Genetic Analyses, BSA-Seq, and Transcriptome Analyses Reveal Candidate Genes Controlling Leaf Plastochron in Rapeseed (Brassica napus L.)
Abstract
1. Introduction
2. Results
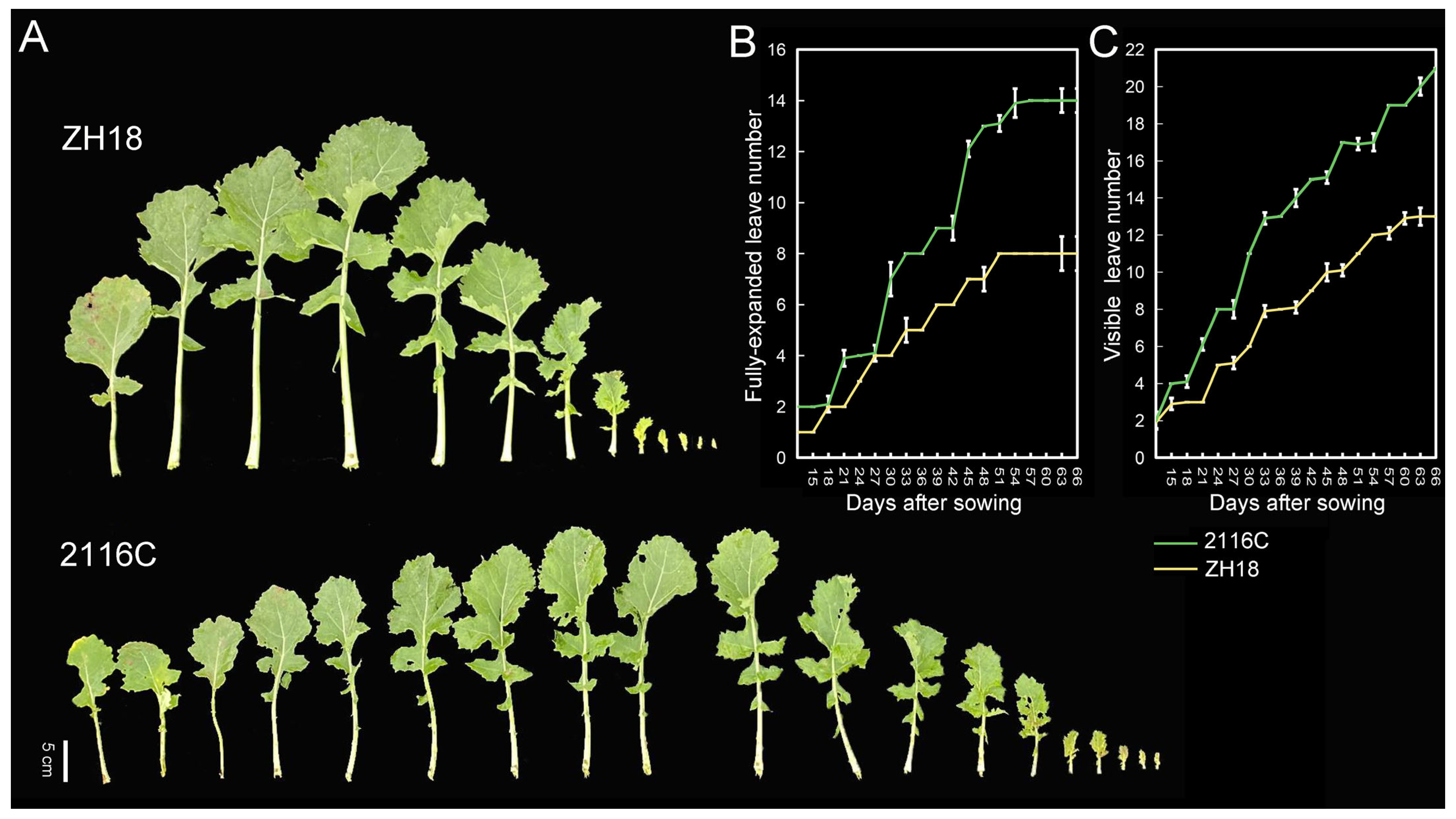
2.1. Investigation of Plastochron for the Parents
2.2. Phenotype Investigation for the Parents and Genetic Population
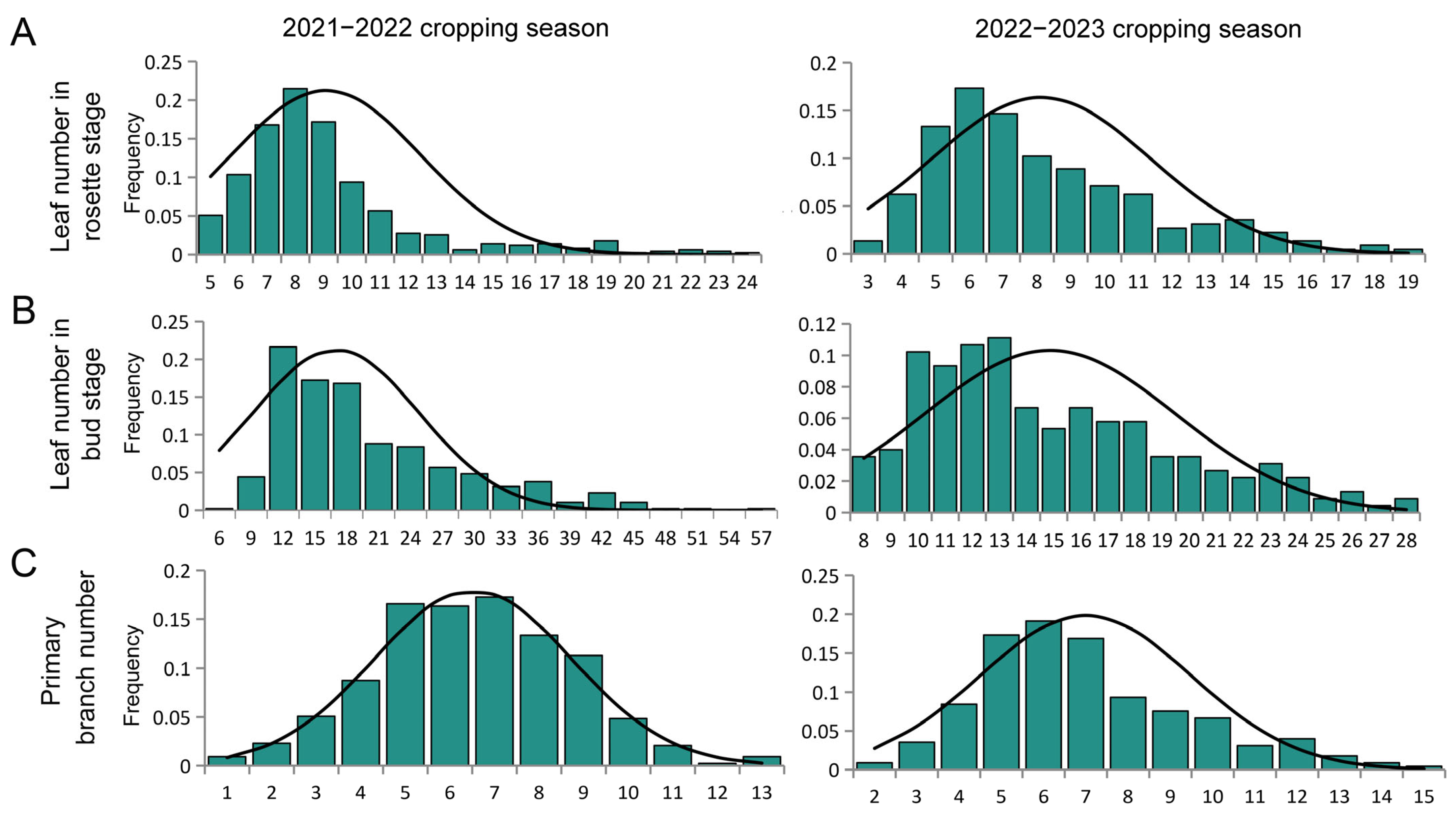
2.3. Genetic Analysis of Leaf Plastochron in Rapeseed
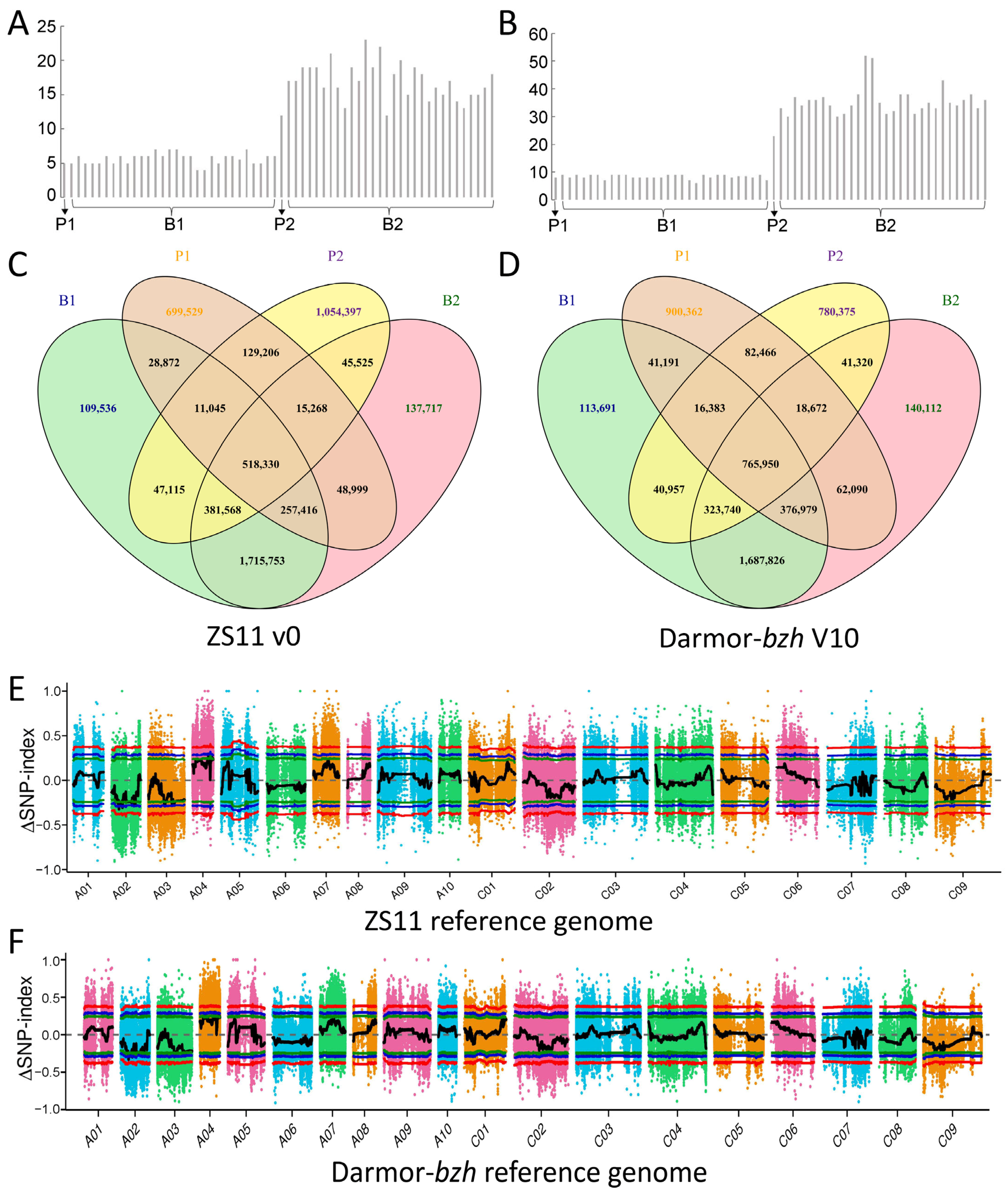
2.4. Primary QTL Identification Based on BSA Analyses
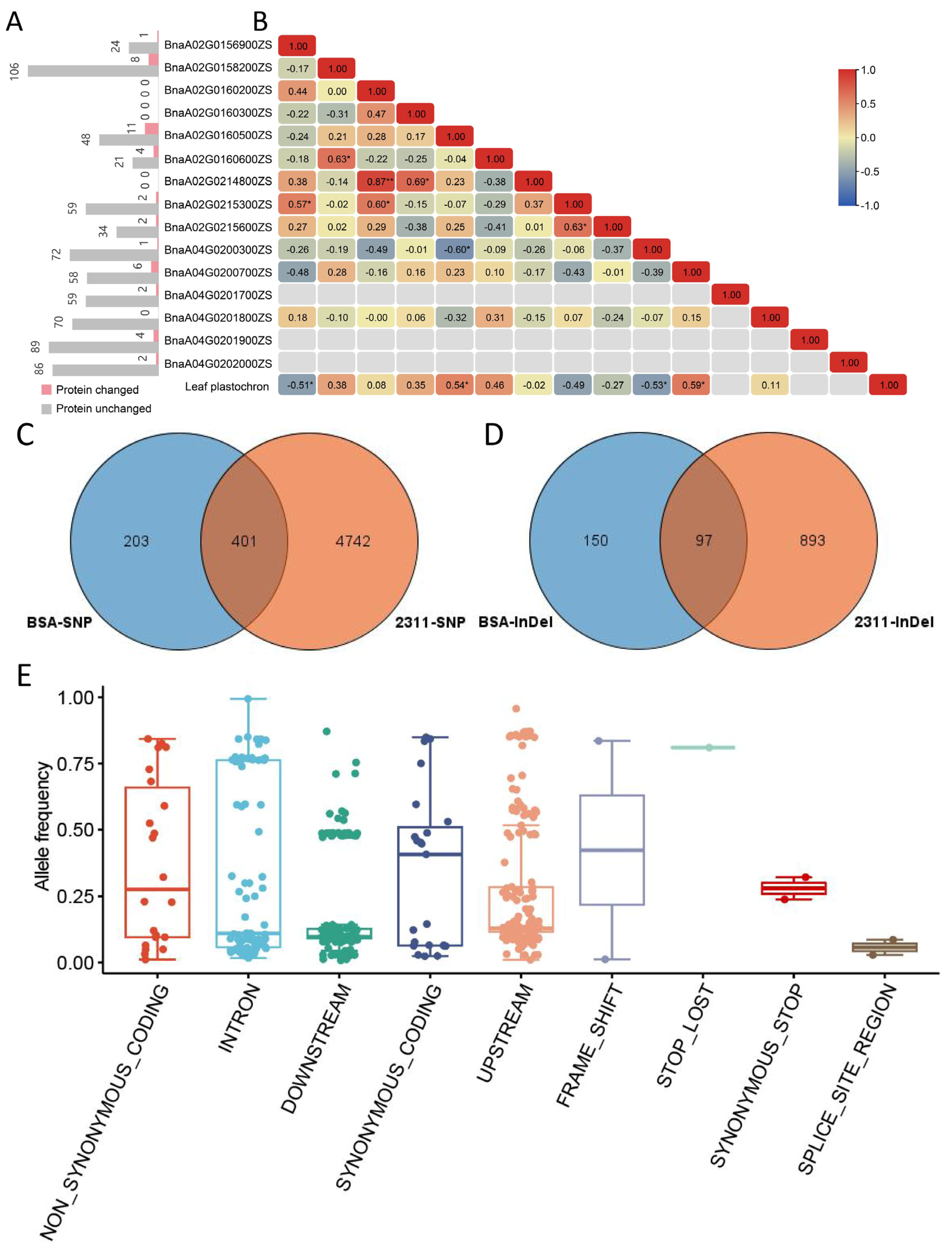
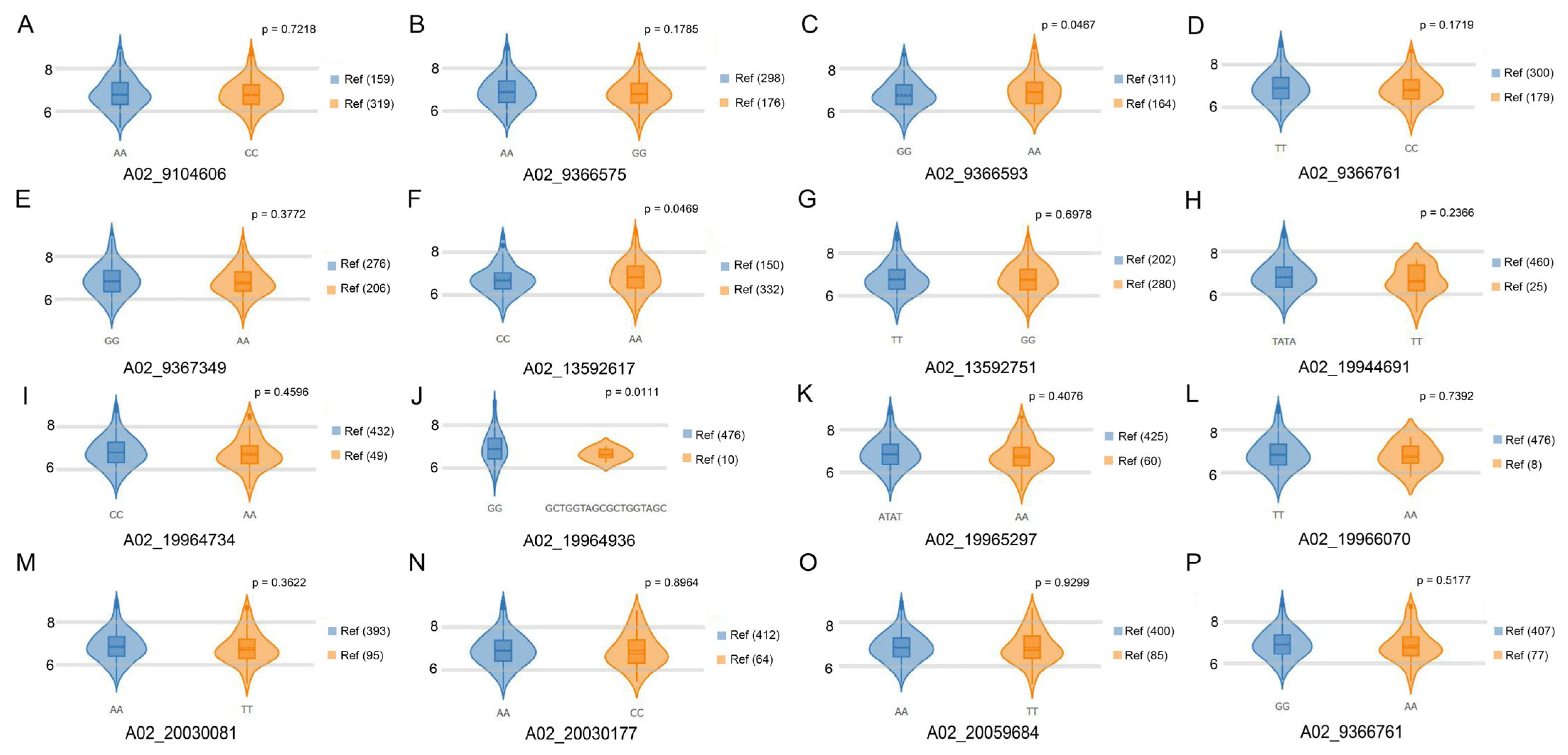
2.5. Gene Annotations and Sequence Variants Analyses in Candidate Genes
2.6. Gene Expression Analyses in Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Plant Plantation and Phenotype Collection
4.2. Segregation Analysis for Leaf Plastochron-Related Traits
4.3. DNA Isolation and BSA-Sequencing
4.4. SNP-Index Calculation and QTL Detection
4.5. RNA Isolation and RNA-Seq Analyses
4.6. Gene Prediction in the Mapping Interval
4.7. Candidate Gene Expression and Variants in the Public Database
4.8. Data Analyses and Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kawakatsu, T.; Itoh, J.-I.; Miyoshi, K.; Kurata, N.; Alvarez, N.; Veit, B.; Nagato, Y. PLASTOCHRON2 Regulates Leaf Initiation and Maturation in Rice. Plant Cell 2006, 18, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Ahn, B.-O.; Kawakatsu, T.; Ito, Y.; Itoh, J.-I.; Nagato, Y.; Kurata, N. PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc. Natl. Acad. Sci. USA 2004, 101, 875–880. [Google Scholar] [CrossRef]
- Padilla, J.M.; Otegui, M.E. Co-ordination between Leaf Initiation and Leaf Appearance in Field-grown Maize (Zea mays): Genotypic Differences in Response of Rates to Temperature. Ann. Bot. 2005, 96, 997–1007. [Google Scholar] [CrossRef]
- Strable, J.; Nelissen, H. The dynamics of maize leaf development: Patterned to grow while growing a pattern. Curr. Opin. Plant Biol. 2021, 63, 102038. [Google Scholar] [CrossRef] [PubMed]
- Prusinkiewicz, P.; Crawfordb, S.; Smitha, R.S.; Ljungc, K.; Bennett, T.; Ongaro, V.; Leyser, O. Control of bud activation by an auxin transport switch. Proc. Natl. Acad. Sci. USA 2009, 106, 17431–17436. [Google Scholar] [CrossRef]
- Li, Z.; Ding, Y.; Xie, L.; Jian, H.; Gao, Y.; Yin, J.; Li, J.; Liu, L. Regulation by sugar and hormone signaling of the growth of Brassica napus L. axillary buds at the transcriptome level. Plant Growth Regul. 2020, 90, 571–584. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, Y.; Shi, L.; Wang, L.; Li, W. Interaction of Phytohormones and External Environmental Factors in the Regulation of the Bud Dormancy in Woody Plants. Int. J. Mol. Sci. 2023, 24, 17200. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wagner, D. Plant Inflorescence Architecture: The Formation, Activity, and Fate of Axillary Meristems. Cold Spring Harb. Perspect. Biol. 2020, 12, a034652. [Google Scholar] [CrossRef]
- Wang, C.; Yang, J.; Chen, W.; Zhao, X.; Wang, Z. Contribution of the leaf and silique photosynthesis to the seeds yield and quality of oilseed rape (Brassica napus L.) in reproductive stage. Sci. Rep. 2023, 13, 4721. [Google Scholar] [CrossRef]
- Somssich, M.; Je, B.I.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef]
- Ahn, B.; Miyoshi, K.; Itoh, J.-I.; Nagato, Y.; Kurata, N. A Recessive Heterochronic Mutation, plastochron1, Shortens the Plastochron and Elongates the Vegetative Phase in Rice. Plant Cell 2002, 105, 654–659. [Google Scholar] [CrossRef]
- Mimura, M.; Nagato, Y.; Itoh, J. Rice PLASTOCHRON genes regulate leaf maturation downstream of the gibberellin signal transduction pathway. Planta 2012, 235, 1081–1089. [Google Scholar] [CrossRef]
- Xiong, Y.; Xie, J.; Zhang, X.; Li, Y.; Tian, W.; Ni, J.; Zhu, Z.; Wang, Y.; Wen, X.; Sang, X. PLASTOCHRON1 regulates leaf inclination through Brassinolide pathway in Oryza sativa. Crop Sci. 2021, 61, 1280–1288. [Google Scholar] [CrossRef]
- Sun, X.; Cahill, J.; Van Hautegem, T.; Feys, K.; Whipple, C.; Novak, O.; Delbare, S.; Versteele, C.; Demuynck, K.; De Block, J.; et al. Altered expression of maize PLASTOCHRON1 enhances biomass and seed yield by extending cell division duration. Nat. Commun. 2017, 8, 14752. [Google Scholar] [CrossRef]
- Dhami, N.; Cazzonelli, C.I. Short photoperiod attenuates CO2 fertilization effect on shoot biomass in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2021, 27, 825–834. [Google Scholar] [CrossRef]
- Duan, T.; Chapman, S.C.; Holland, E.; Rebetzke, G.J.; Guo, Y.; Zheng, B. Dynamic quantification of canopy structure to characterize early plant vigour in wheat genotypes. J. Exp. Bot. 2016, 67, 4523–4534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Christensen, M.; Nan, Z.; Whish, J.P.M.; Bell, L.W.; Wang, J.; Wang, Z.; Sim, R. Plant development and solar radiation interception of four annual forage plants in response to sowing date in a semi-arid environment. Ind. Crops Prod. 2019, 131, 41–53. [Google Scholar] [CrossRef]
- Banaś, K.; Piwowar, A.; Harasym, J. The potential of rapeseed (canola) oil nutritional benefits wide spreading via oleogelation. Food Biosci. 2023, 56, 103162. [Google Scholar] [CrossRef]
- Liu, G.; Yan, L.; Wang, S.; Yuan, H.; Zhu, Y.; Xie, C.; Wang, P.; Yang, R. A novel type of sprout food development: Effects of germination on phytic acid, glucosinolates, and lipid profiles in rapeseed. Food Biosci. 2023, 55, 102893. [Google Scholar] [CrossRef]
- Huang, H.; Shi, Y.; Liu, T.; Zhou, Y. Oilseed-Vegetable-Dual-Purpose Rape Key Technology Research and Its Application Prospect Analysis. Agric. Sci. 2014, 5, 1291–1295. [Google Scholar] [CrossRef][Green Version]
- McMASTER, G.S. Phytomers, phyllochrons, phenology and temperate cereal development. J. Agric. Sci. 2005, 143, 137–150. [Google Scholar] [CrossRef]
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef]
- Luo, X.; Ma, C.; Yue, Y.; Hu, K.; Li, Y.; Duan, Z.; Wu, M.; Tu, J.; Shen, J.; Yi, B.; et al. Unravelling the complex trait of harvest index in rapeseed (Brassica napus L.) with association mapping. BMC Genomics 2015, 16, 379. [Google Scholar] [CrossRef]
- Lu, K.; Xiao, Z.; Jian, H.; Peng, L.; Qu, C.; Fu, M.; He, B.; Tie, L.; Liang, Y.; Xu, X.; et al. A combination of genome-wide association and transcriptome analysis reveals candidate genes controlling harvest index-related traits in Brassica napus. Sci. Rep. 2016, 6, 36452. [Google Scholar] [CrossRef]
- Hu, J.; Chen, B.; Zhao, J.; Zhang, F.; Xie, T.; Xu, K.; Gao, G.; Yan, G.; Li, H.; Li, L.; et al. Genomic selection and genetic architecture of agronomic traits during modern rapeseed breeding. Nat. Genet. 2022, 54, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.; Song, J.; Guo, N.; Zhang, M.; Zhu, Y.; Huang, Z.; Xu, A. Genome-Wide Association Analyses Reveal Candidate Genes Controlling Harvest Index and Related Agronomic Traits in Brassica napus L. Agronomy 2022, 12, 814. Agronomy 2022, 12, 814. [Google Scholar] [CrossRef]
- Zheng, M.; Zhang, L.; Tang, M.; Liu, J.; Liu, H.; Yang, H.; Fan, S.; Terzaghi, W.; Wang, H.; Hua, W. Knockout of two BnaMAX1 homologs by CRISPR/Cas9 targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 2020, 18, 644–654. [Google Scholar] [CrossRef]
- Zhang, C.; Gong, R.; Zhong, H.; Dai, C.; Zhang, R.; Dong, J.; Li, Y.; Liu, S.; Hu, J. Integrated multi-locus genome-wide association studies and transcriptome analysis for seed yield and yield-related traits in Brassica napus. Front. Plant Sci. 2023, 14, 1153000. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Wigge, P.A. FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 2007, 17, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.D.; Hong, Y. Systemic movement of FT mRNA and a possible role in floral induction. Front. Plant Sci. 2012, 3, 127. [Google Scholar] [CrossRef]
- Mao, Y.; Sun, J.; Cao, P.; Zhang, R.; Fu, Q.; Chen, S.; Chen, F.; Jiang, J. Functional analysis of alternative splicing of the FLOWERING LOCUS T orthologous gene in Chrysanthemum morifolium. Hortic. Res. 2016, 3, 16058. [Google Scholar] [CrossRef]
- Nelissen, H.; Rymen, B.; Jikumaru, Y.; Demuynck, K.; Van Lijsebettens, M.; Kamiya, Y.; Inzé, D.; Beemster, G.T.S. A Local Maximum in Gibberellin Levels Regulates Maize Leaf Growth by Spatial Control of Cell Division. Curr. Biol. 2012, 22, 1183–1187. [Google Scholar] [CrossRef]
- Wen, C.K.; Chang, C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 2002, 14, 87–100. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 Interacts with the DELLA Protein RGL1 to Positively Regulate Age-Triggered Leaf Senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Maodzeka, A.; Zhou, L.; Ali, E.; Wang, Z.; Jiang, L. Removal of DELLA repression promotes leaf senescence in Arabidopsis. Plant Sci. 2014, 219–220, 26–34. [Google Scholar] [CrossRef]
- Muller, D.; Schmitz, G.; Theres, K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 2006, 18, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Cheng, L.; Li, Z.; Zhao, Y.; Wang, Y. Over-expression of CcMYB24, encoding a R2R3-MYB transcription factor from a high-leaf-number mutant of Cymbidium, increases the number of leaves in Arabidopsis. PeerJ 2023, 11, e15490. [Google Scholar] [CrossRef] [PubMed]
- Rutjens, B.; Bao, D.; van Eck-Stouten, E.; Brand, M.; Smeekens, S.; Proveniers, M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 2009, 58, 641–654. [Google Scholar] [CrossRef]
- Smith, H.M.; Hake, S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 2003, 15, 1717–1727. [Google Scholar] [CrossRef]
- Wang, L.; Ming, L.; Liao, K.; Xia, C.; Sun, S.; Chang, Y.; Wang, H.; Fu, D.; Xu, C.; Wang, Z.; et al. Bract suppression regulated by the miR156/529-SPLs-NL1-PLA1 module is required for the transition from vegetative to reproductive branching in rice. Mol. Plant 2021, 14, 1168–1184. [Google Scholar] [CrossRef]
- Booker, J.; Sieberer, T.; Wright, W.; Williamson, L.; Willett, B.; Stirnberg, P.; Turnbull, C.; Srinivasan, M.; Goddard, P.; Leyser, O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 2005, 8, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J.; Grisafi, P.; Fink, G.R.; Bartel, B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 1997, 9, 1781–1790. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, T.; Wang, Z.; Zhao, X.; Li, R.; Li, J. Nitrilases NIT1/2/3 Positively Regulate Flowering by Inhibiting MAF4 Expression in Arabidopsis. Front. Plant Sci. 2022, 13, 889460. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Cho, H.; Noh, J.; Qi, J.; Jung, H.-J.; Nam, H.; Lee, S.; Hwang, D.; Greb, T.; Hwang, I. BIL1-mediated MP phosphorylation integrates PXY and cytokinin signalling in secondary growth. Nat. Plants 2018, 4, 605–614. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Gampala, S.S.; Kim, T.W.; He, J.X.; Tang, W.; Deng, Z.; Bai, M.Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef]
- Leibfried, A.; To, J.P.C.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005, 438, 1172–1175. [Google Scholar] [CrossRef]
- Wang, J.-T.; Zhang, Y.-W.; Du, Y.-W.; Ren, W.-L.; Li, H.-F.; Sun, W.-X.; Ge, C.; Zhang, Y.-M. SEA v2.0: An R software package for mixed major genes plus polygenes inheritance analysis of quantitative traits. Acta Agron. Sin. 2022, 48, 1416–1424. [Google Scholar] [CrossRef]
- Doyle, J.J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Song, J.M.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R.; et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Rousseau-Gueutin, M.; Belser, C.; Da Silva, C.; Richard, G.; Istace, B.; Cruaud, C.; Falentin, C.; Boideau, F.; Boutte, J.; Delourme, R.; et al. Long-read assembly of the Brassica napus reference genome Darmor-bzh. Gigascience 2020, 9, giaa137. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2014, 6, 80–92. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, S.; Wei, L.; Huang, Y.; Liu, D.; Jia, Y.; Luo, C.; Lin, Y.; Liang, C.; Hu, Y.; et al. BnIR: A multi-omics database with various tools for Brassica napus research and breeding. Mol. Plant 2023, 16, 775–789. [Google Scholar] [CrossRef]
- Li, F.; Chen, B.; Xu, K.; Gao, G.; Yan, G.; Qiao, J.; Li, J.; Li, H.; Li, L.; Xiao, X.; et al. A genome-wide association study of plant height and primary branch number in rapeseed (Brassica napus). Plant Sci. 2016, 242, 169–177. [Google Scholar] [CrossRef]
- Zhang, G.; Peng, Y.; Zhou, J.; Tan, Z.; Jin, C.; Fang, S.; Zhong, S.; Jin, C.; Wang, R.; Wen, X.; et al. Genome-Wide Association Studies of Salt-Alkali Tolerance at Seedling and Mature Stages in Brassica napus. Front. Plant Sci. 2022, 13, 857149. [Google Scholar] [CrossRef]
| Trait | Model of (2021–2022) | AIC Value | Log Max Likelihood Value | Model (2022–2023) | AIC Value | Log Max Likelihood Value |
|---|---|---|---|---|---|---|
| Leaf number in the rosette stage | 1MG-EAD | 2516.879 | −1253.44 | 2MG-EAD | 1210.203 | −601.102 |
| 1MG-AD | 2518.637 | −1253.319 | 1MG-AD | 1212.56 | −600.28 | |
| 2MG-EAD | 2525.118 | −1258.559 | MX2-CD-AD | 1217.865 | −605.933 | |
| 2MG-EA | 2534.13 | −1263.065 | 2MG-A | 1218.415 | −604.208 | |
| MX2-ADI-ADI | 2546.923 | −1261.461 | 1MG-EAD | 1218.642 | −604.321 | |
| Leaf number in the bud stage | 2MG-EAD | 2936.697 | −1464.349 | MX2-CD-AD | 1352.254 | −673.127 |
| 2MG-EA | 2942.665 | −1467.333 | MX2-EAD-AD | 1352.505 | −674.253 | |
| 1MG-AD | 2946.55 | −1467.275 | 2MG-EAD | 1353.224 | −672.612 | |
| MX2-ADI-AD | 2969.929 | −1475.965 | MX2-AD-AD | 1354.104 | −672.052 | |
| MX2-ADI-ADI | 2975.617 | −1475.808 | 1MG-AD | 1357.429 | −672.715 | |
| Primary branch number | 2MG-EA | 1959.893 | −975.9467 | 2MG-EAD | 1089.852 | −540.926 |
| 2MG-EAD | 1962.112 | −977.0562 | 1MG-EAD | 1091.632 | −540.816 | |
| 1MG-EAD | 1964.117 | −977.0583 | 1MG-AD | 1093.476 | −540.738 | |
| 1MG-NCD | 1964.168 | −977.0842 | 1MG-NCD | 1098.71 | −544.355 | |
| 1MG-AD | 1966.149 | −977.0747 | 2MG-A | 1099.805 | −544.902 |
| QTL Label | Chr | Start | End | Interval Length (Mb) | No. of Gene | Reference Genome |
|---|---|---|---|---|---|---|
| qPLA.A02-1 | A02 | 9,045,489 | 9,478,410 | 0.43 | 53 | ZS11 v0 |
| qPLA.A02-2 | A02 | 13,517,525 | 13,661,121 | 0.14 | 19 | ZS11 v0 |
| qPLA.A04 | A04 | 19,839,469 | 20,142,478 | 0.30 | 35 | ZS11 v0 |
| qPLA.A02-1 | A02 | 9,066,916 | 9,477,102 | 0.41 | 46 | Darmor-bzh V10 |
| qPLA.A02-2 | A02 | 13,609,227 | 13,728,641 | 0.12 | 14 | Darmor-bzh V10 |
| qPLA.A04 | A04 | 17,128,404 | 17,432,209 | 0.30 | 53 | Darmor-bzh V10 |
| Gene ID | Genomic Position | Best Hit to A. thaliana | Function Description |
|---|---|---|---|
| BnaA02G0156900ZS | A02:9104462−9107270 | AT1G65480 | Protein FLOWERING LOCUS T |
| BnaA02G0160500ZS | A02:9357771−9367517 | AT1G66350 | DELLA protein RGL1 |
| BnaA02G0160600ZS | A02:9375716−9377422 | AT1G56650 | MYB-like, a putative MYB domain-containing transcription factor |
| BnaA02G0215300ZS | A02:13571765−13573401 | AT1G75410 | BLH3, BEL1-like homeodomain protein 3 |
| BnaA02G0215600ZS | A02:13592588−13594117 | AT1G47620 | CYP96A8, a member of CYP96A |
| BnaA04G0200300ZS | A04:19944280−19946261 | AT3G44300 | NIT2, nitrilase 2 |
| BnaA04G0200700ZS | A04:19964345−19967629 | AT2G30980 | ASK6, a shaggy-related protein kinase |
| BnaA04G0201700ZS | A04:20020034−20020282 | AT2G31081 | CLAVATA3/ESR (CLE)-related protein 4 |
| BnaA04G0201900ZS | A04:20029992−20030249 | AT2G31082 | CLAVATA3/ESR (CLE)-related protein 7 |
| BnaA04G0202000ZS | A04:20059635−20059880 | AT2G31085 | CLAVATA3/ESR (CLE)-related protein 6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, M.; Liu, X.; Song, J.; Zhao, F.; Shi, Y.; Xu, Y.; Guo, Z.; Zhang, T.; Wu, J.; Wang, J.; et al. Genetic Analyses, BSA-Seq, and Transcriptome Analyses Reveal Candidate Genes Controlling Leaf Plastochron in Rapeseed (Brassica napus L.). Plants 2025, 14, 1719. https://doi.org/10.3390/plants14111719
Qin M, Liu X, Song J, Zhao F, Shi Y, Xu Y, Guo Z, Zhang T, Wu J, Wang J, et al. Genetic Analyses, BSA-Seq, and Transcriptome Analyses Reveal Candidate Genes Controlling Leaf Plastochron in Rapeseed (Brassica napus L.). Plants. 2025; 14(11):1719. https://doi.org/10.3390/plants14111719
Chicago/Turabian StyleQin, Mengfan, Xiang Liu, Jia Song, Feixue Zhao, Yiji Shi, Yu Xu, Zhiting Guo, Tianye Zhang, Jiapeng Wu, Jinxiong Wang, and et al. 2025. "Genetic Analyses, BSA-Seq, and Transcriptome Analyses Reveal Candidate Genes Controlling Leaf Plastochron in Rapeseed (Brassica napus L.)" Plants 14, no. 11: 1719. https://doi.org/10.3390/plants14111719
APA StyleQin, M., Liu, X., Song, J., Zhao, F., Shi, Y., Xu, Y., Guo, Z., Zhang, T., Wu, J., Wang, J., Li, W., Li, K., Li, S., Huang, Z., & Xu, A. (2025). Genetic Analyses, BSA-Seq, and Transcriptome Analyses Reveal Candidate Genes Controlling Leaf Plastochron in Rapeseed (Brassica napus L.). Plants, 14(11), 1719. https://doi.org/10.3390/plants14111719






