Selection and Validation of Reference Genes for RT-qPCR in Protonemal Tissue of the Desiccation-Tolerant Moss Pseudocrossidium replicatum Under Multiple Abiotic Stress Conditions
Abstract
1. Introduction
2. Results
2.1. Selection of Candidate Reference Genes from Transcriptome Data
2.2. Verification of Primer Specificity
2.3. Expression Profile of Candidate Reference Genes in Response to ABA and Abiotic Stresses via RT-qPCR
2.4. Evaluation of the Expression Stability of Candidate Reference Genes Using the ΔΔCt Method
2.5. Evaluation of the Ideal Number of Reference Genes for Normalisation
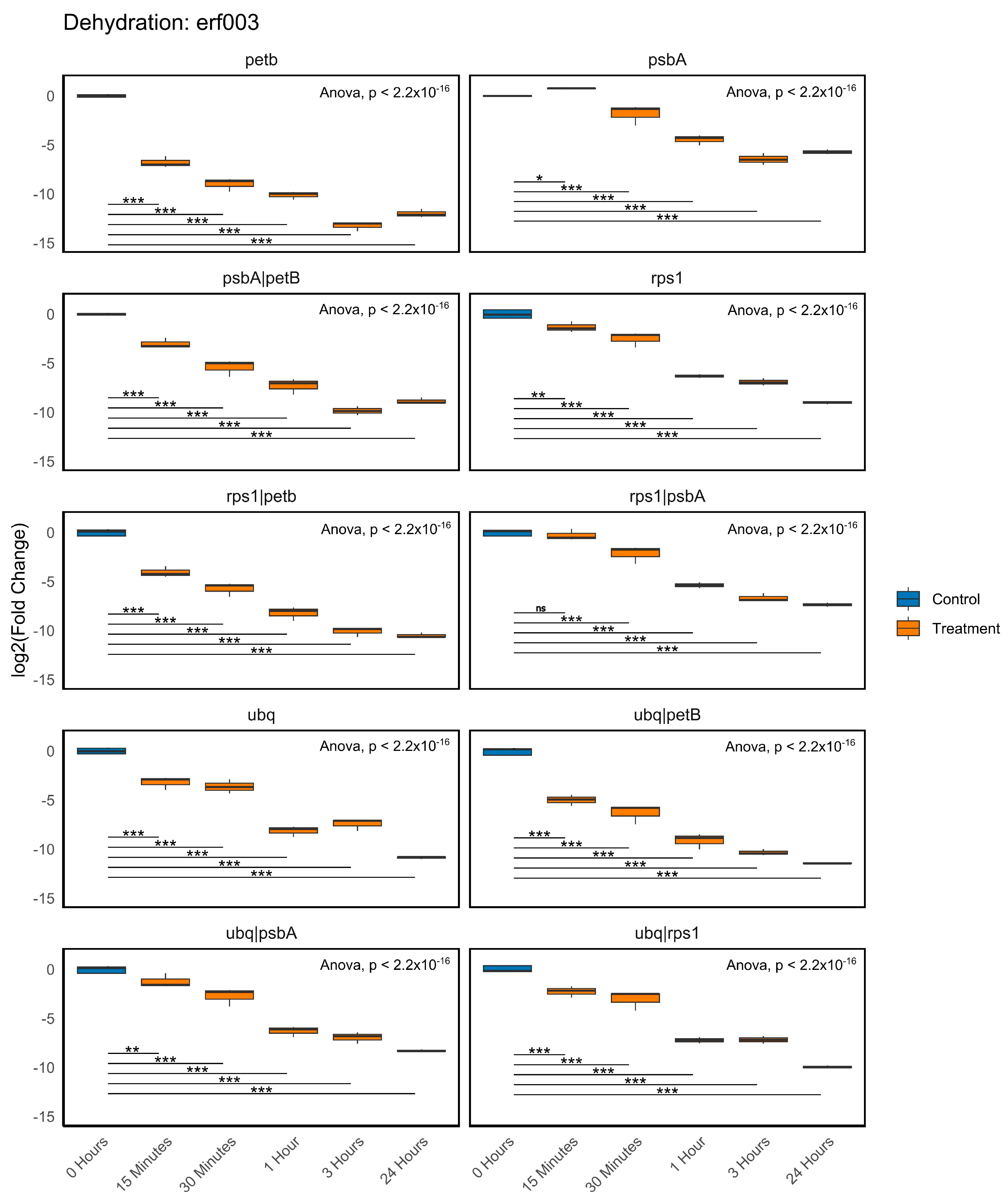
2.6. Validation of the Recommended Reference Genes
3. Discussion
4. Materials and Methods
4.1. Biological Material and Stress Treatments
4.2. RNA-Seq Analyses and De Novo Transcriptome Assembly
4.3. Analysis of Differentially Expressed Genes (DEGs)
4.4. Selection of Candidate Reference Genes
4.5. Primer Design and Evaluation of Candidate Reference Genes
4.6. Total RNA Extraction and cDNA Synthesis
4.7. RT-qPCR Analysis
4.8. RT-qPCR Data Analysis of Reference Gene Stability
4.9. Normalisation of the Expression of P. replicatum Dehydration-Responsive Genes in RT-qPCR Experiments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lai, J.; Kooijmans, L.M.J.; Sun, W.; Lombardozzi, D.; Campbell, J.E.; Gu, L.; Luo, Y.; Kuai, L.; Sun, Y. Terrestrial photosynthesis inferred from plant carbonyl sulfide uptake. Nature 2024, 634, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Walter, V.R. Ecosystems and Human Well-Being: Synthesis; Island Press: Washington, DC, USA, 2005; Available online: www.millenniumassessment.org/documents/document.356.aspx.pdf (accessed on 23 March 2025).
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef] [PubMed]
- The State of Food and Agriculture 2023; FAO: Rome, Italy, 2023. [CrossRef]
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.M.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobell, D.; Mazur, B.J.; Molden, D.; et al. Radically Rethinking Agriculture for the 21st Century. Science (1979) 2010, 327, 833–834. [Google Scholar] [CrossRef] [PubMed]
- Oshunsanya, S.O.; Nwosu, N.J.; Li, Y. Abiotic Stress in Agricultural Crops Under Climatic Conditions. In Sustainable Agriculture, Forest and Environmental Management; Springer: Singapore, 2019; pp. 71–100. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Eckardt, N.A.; Cutler, S.; Juenger, T.E.; Marshall-Colon, A.; Udvardi, M.; Verslues, P.E. Focus on climate change and plant abiotic stress biology. Plant Cell 2023, 35, 1–3. [Google Scholar] [CrossRef]
- Boyer, J.S. Plant Productivity and Environment. Science (1979) 1982, 218, 443–448. [Google Scholar] [CrossRef]
- Araus, J.L.; Slafer, G.; Reynolds, M.P.; Royo, C. Plant breeding and drought in C3 cereals: What should we breed for? Ann. Bot. 2002, 89, 925–940. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science (1979) 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science (1979) 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- The State of the World’s Land and Water Resources for Food and Agriculture—Systems at Breaking Point (SOLAW 2021); FAO: Rome, Italy, 2021. [CrossRef]
- Villalobos-López, M.A.; Arroyo-Becerra, A.; Quintero-Jiménez, A.; Iturriaga, G. Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops. Int. J. Mol. Sci. 2022, 23, 12053. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, J.; Li, R.; Ge, Y.; Li, Y.; Li, R. Plants’ Response to Abiotic Stress: Mechanisms and Strategies. Int. J. Mol. Sci. 2023, 24, 10915. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, S. Selection of the Reference Gene for Expression Normalization in Salsola ferganica under Abiotic Stress. Genes 2022, 13, 571. [Google Scholar] [CrossRef]
- Kulshrestha, S.; Jibran, R.; van Klink, J.W.; Zhou, Y.; Brummell, D.A.; Albert, N.W.; Schwinn, K.E.; Chagné, D.; Landi, M.; Bowman, J.L.; et al. Stress, senescence, and specialized metabolites in bryophytes. J. Exp. Bot. 2022, 73, 4396–4411. [Google Scholar] [CrossRef]
- Lambris, J.D. Advances in Experimental Medicine and Biology. Available online: http://www.springer.com/series/5584 (accessed on 23 March 2025).
- Ríos-Meléndez, S.; Valadez-Hernández, E.; Delgadillo, C.; Luna-Guevara, M.L.; Martínez-Núñez, M.A.; Sánchez-Pérez, M.; Martínez-Y-Pérez, J.L.; Arroyo-Becerra, A.; Cárdenas, L.; Bibbins-Martínez, M.; et al. Pseudocrossidium replicatum (Taylor) R.H. Zander is a fully desiccation-tolerant moss that expresses an inducible molecular mechanism in response to severe abiotic stress. Plant Mol. Biol. 2021, 107, 387–404. [Google Scholar] [CrossRef]
- Wood, A.J. Invited essay: New frontiers in bryology and lichenology: The nature and distribution of vegetative desiccation-tolerance in hornworts, liverworts and mosses. Bryologist 2007, 110, 163–177. [Google Scholar] [CrossRef]
- Oliver, M.J.; Velten, J.; Wood, A.J. Bryophytes as experimental models for the study of environmental stress tolerance: Tortula ruralis and desiccation-tolerance in mosses. Plant Ecol. 2000, 151, 73–84. [Google Scholar] [CrossRef]
- Yin, B.-F.; Zhang, Y.-M.; Lou, A.-R. Impacts of the removal of shrubs on the physiological and biochemical characteristics of Syntrichia caninervis Mitt: In a temperate desert. Sci. Rep. 2017, 7, 45268. [Google Scholar] [CrossRef]
- Ekwealor, J.T.B.; Mishler, B.D. Transcriptomic Effects of Acute Ultraviolet Radiation Exposure on Two Syntrichia Mosses. Front. Plant Sci. 2021, 12, 752913. [Google Scholar] [CrossRef]
- Li, X.; Bai, W.; Yang, Q.; Yin, B.; Zhang, Z.; Zhao, B.; Kuang, T.; Zhang, Y.; Zhang, D. The extremotolerant desert moss Syntrichia caninervis is a promising pioneer plant for colonizing extraterrestrial environments. Innovation 2024, 5, 100657. [Google Scholar] [CrossRef] [PubMed]
- Teng, P.; Liu, W.; Yang, Y. Syntrichia Caninervis Adapt to Salt Stress by Altering Physiological Propertie. 2023. Available online: https://doi.org/10.2139/ssrn.4600956 (accessed on 23 March 2025).
- Koster, K.L.; Balsamo, R.A.; Espinoza, C.; Oliver, M.J. Desiccation sensitivity and tolerance in the moss Physcomitrella patens: Assessing limits and damage. Plant Growth Regul. 2010, 62, 293–302. [Google Scholar] [CrossRef]
- Xiao, L.; Yobi, A.; Koster, K.L.; He, Y.; Oliver, M.J. Desiccation tolerance in Physcomitrella patens: Rate of dehydration and the involvement of endogenous abscisic acid (ABA). Plant Cell Environ. 2018, 41, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Minami, A.; Nagao, M.; Arakawa, K.; Fujikawa, S.; Takezawa, D. Abscisic acid-induced freezing tolerance in the moss Physcomitrella patens is accompanied by increased expression of stress-related genes. J. Plant Physiol. 2003, 160, 475–483. [Google Scholar] [CrossRef]
- Saavedra, L.; Svensson, J.; Carballo, V.; Izmendi, D.; Welin, B.; Vidal, S. A dehydrin gene in Physcomitrella patens is required for salt and osmotic stress tolerance. Plant J. 2006, 45, 237–249. [Google Scholar] [CrossRef]
- Frank, W.; Ratnadewi, D.; Reski, R. Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta 2005, 220, 384–394. [Google Scholar] [CrossRef]
- Cevallos, M.A.; Guerrero, G.; Ríos, S.; Arroyo, A.; Villalobos, M.A.; Porta, H. The chloroplast genome of the desiccation-tolerant moss Pseudocrossidium replicatum (Taylor) R. H. Zander. Genet. Mol. Biol. 2019, 42, 488–493. [Google Scholar] [CrossRef]
- Cevallos, M.A.; Guerrero, G.; Ríos, S.; Arroyo, A.; Villalobos, M.A.; Porta, H. The mitogenome of Pseudocrossidium replicatum, a desiccation-tolerant moss. Mitochondrial DNA B Resour. 2020, 5, 2339–2341. [Google Scholar] [CrossRef]
- Timmerhaus, G.; Hanke, S.T.; Buchta, K.; Rensing, S.A. Prediction and validation of promoters involved in the abscisic acid response in Physcomitrella patens. Mol. Plant 2011, 4, 713–729. [Google Scholar] [CrossRef]
- Tong, J.; Hu, M.; Han, B.; Ji, Y.; Wang, B.; Liang, H.; Liu, M.; Wu, Z.; Liu, N. Determination of reliable reference genes for gene expression studies in Chinese chive (Allium tuberosum) based on the transcriptome profiling. Sci. Rep. 2021, 11, 16558. [Google Scholar] [CrossRef]
- Köhsler, M.; Leitsch, D.; Müller, N.; Walochnik, J. Validation of reference genes for the normalization of RT-qPCR gene expression in Acanthamoeba spp. Sci. Rep. 2020, 10, 10362. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Hu, A.; Yang, X.; Zhu, J.; Wang, X.; Liu, J.; Wang, J.; Wu, H.; Zhang, H.; Zhang, H. Selection and validation of appropriate reference genes for RT–qPCR analysis of Nitraria sibirica under various abiotic stresses. BMC Plant Biol. 2022, 22, 592. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Absolute Quantification of MRNA Using Real-Time Reverse Transcription Polymerase Chain Reaction Assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef]
- Ni, X.; Yang, Y.; Xie, Y.; Li, D.; Xia, X.; Zhang, Y.; Zheng, C. Selection and verification of reference genes for real-time quantitative PCR in endangered mangrove species Acanthus ebracteatus under different abiotic stress conditions. Mar. Environ. Res. 2025, 204, 106862. [Google Scholar] [CrossRef]
- Walker, J.M. Methods in Molecular Biology TM Series Editor. Available online: http://www.springer.com/series/7651 (accessed on 23 March 2025).
- Hu, X.; Zhang, L.; Nan, S.; Miao, X.; Yang, P.; Duan, G.; Fu, H. Selection and validation of reference genes for quantitative real-time PCR in Artemisia sphaerocephala based on transcriptome sequence data. Gene 2018, 657, 39–49. [Google Scholar] [CrossRef]
- Jia, X.; Xiong, Y.; Xiong, Y.; Li, D.; Yu, Q.; Lei, X.; You, M.; Bai, S.; Zhang, J.; Ma, X. Identification and Validation of Reference Genes for RT-qPCR Analysis in Reed Canary Grass during Abiotic Stress. Genes. 2023, 14, 1790. [Google Scholar] [CrossRef]
- Park, M.; Hong, S.G.; Park, H.; Lee, B.-H.; Lee, H. Identification of reference genes for rt-qpcr in the antarctic moss Sanionia uncinata under abiotic stress conditions. PLoS ONE 2018, 13, e0199356. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of MRNA Using Real-Time Reverse Transcription PCR (RT-PCR): Trends and Problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Li, X.; Yang, Q.; Zhao, M.; Bai, W.; Liang, Y.; Liu, X.; Gao, B.; Zhang, D. Transcriptional profiling analysis providing insights into desiccation tolerance mechanisms of the desert moss Syntrichia caninervis. Front. Plant Sci. 2023, 14, 1127541. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper-Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
- Cheng, S.-S.; Ku, Y.-S.; Cheung, M.-Y.; Lam, H.-M. Identification of stably expressed reference genes for expression studies in Arabidopsis thaliana using mass spectrometry-based label-free quantification. Front. Plant Sci. 2022, 13, 1001920. [Google Scholar] [CrossRef]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Kumar, D.; Das, P.K.; Sarmah, B.K. Reference gene validation for normalization of RT-qPCR assay associated with germination and survival of rice under hypoxic condition. J. Appl. Genet. 2018, 59, 419–430. [Google Scholar] [CrossRef]
- Manoli, A.; Sturaro, A.; Trevisan, S.; Quaggiotti, S.; Nonis, A. Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 2012, 169, 807–815. [Google Scholar] [CrossRef]
- Duan, Y.-K.; Han, R.; Su, Y.; Wang, A.-Y.; Li, S.; Sun, H.; Gong, H.-J. Transcriptional search to identify and assess reference genes for expression analysis in Solanumlycopersicum under stress and hormone treatment conditions. J. Integr. Agric. 2022, 21, 3216–3229. [Google Scholar] [CrossRef]
- Le Bail, A.; Scholz, S.; Kost, B. Evaluation of Reference Genes for RT qPCR Analyses of Structure-Specific and Hormone Regulated Gene Expression in Physcomitrella patens Gametophytes. PLoS ONE 2013, 8, e70998. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, D.; Li, H.; Gao, B.; Yang, H.; Zhang, Y.; Wood, A.J. Characterization of reference genes for RT-qPCR in the desert moss Syntrichia caninervis in response to abiotic stress and desiccation/rehydration. Front. Plant Sci. 2015, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Liu, J.-X.; Xing, G.-M.; Sun, S.; Li, S.; Duan, A.-Q.; Wang, F.; Li, M.-Y.; Xu, Z.-S.; Xiong, A.-S. Selection of appropriate reference genes for RT-qPCR analysis under abiotic stress and hormone treatment in celery. PeerJ 2019, 7, e7925. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, R.; Zhou, Z. Identification and validation of reference genes for gene expression analysis in Schima superba. Genes 2021, 12, 732. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, C.; Yang, S.; Zhong, Q.; Tian, J. Screening and Verification of Reference Genes for Analysis of Gene Expression in Garlic (Allium sativum L.) under Cold and Drought Stress. Plants 2023, 12, 763. [Google Scholar] [CrossRef]
- Wang, F.; Li, P.; Liu, Q.; Nie, G.; Zhu, Y.; Zhang, X. Selection and Validation of Reference Genes in Sudan Grass (Sorghum sudanense (Piper) Stapf) under Various Abiotic Stresses by qRT-PCR. Genes 2024, 15, 210. [Google Scholar] [CrossRef]
- Ferreira, D.B.; Gasparoni, L.M.; Bronzeri, C.F.; Paiva, K.B.S. RPLP0/TBP are the most stable reference genes for human dental pulp stem cells under osteogenic differentiation. World J. Stem Cells 2024, 16, 656–669. [Google Scholar] [CrossRef]
- Wang, X.; Shu, X.; Su, X.; Xiong, Y.; Xiong, Y.; Chen, M.; Tong, Q.; Ma, X.; Zhang, J.; Zhao, J. Selection of Suitable Reference Genes for RT-qPCR Gene Expression Analysis in Centipedegrass under Different Abiotic Stress. Genes 2023, 14, 1874. [Google Scholar] [CrossRef]
- Zhou, Y.; Meng, F.; Han, K.; Zhang, K.; Gao, J.; Chen, F. Screening and validating of endogenous reference genes in Chlorella sp. TLD 6B under abiotic stress. Sci. Rep. 2023, 13, 1555. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Yu, H.; Li, X.; Cui, H.; Chen, J.; Huang, P. Selection and Validation of Reference Genes for RT-qPCR Analysis in Aegilops tauschii (Coss.) under Different Abiotic Stresses. Int. J. Mol. Sci. 2021, 22, 11017. [Google Scholar] [CrossRef] [PubMed]
- Gohain, B. Rubisco-bis-phosphate oxygenase (RuBP)- A potential housekeeping gene forqPCR assays in tea. Afr. J. Biotechnol. 2012, 11, 11193–11199. [Google Scholar] [CrossRef]
- Huang, T.; Long, J.; Liu, S.-W.; Yang, Z.-W.; Zhu, Q.-J.; Zhao, X.-L.; Peng, C. Selection and Validation of Reference Genes for mRNA Expression by Quantitative Real-Time PCR Analysis in Neolamarckia cadamba. Sci. Rep. 2018, 8, 9311. [Google Scholar] [CrossRef]
- Li, T.; Wang, J.; Lu, M.; Zhang, T.; Qu, X.; Wang, Z. Selection and Validation of Appropriate Reference Genes for qRT-PCR Analysis in Isatis indigotica Fort. Front. Plant Sci. 2017, 8, 1139. [Google Scholar] [CrossRef]
- Li, G.; Sun, X.; Zhu, X.; Wu, B.; Hong, H.; Xin, Z.; Xin, X.; Peng, J.; Jiang, S. Selection and Validation of Reference Genes in Virus-Infected Sweet Potato Plants. Genes 2023, 14, 1477. [Google Scholar] [CrossRef]
- Marzin, S.; Mihaly, R.; Pauk, J.; Schweizer, P. A transient assay system for the assessment of cell-autonomous gene function in dehydration-stressed barley. J. Exp. Bot. 2008, 59, 3359–3369. [Google Scholar] [CrossRef]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K.; et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11178–E11187. [Google Scholar] [CrossRef]
- Villalobos, M.A.; Bartels, D.; Iturriaga, G. Stress Tolerance and Glucose Insensitive Phenotypes in Arabidopsis Overexpressing the CpMYB10 Transcription Factor Gene. Plant Physiol. 2004, 135, 309–324. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Quan, R.; Pan, X.; Wan, L.; Huang, R. EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 2013, 237, 1443–1451. [Google Scholar] [CrossRef]
- Klay, I.; Pirrello, J.; Riahi, L.; Bernadac, A.; Cherif, A.; Bouzayen, M.; Bouzid, S. Ethylene response factor Sl-ERF.B.3 is responsive to abiotic stresses and mediates salt and cold stress response regulation in tomato. Sci. World J. 2014, 2014, 167681. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.; Gilley, J.; Coleman, M.P. Difference Tracker: ImageJ plugins for fully automated analysis of multiple axonal transport parameters. J. Neurosci. Methods 2010, 193, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.-H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
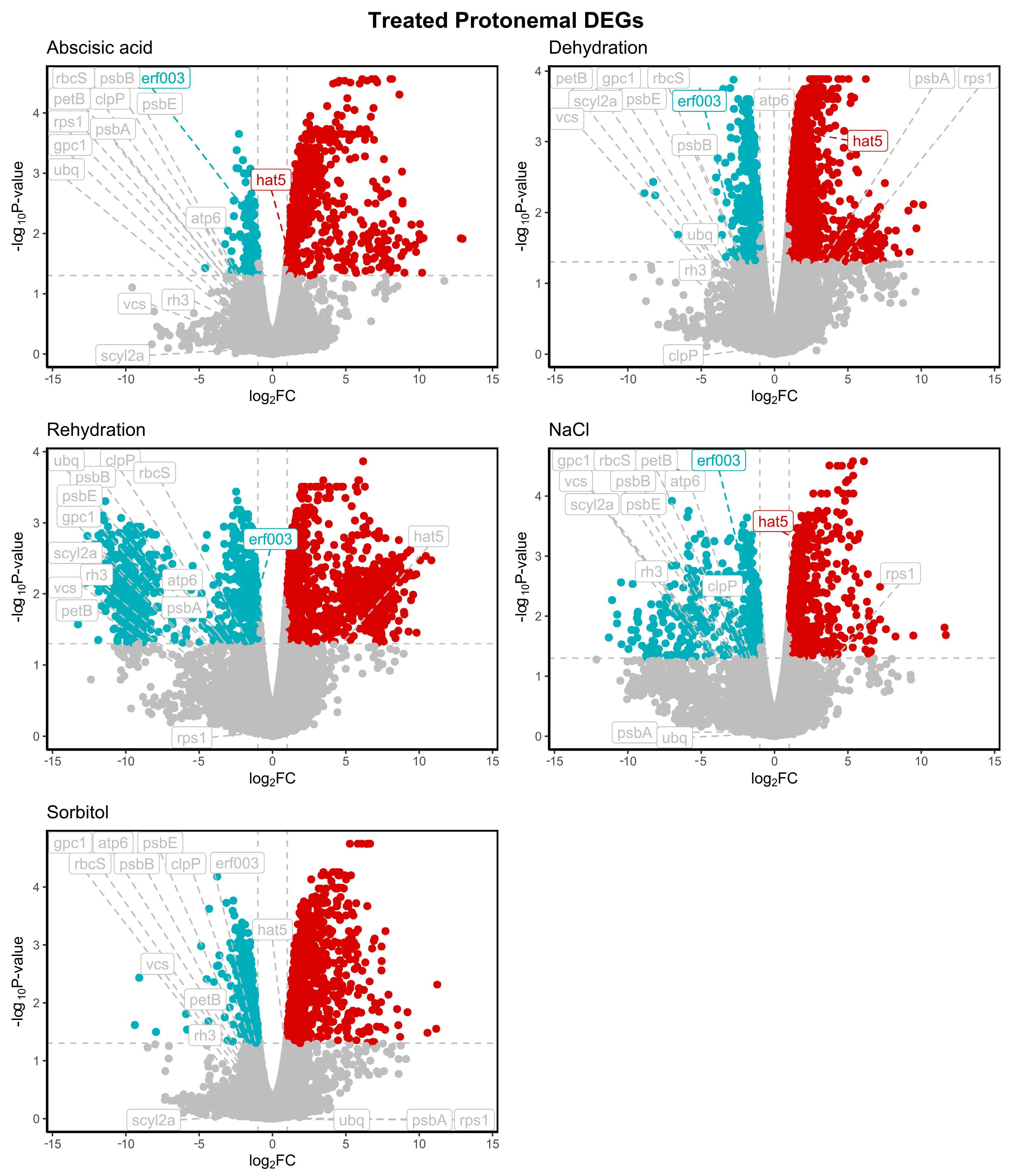

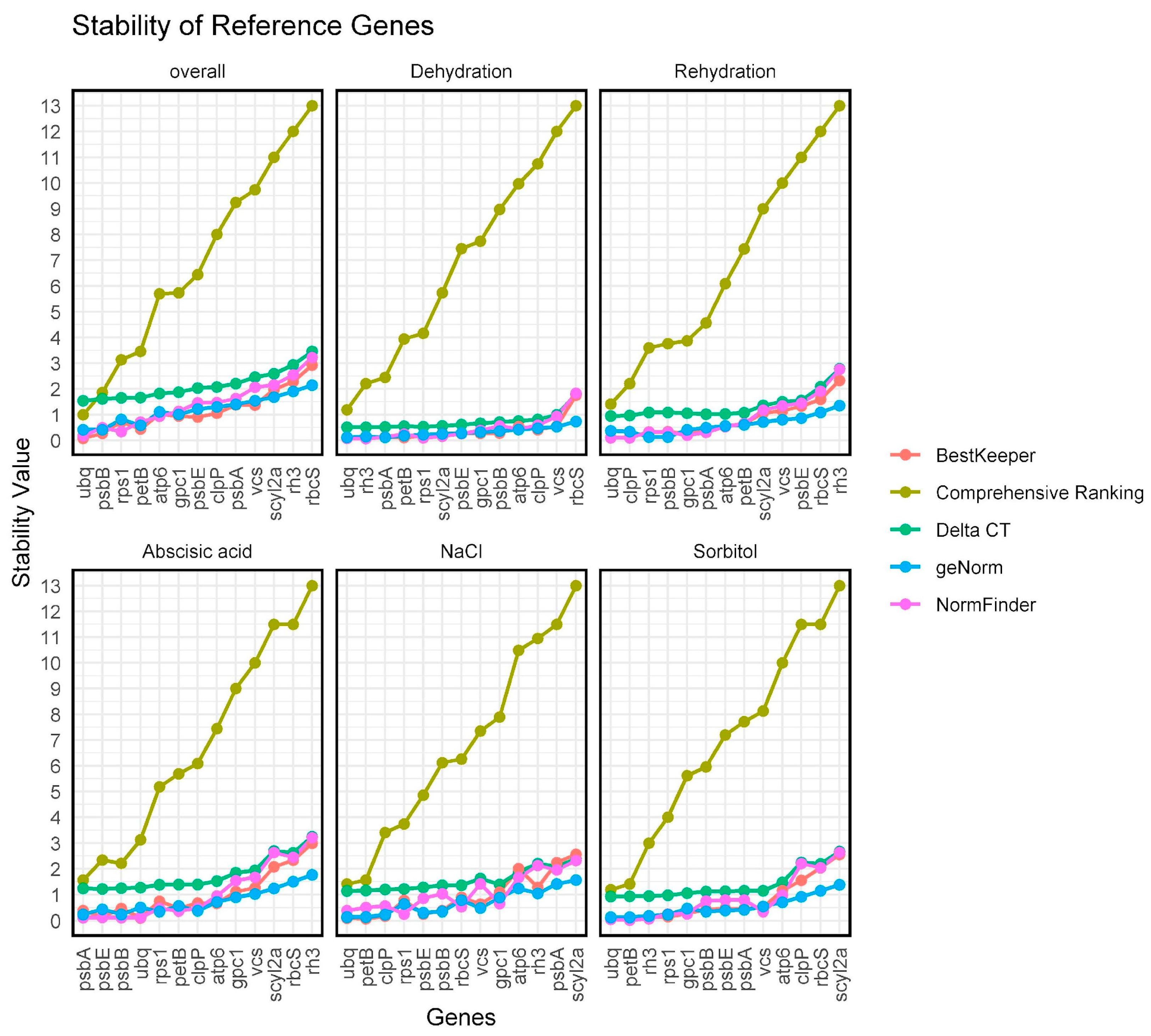
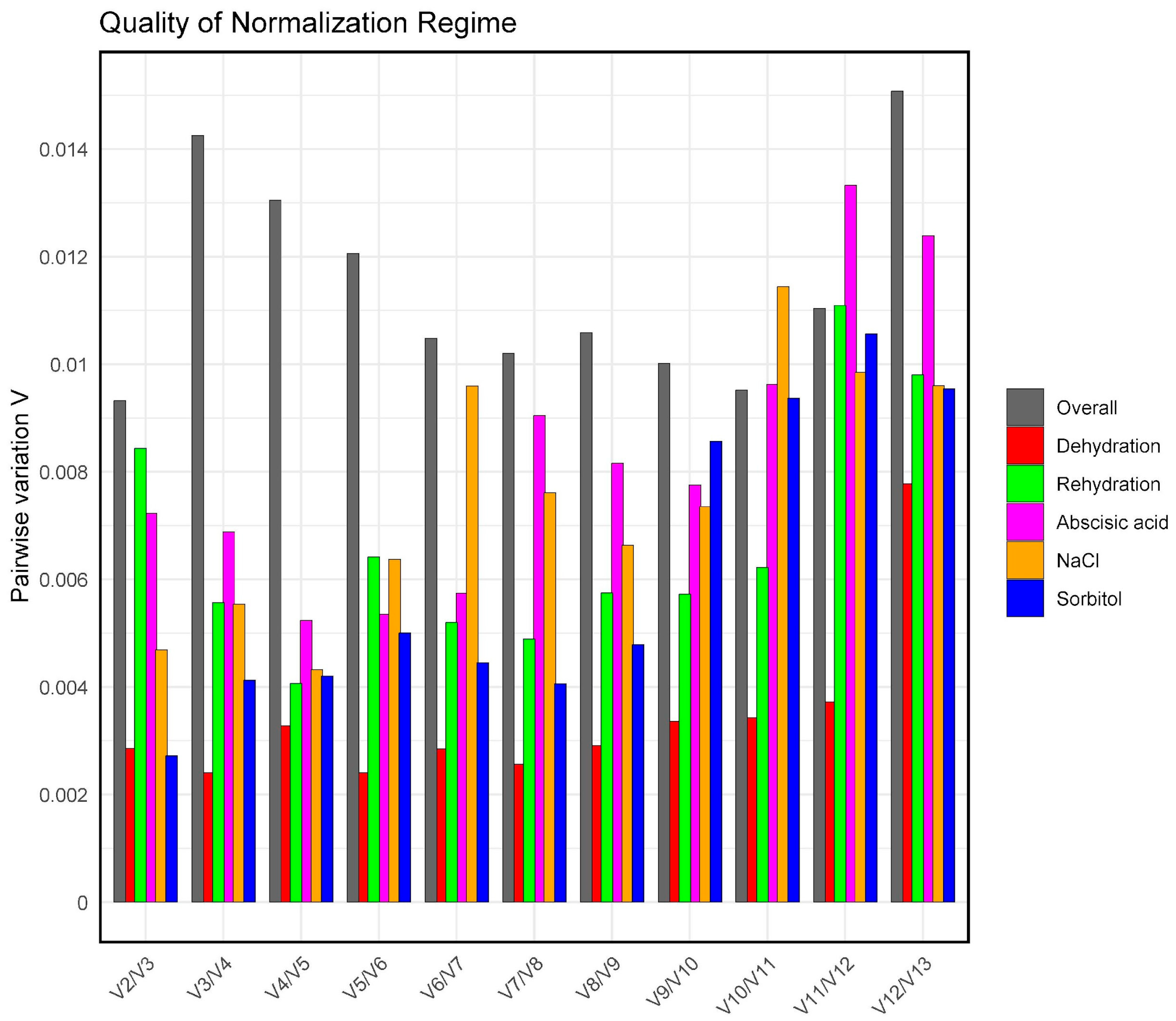
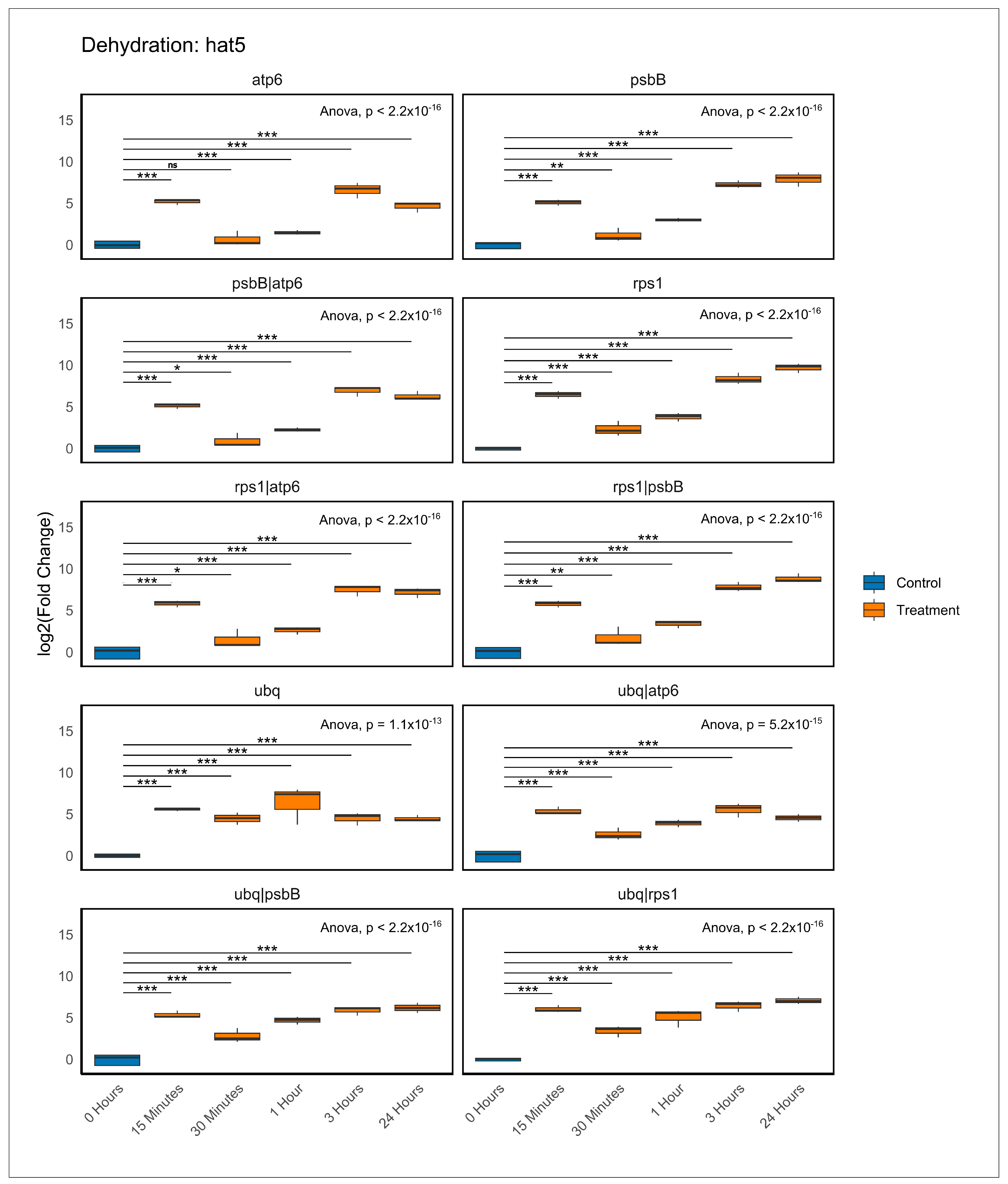
| Category | ID | Gene | Primers Forward 5′ a 3′ | Primers Reverse 5′ a 3′ | Amplicon (bp) |
|---|---|---|---|---|---|
| Putative reference genes | TRINITY_DN1938_c133_g1 | psbA | CATCGTAGCTGCTCATGGTTAC | GAAAGCCATTGTGCTGATACCT | 145 |
| TRINITY_DN9694_c9_g1 | psbB | TGATCCTTTTGTTCCAGGAGGA | CAGTTACACCATTGCCCATACG | 178 | |
| TRINITY_DN1103_c0_g1 | atp6 | CCCTTGTCGTTAGCACCCTTC | GTCCAAGCAAACCCGCTTAGAA | 140 | |
| TRINITY_DN4025_c0_g2_i1 | clpP | TCTCCTGGTGGAGCTGTATTAG | AAAGCAAAGCCGCAAAGC | 140 | |
| TRINITY_DN798_c0_g1_i4 | rps1 | TGGCAATATCGTGGTAAAAGAGA | GCAGGTGTTTCCGCTTTG | 174 | |
| TRINITY_DN269_c0_g1_i28 | rbcS | ATGATTCAGCAGAACCTG | ATCCACATGGTCCAGTAG | 104 | |
| TRINITY_DN1154_c1_g1 | ubq | TCAAGCACAAGAAGAAGAAGG | ATCAGGGTTTGGGCACTC | 116 | |
| TRINITY_DN2574_c0_g1 | vcs | CTGTTGAGACTTGTGAAGAC | GCGGAGCAATCTGTTAAG | 148 | |
| TRINITY_DN224_c0_g1 | scyl2a | CCATCATACTCGCGGACCATC | TCGCCGCTGTTAGAGTA | 174 | |
| TRINITY_DN5925_c0_g1 | gpc1 | CGGCGGAGATTCAGTAGA | CATGGAGTTGCTCATCAGATT | 137 | |
| TRINITY_DN5657_c0_g1 | rh3 | CAATGTCGGCAAGATTCGTA | CACCAACTTCGGCAACTT | 148 | |
| TRINITY_DN717_c2_g1_i1 | petB | AGGAGGCATCACCTTAACTTGTTT | CCATCATACTCGCGGACCATC | 173 | |
| TRINITY_DN5651_c0_g3 | psbE | AGGAGAGCGCCCTTTTGCTGATA | AGGCACTTCTTACCGGCTTACTGT | 178 | |
| Abiotic stress responsive genes | TRINITY_DN80_c0_g3 | hat5 | TGGAAGACGAAGCAACTG | AGTGGACTCACCTGACAG | 138 |
| TRINITY_DN2254_c0_g1 | erf003 | CTTCTTCGACTGCCAGAT | GAAGGAGTCTTGCGTGAA | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nava-Nolazco, R.M.; Ríos-Melendez, S.; Galván-Gordillo, S.V.; Martínez-Navarro, A.C.; Sánchez-Pérez, M.; Chavez-Santoscoy, R.A.; Bibbins-Martínez, M.; Maldonado-Mendoza, I.E.; Arroyo-Becerra, A.; Villalobos-López, M.A. Selection and Validation of Reference Genes for RT-qPCR in Protonemal Tissue of the Desiccation-Tolerant Moss Pseudocrossidium replicatum Under Multiple Abiotic Stress Conditions. Plants 2025, 14, 1752. https://doi.org/10.3390/plants14121752
Nava-Nolazco RM, Ríos-Melendez S, Galván-Gordillo SV, Martínez-Navarro AC, Sánchez-Pérez M, Chavez-Santoscoy RA, Bibbins-Martínez M, Maldonado-Mendoza IE, Arroyo-Becerra A, Villalobos-López MA. Selection and Validation of Reference Genes for RT-qPCR in Protonemal Tissue of the Desiccation-Tolerant Moss Pseudocrossidium replicatum Under Multiple Abiotic Stress Conditions. Plants. 2025; 14(12):1752. https://doi.org/10.3390/plants14121752
Chicago/Turabian StyleNava-Nolazco, Rosa María, Selma Ríos-Melendez, Santiago Valentín Galván-Gordillo, Angélica C. Martínez-Navarro, Mishael Sánchez-Pérez, Rocio Alejandra Chavez-Santoscoy, Martha Bibbins-Martínez, Ignacio Eduardo Maldonado-Mendoza, Analilia Arroyo-Becerra, and Miguel Angel Villalobos-López. 2025. "Selection and Validation of Reference Genes for RT-qPCR in Protonemal Tissue of the Desiccation-Tolerant Moss Pseudocrossidium replicatum Under Multiple Abiotic Stress Conditions" Plants 14, no. 12: 1752. https://doi.org/10.3390/plants14121752
APA StyleNava-Nolazco, R. M., Ríos-Melendez, S., Galván-Gordillo, S. V., Martínez-Navarro, A. C., Sánchez-Pérez, M., Chavez-Santoscoy, R. A., Bibbins-Martínez, M., Maldonado-Mendoza, I. E., Arroyo-Becerra, A., & Villalobos-López, M. A. (2025). Selection and Validation of Reference Genes for RT-qPCR in Protonemal Tissue of the Desiccation-Tolerant Moss Pseudocrossidium replicatum Under Multiple Abiotic Stress Conditions. Plants, 14(12), 1752. https://doi.org/10.3390/plants14121752










