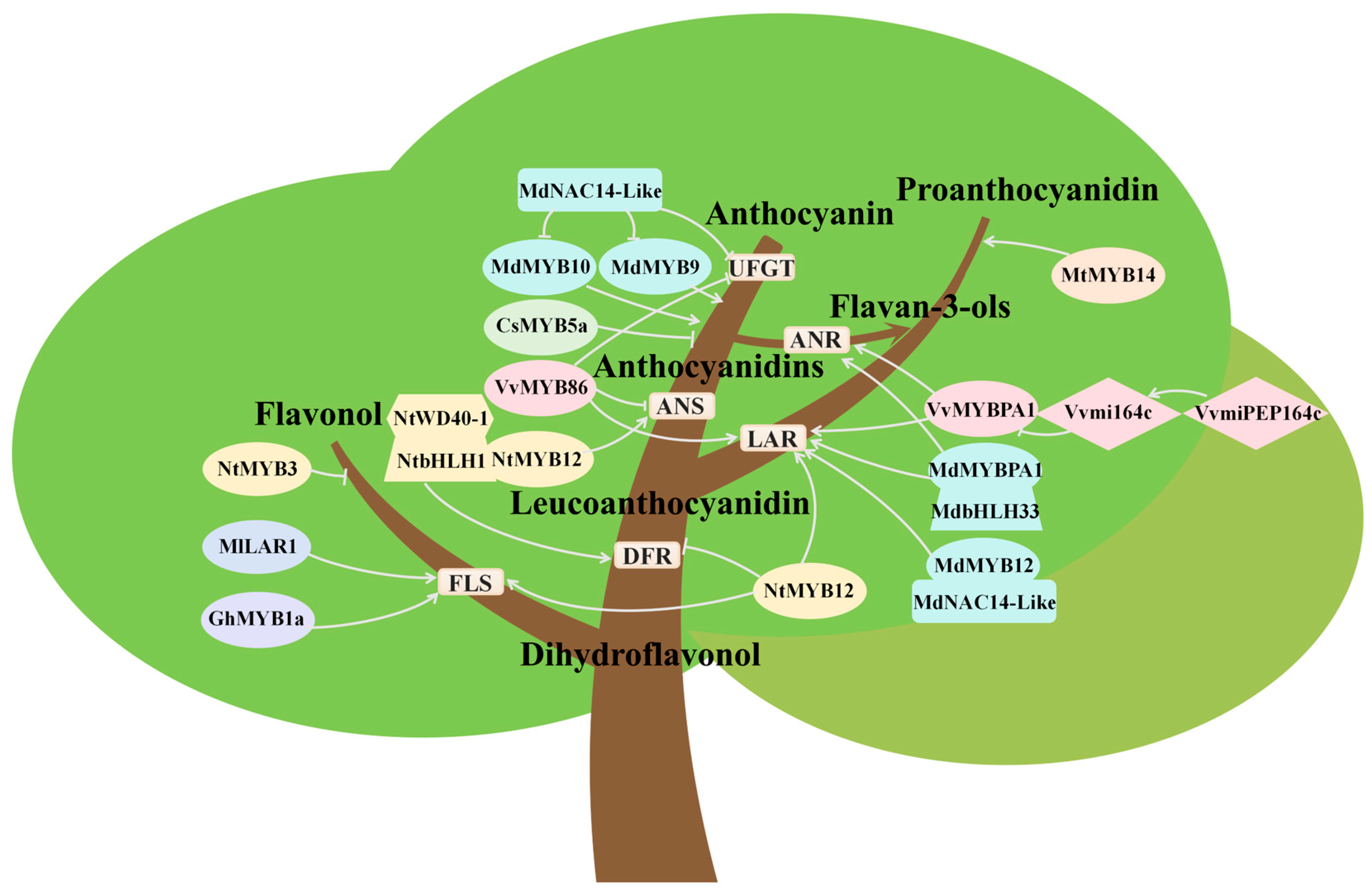
5. Competitive Relationships in Biosynthesis Among Different Flavonoids
5.1. Competition Between Anthocyanin and Flavonol Biosynthesis
5.2. Competition Between Anthocyanin and Proanthocyanidin Biosynthesis
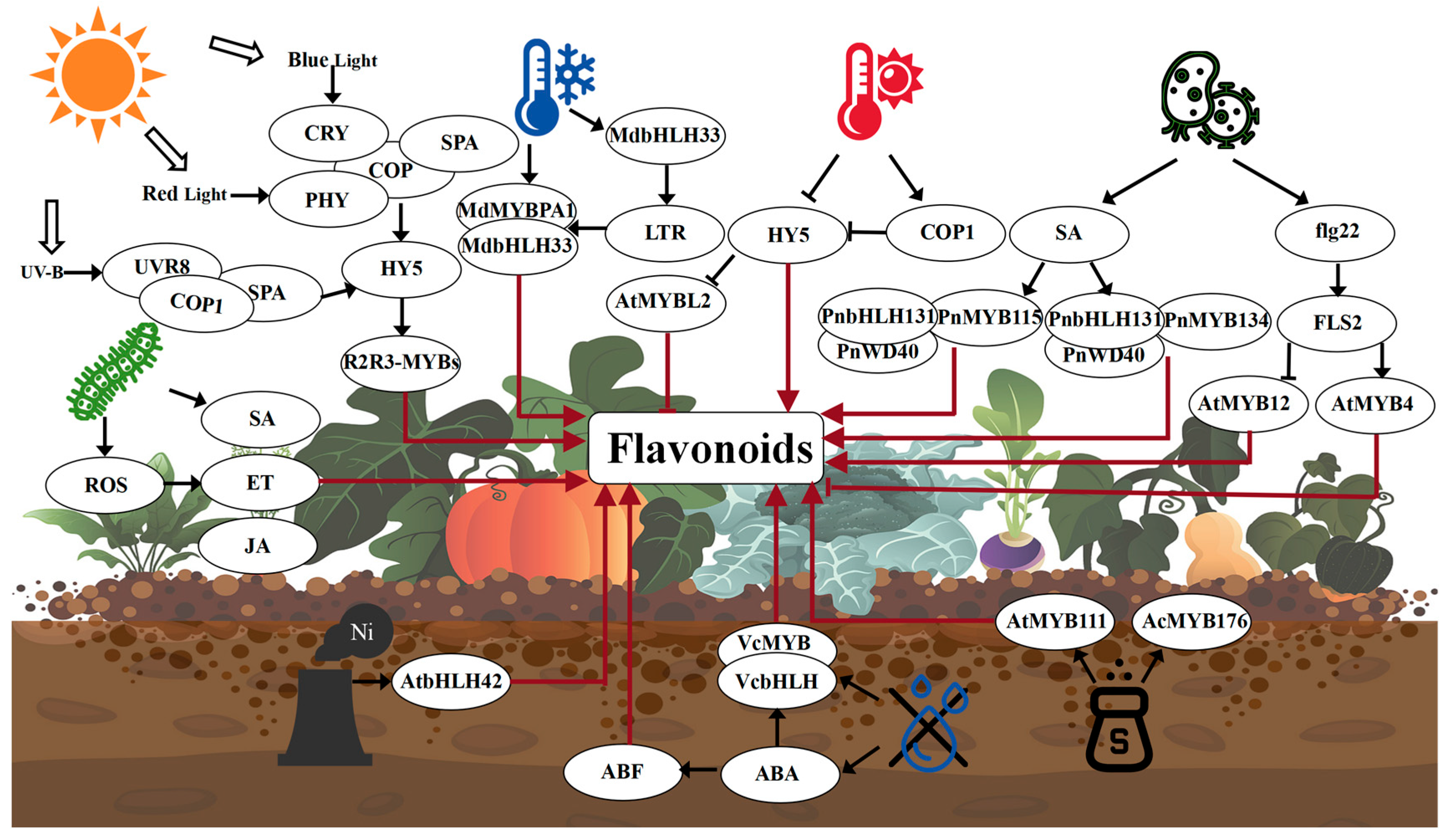
6. Environmental Factors Affecting Flavonoid Biosynthesis
6.1. Biological Factors
6.2. Abiotic Factors
7. Advances in Flavonoid Synthetic Biology
7.1. CRISPR/Cas9 Gene Editing Technology
7.2. Transgenic Engineering
7.3. Microbial Synthesis
8. Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Stafford, H.A. Flavonoid Evolution: An Enzymic Approach. Plant Physiol. 1991, 96, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The Origin and Evolution of Plant Flavonoid Metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lv, S.; Zhao, L.; Gao, T.; Yu, C.; Hu, J.; Ma, F. Advances in the study of the function and mechanism of the action of flavonoids in plants under environmental stresses. Planta 2023, 257, 108. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Andre, C.M.; Kulshrestha, S.; Zhou, Y.; Schwinn, K.E.; Albert, N.W.; Chagné, D.; van Klink, J.W.; Landi, M.; Bowman, J.L. The evolution of flavonoid biosynthesis. Philos. Trans. R. Soc. B Biol. Sci. 2024, 379, 20230361. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef]
- Procházková, D.; Boušová, I.; Wilhelmová, N. Antioxidant and prooxidant properties of flavonoids. Fitoterapia 2011, 82, 513–523. [Google Scholar] [CrossRef]
- Hu, L.; Luo, Y.; Yang, J.; Cheng, C. Botanical Flavonoids: Efficacy, Absorption, Metabolism and Advanced Pharmaceutical Technology for Improving Bioavailability. Molecules 2025, 30, 1184. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, M.; Dong, H.; Liu, W.; Guo, L.; Wang, X. A spatially-resolved approach to visualize the distribution and biosynthesis of flavones in Scutellaria baicalensis Georgi. J. Pharm. Biomed. Anal. 2020, 179, 113014. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Wang, T.; Liu, H.; Xu, X.; Chen, K.; Zhang, P. The moss flavone synthase I positively regulates the tolerance of plants to drought stress and UV-B radiation. Plant Sci. 2020, 298, 110591. [Google Scholar] [CrossRef]
- Jaakola, L.; Hohtola, A. Effect of latitude on flavonoid biosynthesis in plants. Plant Cell Environ. 2010, 33, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Teale, W.D.; Pasternak, T.; Bosco, C.D.; Dovzhenko, A.; Kratzat, K.; Bildl, W.; Schwörer, M.; Falk, T.; Ruperti, B.; Schaefer, J.V.; et al. Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J. 2020, 40, e104416. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Han, Y.; Han, B.; Qi, X.; Cai, X.; Ge, S.; Xue, H. Extraction and purification of anthocyanins: A review. J. Agric. Food Res. 2022, 8, 100306. [Google Scholar] [CrossRef]
- Teles, Y.C.F.; Souza, M.S.R.; Souza, M.D.F.V.d. Sulphated Flavonoids: Biosynthesis, Structures, and Biological Activities. Molecules 2018, 23, 480. [Google Scholar] [CrossRef]
- Liu, A.; Zhu, Y.; Wang, Y.; Wang, T.; Zhao, S.; Feng, K.; Li, L.; Wu, P. Molecular identification of phenylalanine ammonia lyase-encoding genes EfPALs and EfPAL2-interacting transcription factors in Euryale ferox. Front. Plant Sci. 2023, 14, 1114345. [Google Scholar] [CrossRef]
- Ferreyra, M.L.F.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef]
- Liu, T.; Liu, T.; Zhang, X.; Song, J.; Qiu, Y.; Yang, W.; Jia, H.; Wang, H.; Li, X. Combined widely targeted metabolomics and transcriptomics analysis reveals differentially accumulated metabolites and the underlying molecular bases in fleshy taproots of distinct radish genotypes. Plant Physiol. Biochem. 2023, 195, 351–361. [Google Scholar] [CrossRef]
- Lapcik, O.; Honys, D.; Koblovska, R.; Mackova, Z.; Vitkova, M.; Klejdus, B. Isoflavonoids are present in Arabidopsis thaliana despite the absence of any homologue to known isoflavonoid synthases. Plant Physiol. Biochem. 2006, 44, 106–114. [Google Scholar] [CrossRef]
- Xie, C.; Zhan, T.; Huang, J.; Lan, J.; Shen, L.; Wang, H.; Zheng, X. Functional characterization of nine critical genes encoding rate-limiting enzymes in the flavonoid biosynthesis of the medicinal herb Grona styracifolia. BMC Plant Biol. 2023, 23, 299. [Google Scholar] [CrossRef]
- Leonard, E.; Yan, Y.; Lim, K.H.; Koffas, M.A.G. Investigation of Two Distinct Flavone Synthases for Plant-Specific Flavone Biosynthesis inSaccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 8241–8248. [Google Scholar] [CrossRef]
- Feng, Y.; Tian, X.; Liang, W.; Nan, X.; Zhang, A.; Li, W.; Ma, Z. Genome-wide identification of grape ANS gene family and expression analysis at different fruit coloration stages. BMC Plant Biol. 2023, 23, 632. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wen, J.; Xia, Y.; Zhang, L.; Du, H. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic. Res. 2022, 9, uhac058. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Chezem, W.R.; Memon, A.; Li, F.-S.; Weng, J.-K.; Clay, N.K. SG2-Type R2R3-MYB Transcription Factor MYB15 Controls Defense-Induced Lignification and Basal Immunity in Arabidopsis. Plant Cell 2017, 29, 1907–1926. [Google Scholar] [CrossRef]
- Lotkowska, M.E.; Tohge, T.; Fernie, A.R.; Xue, G.-P.; Balazadeh, S.; Mueller-Roeber, B. The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 2015, 169, 1862–1880. [Google Scholar] [CrossRef]
- Shi, M.-Z.; Xie, D.-Y. Biosynthesis and Metabolic Engineering of Anthocyanins in Arabidopsis thaliana. Recent Pat. Biotechnol. 2014, 8, 47–60. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef]
- Zhang, X.; He, Y.; Li, L.; Liu, H.; Hong, G.; Hancock, R. Involvement of the R2R3-MYB transcription factor MYB21 and its homologs in regulating flavonol accumulation in Arabidopsis stamen. J. Exp. Bot. 2021, 72, 4319–4332. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. MYB46 and MYB83 Bind to the SMRE Sites and Directly Activate a Suite of Transcription Factors and Secondary Wall Biosynthetic Genes. Plant Cell Physiol. 2012, 53, 368–380. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Battat, M.; Eitan, A.; Rogachev, I.; Hanhineva, K.; Fernie, A.; Tohge, T.; Beekwilder, J.; Aharoni, A. A MYB Triad Controls Primary and Phenylpropanoid Metabolites for Pollen Coat Patterning. Plant Physiol. 2019, 180, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Mendenhall, J.; Huo, Y.; Lloyd, A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009, 325, 412–421. [Google Scholar] [CrossRef]
- Rowan, D.D.; Cao, M.; Lin-Wang, K.; Cooney, J.M.; Jensen, D.J.; Austin, P.T.; Hunt, M.B.; Norling, C.; Hellens, R.P.; Schaffer, R.J.; et al. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 2009, 182, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Luo, Q.; Li, Y.; Yin, L.; Zhou, N.; Li, X.; Gan, J.; Dong, A. Structural insights into target DNA recognition by R2R3-MYB transcription factors. Nucleic Acids Res. 2020, 48, 460–471. [Google Scholar] [CrossRef]
- Prouse, M.B.; Campbell, M.M.; Unver, T. Interactions between the R2R3-MYB transcription factor, AtMYB61, and target DNA binding sites. PLoS ONE 2013, 8, e65132. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wei, L.; Sun, Z.; Gao, L.; Meng, Y.; Tang, Y.; Wu, Y. Production and transcriptional regulation of proanthocyanidin biosynthesis in forage legumes. Appl. Microbiol. Biotechnol. 2015, 99, 3797–3806. [Google Scholar] [CrossRef]
- Solfanelli, C.; Poggi, A.; Loreti, E.; Alpi, A.; Perata, P. Sucrose-Specific Induction of the Anthocyanin Biosynthetic Pathway in Arabidopsis. Plant Physiol. 2006, 140, 637–646. [Google Scholar] [CrossRef]
- Jeong, C.Y.; Kim, J.H.; Lee, W.J.; Jin, J.Y.; Kim, J.; Hong, S.-W.; Lee, H. AtMyb56 Regulates Anthocyanin Levels via the Modulation of AtGPT2 Expression in Response to Sucrose in Arabidopsis. Mol. Cells 2018, 41, 351–361. [Google Scholar] [CrossRef]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef]
- Fornalé, S.; Lopez, E.; Salazar-Henao, J.E.; Fernández-Nohales, P.; Rigau, J.; Caparros-Ruiz, D. AtMYB7, a New Player in the Regulation of UV-Sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.; Wheeler, J.; Heazlewood, J.; Li, S.F.; Parish, R.W. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 2004, 40, 979–995. [Google Scholar] [CrossRef]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.-M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Transcriptional control of flavonoid biosynthesis. Plant Signal. Behav. 2014, 9, e27522. [Google Scholar] [CrossRef]
- Viola, I.L.; Camoirano, A.; Gonzalez, D.H. Redox-Dependent Modulation of Anthocyanin Biosynthesis by the TCP Transcription Factor TCP15 during Exposure to High Light Intensity Conditions in Arabidopsis. Plant Physiol. 2016, 170, 74–85. [Google Scholar] [CrossRef]
- Li, S.; Zachgo, S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013, 76, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.-Y.; Felippes, F.F.; Liu, C.-J.; Weigel, D.; Wang, J.-W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef]
- Morishita, T.; Kojima, Y.; Maruta, T.; Nishizawa-Yokoi, A.; Yabuta, Y.; Shigeoka, S. Arabidopsis NAC Transcription Factor, ANAC078, Regulates Flavonoid Biosynthesis under High-light. Plant Cell Physiol. 2009, 50, 2210–2222. [Google Scholar] [CrossRef]
- Li, S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal Behav. 2015, 10, e1044192. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, T.; Guo, S.; Hu, J.; Zhan, Y. Ectopic Expression of AeNAC83, a NAC Transcription Factor from Abelmoschus esculentus, Inhibits Growth and Confers Tolerance to Salt Stress in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 10182. [Google Scholar] [CrossRef]
- Iwakawa, H.-o.; Tomari, Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef]
- Bonar, N.; Liney, M.; Zhang, R.; Austin, C.; Dessoly, J.; Davidson, D.; Stephens, J.; McDougall, G.; Taylor, M.; Bryan, G.J.; et al. Potato miR828 Is Associated With Purple Tuber Skin and Flesh Color. Front. Plant Sci. 2018, 9, 1742. [Google Scholar] [CrossRef]
- Luo, Q.-J.; Mittal, A.; Jia, F.; Rock, C.D. An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 2011, 80, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Cao, A.; Wen, Y.; Bao, W.; She, F.; Wu, W.; Zheng, S.; Yang, N. Characteristics and Functions of MYB (v-Myb avivan myoblastsis virus oncogene homolog)-Related Genes in Arabidopsis thaliana. Genes 2023, 14, 2026. [Google Scholar] [CrossRef]
- Loreti, E.; Povero, G.; Novi, G.; Solfanelli, C.; Alpi, A.; Perata, P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008, 179, 1004–1016. [Google Scholar] [CrossRef]
- Song, R.-F.; Hu, X.-Y.; Liu, W.-C.; Yuan, H.-M. ABA functions in low phosphate-induced anthocyanin accumulation through the transcription factor ABI5 in Arabidopsis. Plant Cell Rep. 2024, 43, 55. [Google Scholar] [CrossRef]
- Song, S.; Liu, B.; Song, J.; Pang, S.; Song, T.; Gao, S.; Zhang, Y.; Huang, H.; Qi, T. A molecular framework for signaling crosstalk between jasmonate and ethylene in anthocyanin biosynthesis, trichome development, and defenses against insect herbivores in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 1770–1788. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Kim, S.-H.; Park, T.-K.; Kim, Y.-P.; Lee, J.-W.; Kim, T.-W. Transcription factors BZR1 and PAP1 cooperate to promote anthocyanin biosynthesis in Arabidopsis shoots. Plant Cell 2024, 36, 3654–3673. [Google Scholar] [CrossRef]
- Bühler, E.; Fahrbach, E.; Schaller, A.; Stührwohldt, N. Sulfopeptide CLEL6 inhibits anthocyanin biosynthesis in Arabidopsis thaliana. Plant Physiol. 2023, 193, 809–820. [Google Scholar] [CrossRef]
- Cheng, C.; Guo, Z.; Li, H.; Mu, X.; Wang, P.; Zhang, S.; Yang, T.; Cai, H.; Wang, Q.; Lü, P.; et al. Integrated metabolic, transcriptomic and chromatin accessibility analyses provide novel insights into the competition for anthocyanins and flavonols biosynthesis during fruit ripening in red apple. Front. Plant Sci. 2022, 13, 975356. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Tang, Y.; Pang, B.; Li, X.; Yang, Y.; Deng, J.; Feng, C.; Li, L.; Ren, G.; Wang, Y.; et al. The R2R3-MYB transcription factor GhMYB1a regulates flavonol and anthocyanin accumulation in Gerbera hybrida. Hortic. Res. 2020, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, L.-Y.; Wang, S.-L.; Niu, X.-Y. Analysis of anthocyanins and flavonols in petals of 10 Rhododendron species from the Sygera Mountains in Southeast Tibet. Plant Physiol. Biochem. 2016, 104, 250–256. [Google Scholar] [CrossRef]
- Chen, W.; Xiao, Z.; Wang, Y.; Wang, J.; Zhai, R.; Lin-Wang, K.; Espley, R.; Ma, F.; Li, P. Competition between anthocyanin and kaempferol glycosides biosynthesis affects pollen tube growth and seed set of Malus. Hortic. Res. 2021, 8, 173. [Google Scholar] [CrossRef]
- Yuan, Y.-W.; Rebocho, A.B.; Sagawa, J.M.; Stanley, L.E.; Bradshaw, H.D., Jr. Competition between anthocyanin and flavonol biosynthesis produces spatial pattern variation of floral pigments between Mimulus species. Proc. Natl. Acad. Sci. USA 2016, 113, 2448–2453. [Google Scholar] [CrossRef]
- Maloney, G.S.; DiNapoli, K.T.; Muday, G.K. The anthocyanin reduced Tomato Mutant Demonstrates the Role of Flavonols in Tomato Lateral Root and Root Hair Development. Plant Physiol. 2014, 166, 614–631. [Google Scholar] [CrossRef]
- Luo, P.; Ning, G.; Wang, Z.; Shen, Y.; Jin, H.; Li, P.; Huang, S.; Zhao, J.; Bao, M. Disequilibrium of Flavonol Synthase and Dihydroflavonol-4-Reductase Expression Associated Tightly to White vs. Red Color Flower Formation in Plants. Front. Plant Sci. 2016, 6, 1257. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, X.; Aucapiña, C.b.; Zhang, D.; Huang, J.; Hao, Z.; Zhang, Y.; Ren, Y.; Miao, Y. NtMYB12 requires for competition between flavonol and (pro)anthocyanin biosynthesis in Narcissus tazetta tepals. Mol. Hortic. 2023, 3, 2. [Google Scholar] [CrossRef]
- Anwar, M.; Yu, W.; Yao, H.; Zhou, P.; Allan, A.C.; Zeng, L. NtMYB3, an R2R3-MYB from Narcissus, Regulates Flavonoid Biosynthesis. Int. J. Mol. Sci. 2019, 20, 5456. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Saito, M.; Yamada, E.; Fujita, K.; Kakizaki, Y.; Nishihara, M. Isolation and characterization of GtMYBP3 and GtMYBP4, orthologues of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J. Exp. Bot. 2012, 63, 6505–6517. [Google Scholar] [CrossRef]
- Xu, T.; Yu, L.; Huang, N.; Liu, W.; Fang, Y.; Chen, C.; Jiang, L.; Wang, T.; Zhao, J.; Zhang, Z.; et al. The regulatory role of MdNAC14-Like in anthocyanin synthesis and proanthocyanidin accumulation in red-fleshed apples. Plant Physiol. Biochem. 2023, 204, 108068. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yu, K.; Shi, Y.; Wang, J.; Duan, C. Transcription Factor VviMYB86 Oppositely Regulates Proanthocyanidin and Anthocyanin Biosynthesis in Grape Berries. Front. Plant Sci. 2021, 11, 613677. [Google Scholar] [CrossRef] [PubMed]
- Vale, M.; Rodrigues, J.; Badim, H.; Gerós, H.; Conde, A. Exogenous Application of Non-mature miRNA-Encoded miPEP164c Inhibits Proanthocyanidin Synthesis and Stimulates Anthocyanin Accumulation in Grape Berry Cells. Front. Plant Sci. 2021, 12, 706679. [Google Scholar] [CrossRef]
- Wang, N.; Qu, C.; Jiang, S.; Chen, Z.; Xu, H.; Fang, H.; Su, M.; Zhang, J.; Wang, Y.; Liu, W.; et al. The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 2018, 96, 39–55. [Google Scholar] [CrossRef]
- Liu, C.; Jun, J.H.; Dixon, R.A. MYB5 and MYB14 Play Pivotal Roles in Seed Coat Polymer Biosynthesis in Medicago truncatula. Plant Physiol. 2014, 165, 1424–1439. [Google Scholar] [CrossRef]
- Jiang, X.; Huang, K.; Zheng, G.; Hou, H.; Wang, P.; Jiang, H.; Zhao, X.; Li, M.; Zhang, S.; Liu, Y.; et al. CsMYB5a and CsMYB5e from Camellia sinensis differentially regulate anthocyanin and proanthocyanidin biosynthesis. Plant Sci. 2018, 270, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Shamsudin, N.F.; Ahmed, Q.U.; Mahmood, S.; Shah, S.A.A.; Khatib, A.; Mukhtar, S.; Alsharif, M.A.; Parveen, H.; Zakaria, Z.A. Antibacterial Effects of Flavonoids and Their Structure-Activity Relationship Study: A Comparative Interpretation. Molecules 2022, 27, 1149. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, K.; Mandal, A.K.; Afzal, O.; Altamimi, A.S.A.; Sahoo, A.; Alossaimi, M.A.; Almalki, W.H.; Alzahrani, A.; Barkat, A.; Almeleebia, T.M.; et al. Emergence of Nano-Based Formulations for Effective Delivery of Flavonoids against Topical Infectious Disorders. Gels 2023, 9, 671. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Lu, H.-Q.; Jiang, K.-X.; Wang, Y.-R.; Wang, Y.-P.; Jiang, J.-J. The Flavonoid Biosynthesis and Regulation in Brassica napus: A Review. Int. J. Mol. Sci. 2022, 24, 357. [Google Scholar] [CrossRef]
- Ullah, C.; Tsai, C.-J.; Unsicker, S.B.; Xue, L.; Reichelt, M.; Gershenzon, J.; Hammerbacher, A. Salicylic acid activates poplar defense against the biotrophic rust fungusMelampsora larici-populinavia increased biosynthesis of catechin and proanthocyanidins. New Phytol. 2018, 221, 960–975. [Google Scholar] [CrossRef]
- Montesinos, Á.; Sacristán, S.; del Prado-Polonio, P.; Arnaiz, A.; Díaz-González, S.; Diaz, I.; Santamaria, M.E. Contrasting plant transcriptome responses between a pierce-sucking and a chewing herbivore go beyond the infestation site. BMC Plant Biol. 2024, 24, 120. [Google Scholar] [CrossRef] [PubMed]
- Chin, S.; Behm, C.A.; Mathesius, U. Functions of Flavonoids in Plant–Nematode Interactions. Plants 2018, 7, 85. [Google Scholar] [CrossRef]
- Schenke, D.; BÖTtcher, C.; Scheel, D. Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ. 2011, 34, 1849–1864. [Google Scholar] [CrossRef]
- Ye, J.-H.; Lv, Y.-Q.; Liu, S.-R.; Jin, J.; Wang, Y.-F.; Wei, C.-L.; Zhao, S.-Q. Effects of Light Intensity and Spectral Composition on the Transcriptome Profiles of Leaves in Shade Grown Tea Plants (Camellia sinensis L.) and Regulatory Network of Flavonoid Biosynthesis. Molecules 2021, 26, 5836. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.; She, G.; Zhang, X.; Jordan, B.; Chen, Q.; Zhao, J.; Wan, X. Metabolite profiling and transcriptomic analyses reveal an essential role of UVR8-mediated signal transduction pathway in regulating flavonoid biosynthesis in tea plants (Camellia sinensis) in response to shading. BMC Plant Biol. 2018, 18, 233. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Ma, J.-Q.; Ma, C.-L.; Shen, S.-Y.; Liu, Y.-F.; Chen, L. Regulation of Growth and Flavonoid Formation of Tea Plants (Camellia sinensis) by Blue and Green Light. J. Agric. Food Chem. 2019, 67, 2408–2419. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, S.; Gu, M.; Chen, X.; Chen, X.; Yang, J.; Zhao, F.; Ye, N. Exploration of the Effects of Different Blue LED Light Intensities on Flavonoid and Lipid Metabolism in Tea Plants via Transcriptomics and Metabolomics. Int. J. Mol. Sci. 2020, 21, 4606. [Google Scholar] [CrossRef]
- Zoratti, L.; Karppinen, K.; Escobar, A.L.; Häggman, H.; Jaakola, L. Light-controlled flavonoid biosynthesis in fruits. Front. Plant Sci. 2014, 5, 534. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Chen, X.-R.; Wang, J.-P.; Cui, W.-Q.; Xing, X.-X.; Chen, X.-Y.; Ding, W.-Y.; God’spower, B.-O.; Eliphaz, N.; Sun, M.-Q.; et al. Transcriptomic analysis reveals flavonoid biosynthesis of Syringa oblata Lindl. in response to different light intensity. BMC Plant Biol. 2019, 19, 487. [Google Scholar] [CrossRef]
- Feng, X.; Bai, S.; Zhou, L.; Song, Y.; Jia, S.; Guo, Q.; Zhang, C. Integrated Analysis of Transcriptome and Metabolome Provides Insights into Flavonoid Biosynthesis of Blueberry Leaves in Response to Drought Stress. Int. J. Mol. Sci. 2024, 25, 11135. [Google Scholar] [CrossRef]
- Perin, E.C.; Messias, R.d.S.; Borowski, J.M.; Crizel, R.L.; Schott, I.B.; Carvalho, I.R.; Rombaldi, C.V.; Galli, V. ABA-dependent salt and drought stress improve strawberry fruit quality. Food Chem. 2019, 271, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2013, 77, 367–379. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, G.; Lee, S.; Zhu, J.-Y.; Paik, I.; Nguyen, T.T.; Kim, J.; Oh, E. High Ambient Temperature Represses Anthocyanin Biosynthesis through Degradation of HY5. Front. Plant Sci. 2017, 8, 1787. [Google Scholar] [CrossRef]
- Li, Z.; Ahammed, G.J. Hormonal regulation of anthocyanin biosynthesis for improved stress tolerance in plants. Plant Physiol. Biochem. 2023, 201, 107835. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Yang, Y. Anthocyanin-mediated arsenic tolerance in plants. Environ. Pollut. 2022, 292, 118475. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Peng, Y.; Qi, X. The TT8 transcription factor alleviates nickel toxicity in Arabidopsis. Biochem. Biophys. Res. Commun. 2025, 757, 151649. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, T.; Niu, K.; Ma, H. Co-expression analyses reveal key Cd stress response-related metabolites and transcriptional regulators in Kentucky bluegrass. Chemosphere 2024, 363, 142937. [Google Scholar] [CrossRef]
- Kim, D.; Jeon, S.J.; Yanders, S.; Park, S.-C.; Kim, H.S.; Kim, S. MYB3 plays an important role in lignin and anthocyanin biosynthesis under salt stress condition in Arabidopsis. Plant Cell Rep. 2022, 41, 1549–1560. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Khan, N.M.; Ali, A.; Zhou, G.; Zhou, Y.; Li, P.; Wan, Y. AcMYB176-Regulated AcCHS5 Enhances Salt Tolerance in Areca catechu by Modulating Flavonoid Biosynthesis and Reactive Oxygen Species Scavenging. Int. J. Mol. Sci. 2025, 26, 3216. [Google Scholar] [CrossRef]
- Cui, J.; Li, X.; Gan, Q.; Lu, Z.; Du, Y.; Noor, I.; Wang, L.; Liu, S.; Jin, B. Flavonoids Mitigate Nanoplastic Stress in Ginkgo biloba. Plant Cell Environ. 2024, 48, 1790–1811. [Google Scholar] [CrossRef]
- Wen, D.; Wu, L.; Wang, M.; Yang, W.; Wang, X.; Ma, W.; Sun, W.; Chen, S.; Xiang, L.; Shi, Y. CRISPR/Cas9-Mediated Targeted Mutagenesis of FtMYB45 Promotes Flavonoid Biosynthesis in Tartary Buckwheat (Fagopyrum tataricum). Front. Plant Sci. 2022, 13, 879390. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, M.; Higuchi, A.; Watanabe, A.; Tasaki, K. Application of the CRISPR/Cas9 system for modification of flower color in Torenia fournieri. BMC Plant Biol. 2018, 18, 331. [Google Scholar] [CrossRef]
- Sun, H.; Wang, S.; Zhu, C.; Yang, K.; Liu, Y.; Gao, Z. A new biotechnology for in-planta gene editing and its application in promoting flavonoid biosynthesis in bamboo leaves. Plant Methods 2023, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Huang, H.; Gao, Q.; Li, Y.; Xiang, H.; Zeng, W.; Xu, L.; Liu, X.; Li, J.; Mi, Q.; et al. Effects of editing DFR genes on flowers, leaves, and roots of tobacco. BMC Plant Biol. 2023, 23, 349. [Google Scholar] [CrossRef]
- Zhang, P.; Du, H.; Wang, J.; Pu, Y.; Yang, C.; Yan, R.; Yang, H.; Cheng, H.; Yu, D. Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 2019, 18, 1384–1395. [Google Scholar] [CrossRef]
- Vaghari-Tabari, M.; Hassanpour, P.; Sadeghsoltani, F.; Malakoti, F.; Alemi, F.; Qujeq, D.; Asemi, Z.; Yousefi, B. CRISPR/Cas9 gene editing: A new approach for overcoming drug resistance in cancer. Cell. Mol. Biol. Lett. 2022, 27, 49. [Google Scholar] [CrossRef]
- Liu, R.; Hu, Y.; Li, J.; Lin, Z. Production of soybean isoflavone genistein in non-legume plants via genetically modified secondary metabolism pathway. Metab. Eng. 2007, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Hu, L.; Lu, H.; Wei, T.; Chen, Q. Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review. Molecules 2021, 26, 4522. [Google Scholar] [CrossRef]
- Eichenberger, M.; Hansson, A.; Fischer, D.; Dürr, L.; Naesby, M. De novo biosynthesis of anthocyanins in Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18, foy046. [Google Scholar] [CrossRef]
- Zhou, S.; Hao, T.; Zhou, J. Fermentation and Metabolic Pathway Optimization to De Novo Synthesize (2S)-Naringenin in Escherichia coli. J. Microbiol. Biotechnol. 2020, 30, 1574–1582. [Google Scholar] [CrossRef]
- Solopova, A.; van Tilburg, A.Y.; Foito, A.; Allwood, J.W.; Stewart, D.; Kulakauskas, S.; Kuipers, O.P. Engineering Lactococcus lactis for the production of unusual anthocyanins using tea as substrate. Metab. Eng. 2019, 54, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Katsuyama, Y.; Miyahisa, I.; Funa, N.; Horinouchi, S. One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 2007, 73, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lyv, Y.; Zhou, S.; Yu, S.; Zhou, J. Microbial cell factories for the production of flavonoids–barriers and opportunities. Bioresour. Technol. 2022, 360, 127538. [Google Scholar] [CrossRef] [PubMed]
| No. | Enzyme | Abbreviation | EC Number | Gene Numbering in A. thaliana |
|---|---|---|---|---|
| 1 | Phenylalanine ammonia-lyase | PAL | 4.3.1.24 | AT2G37040 (PAL1) AT3G53260 (PAL2) AT5G04230 (PAL3) AT3G10340 (PAL4) |
| 2 | Cinnamic acid 4-hydroxylase | C4H | 1.14.14.91 | AT2G30490 (C4H) |
| 3 | 4-Coumarate-CoA ligase | 4CL | 6.2.1.12 | AT1G51680 (4CL1) AT3G21240 (4CL2) AT1G65060 (4CL3) AT3G21230 (4CL5) AT1G62940 (ACOS5) AT4G05160 AT4G19010 AT5G63380 |
| 4 | Chalcone synthase | CHS | 2.3.1.74 | AT5G13930 (TT4) |
| 5 | Chalcone reductase | CHR | None | None |
| 6 | Chalcone isomerase | CHI | 5.5.1.6 | AT3G55120 (TT5) AT5G05270 (CHIL) AT5G66220 |
| 7 | Isoflavonoid synthase | IFS | 1.14.14.87 | None |
| 8 | Flavone synthase | FNS | 1.14.20.5 1.14.19.76 | None |
| 9 | Flavanone 3-hydroxylase | F3H | 1.14.11.9 | AT3G51240 (F3H) |
| 10 | Flavonol synthase | FLS | 1.14.20.6 | AT5G08640 (FLS1) AT5G63580 (FLS2) AT5G63590 (FLS3) AT5G63595 (FLS4) AT5G63600 (FLS5) AT5G43935 (FLS6) |
| 11 | Flavonoid 3′,5′-hydroxylase | F3′5′H | 1.14.14.81 | None |
| 12 | Flavonoid 3′-hydroxylase | F3′H | 1.14.14.82 | AT5G07990 (TT7) |
| 13 | Dihydroflavonol 4-reductase | DFR | 1.1.1.219 | AT5G42800 (DFR) |
| 14 | Anthocyanidin synthase | ANS | 1.14.20.4 | AT4G22870 AT4G22880 (LDOX) |
| 15 | Leucoanthocyanidin reductase | LAR | 1.17.1.3 | None |
| 16 | Anthocyanidin reductase | ANR | 1.3.1.77 | AT1G61720 (BAN) |
| Gene Name | Gene ID | Subgroup | Promote or Inhibit |
|---|---|---|---|
| AtMYB21 | AT3G27810 | R2R3subgroup19 | Promote |
| AtMYB24 | AT5G40350 | R2R3subgroup19 | Promote |
| AtMYB57 | AT3G01530 | R2R3 | Promote |
| AtMYB99/ATMYBCU15 | AT5G62320 | R2R3 | Promote |
| AtMYB11/PFG2 | AT3G62610 | R2R3subgroup7 | Promote |
| AtMYB12/PFG1 | AT2G47460 | R2R3subgroup7 | Promote |
| AtMYB111/PFG3 | AT5G49330 | R2R3subgroup7 | Promote |
| AtMYB123/TT2 | AT5G35550 | R2R3subgroup5 | Promote |
| AtMYB5 | AT3G13540 | R2R3 | Promote |
| AtMYB114 | AT1G66380 | R2R3 | Promote |
| AtMYB113 | AT1G66370 | R2R3subgroup6 | Promote |
| AtMYB75/PAP1 | AT1G56650 | R2R3subgroup6 | Promote |
| AtMYB90/PAP2 | AT1G66390 | R2R3subgroup6 | Promote |
| AtMYB112 | AT1G48000 | R2R3subgroup20 | Promote/Inhibit |
| AtMYB7/ATY49 | AT2G16720 | R2R3subgroup4 | Inhibit |
| AtMYB4 | AT4G38620 | R2R3subgroup4 | Inhibit |
| AtMYB32 | AT4G34990 | R2R3subgroup4 | Inhibit |
| AtMYB3 | AT1G22640 | R2R3subgroup4 | Inhibit |
| AtMYB6 | AT4G09460 | R2R3 | Inhibit |
| AtMYBL2 | AT1G71030 | R3 | Inhibit |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, Y.; Luo, J.; Cai, Z. Biosynthesis and Regulatory Mechanisms of Plant Flavonoids: A Review. Plants 2025, 14, 1847. https://doi.org/10.3390/plants14121847
Mao Y, Luo J, Cai Z. Biosynthesis and Regulatory Mechanisms of Plant Flavonoids: A Review. Plants. 2025; 14(12):1847. https://doi.org/10.3390/plants14121847
Chicago/Turabian StyleMao, Yuye, Jiajia Luo, and Zeping Cai. 2025. "Biosynthesis and Regulatory Mechanisms of Plant Flavonoids: A Review" Plants 14, no. 12: 1847. https://doi.org/10.3390/plants14121847
APA StyleMao, Y., Luo, J., & Cai, Z. (2025). Biosynthesis and Regulatory Mechanisms of Plant Flavonoids: A Review. Plants, 14(12), 1847. https://doi.org/10.3390/plants14121847






