“Secreted in Xylem” Genes (SIX Genes): Relationship to the Aggressiveness of Fusarium oxysporum f. sp. albedinis
Abstract
1. Introduction
2. Material and Methods
2.1. Foa Isolation
2.2. DNA Extraction from Foa
2.3. Molecular Detection of Foa
2.4. Aggressiveness Tests
2.5. Amplification of SIX Genes
3. Results
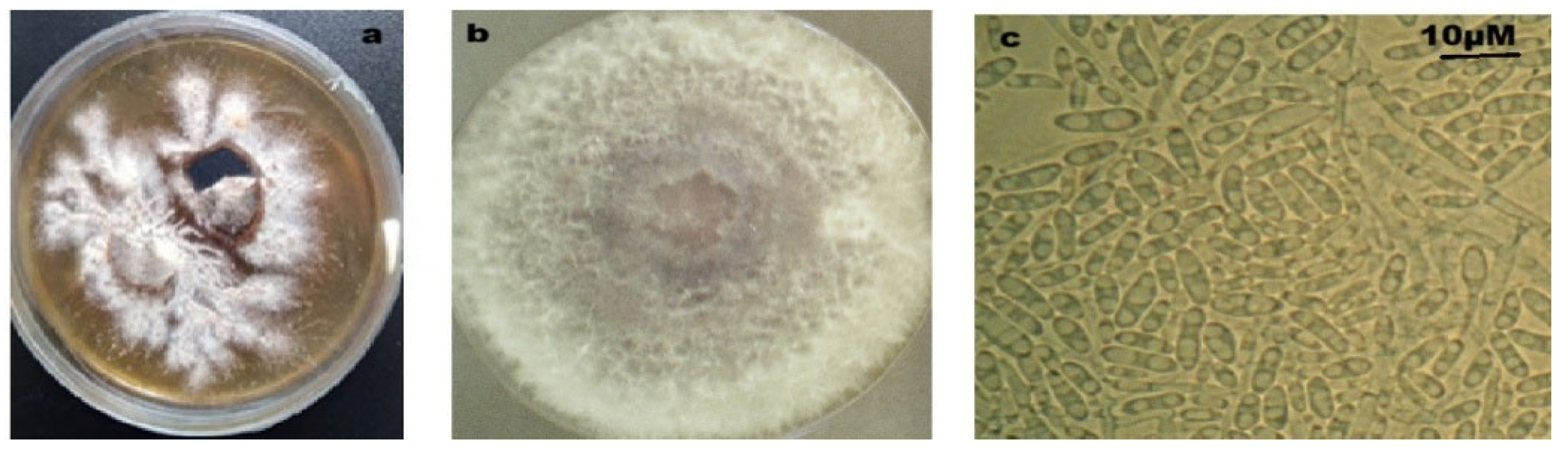
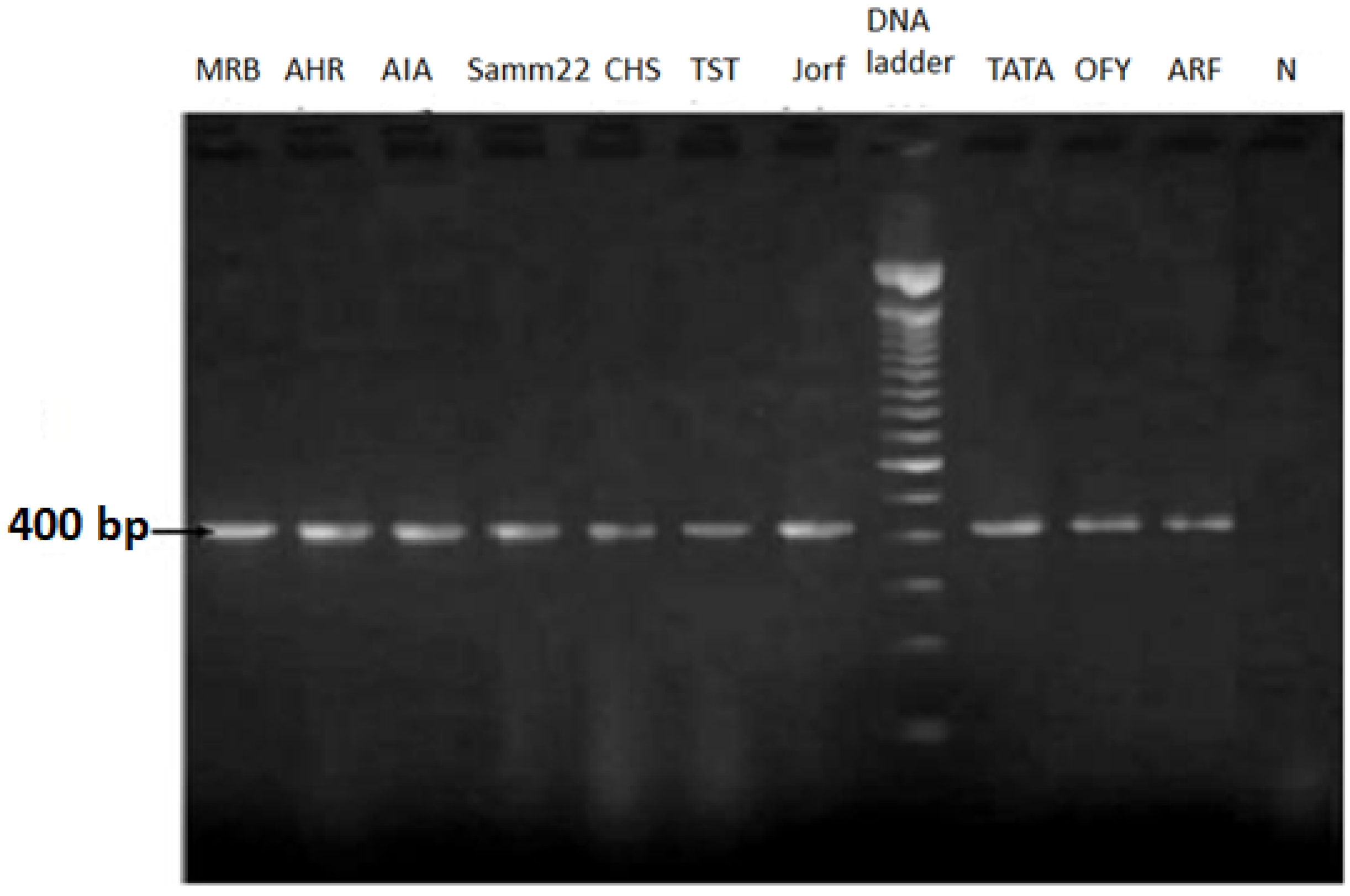
3.1. Foa Isolation and Molecular Detection
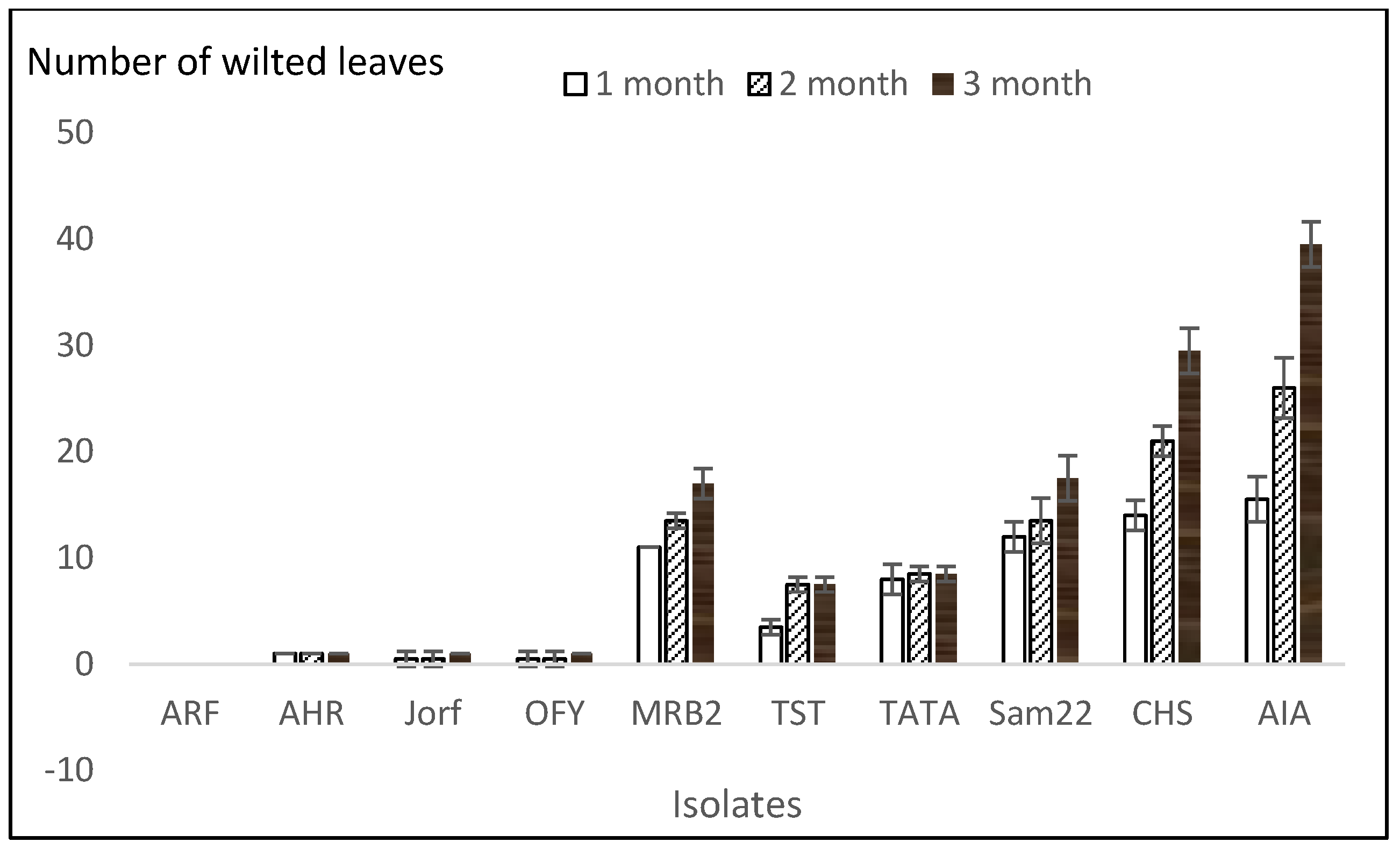
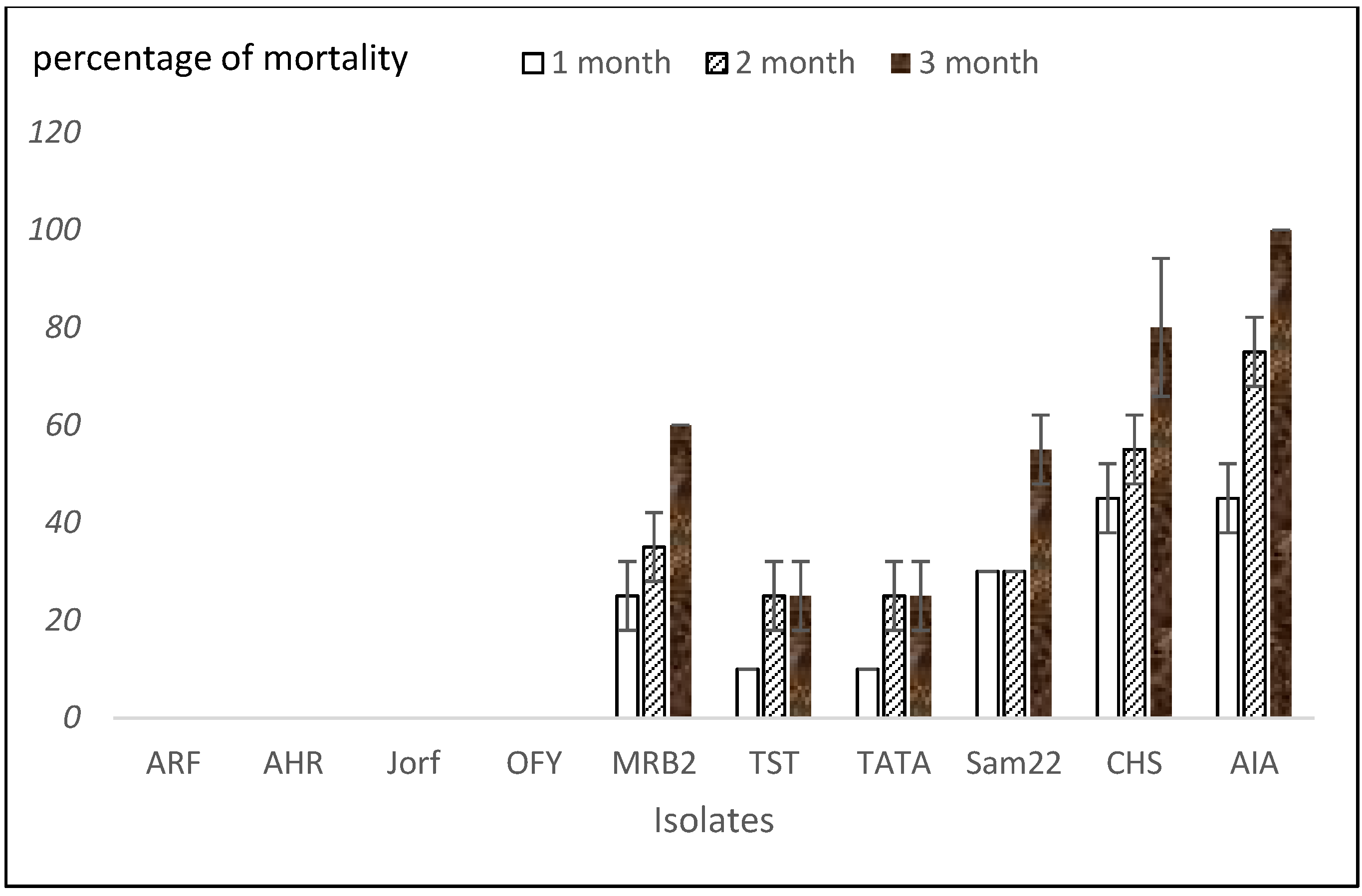
3.2. Aggressiveness Tests
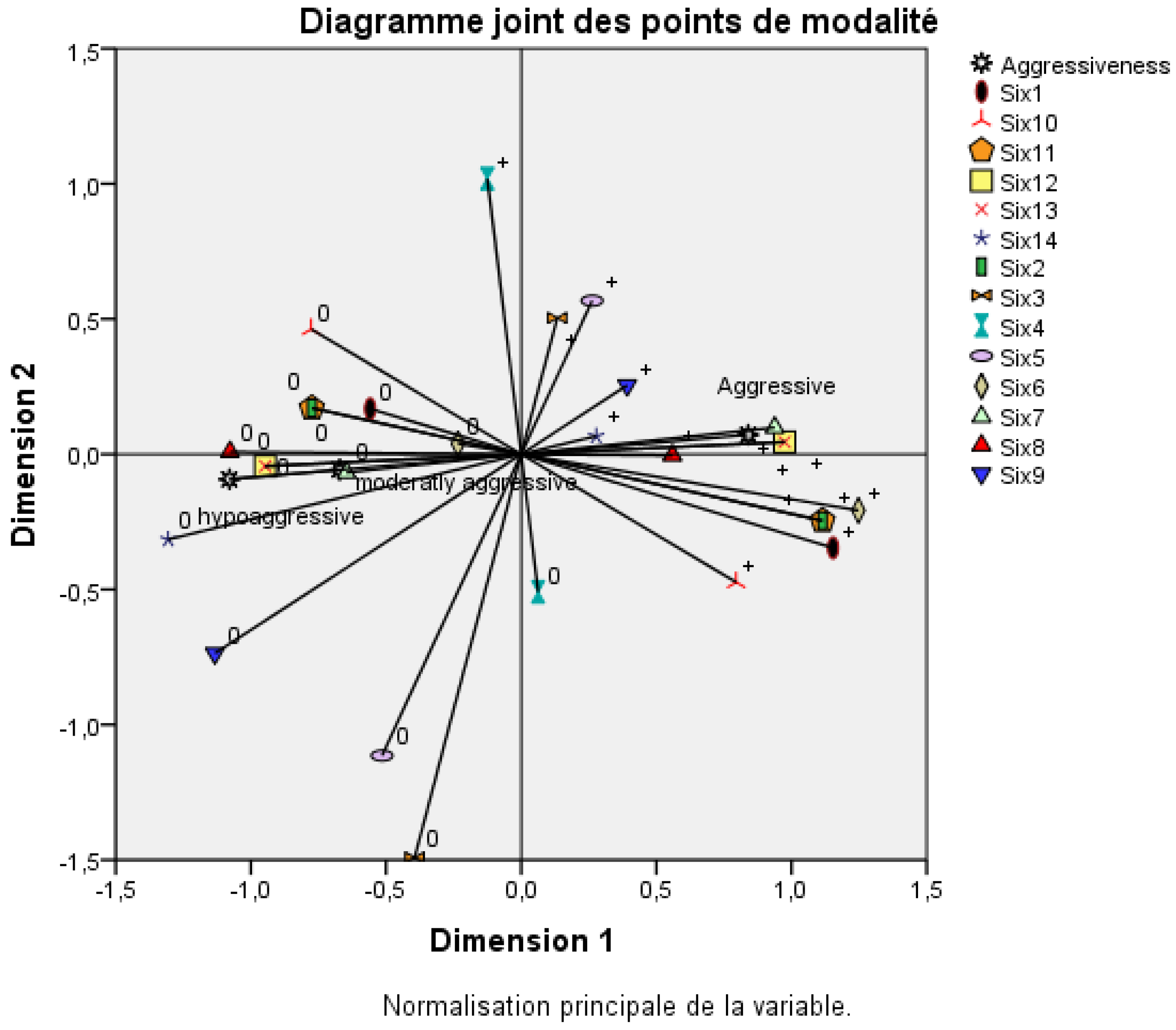
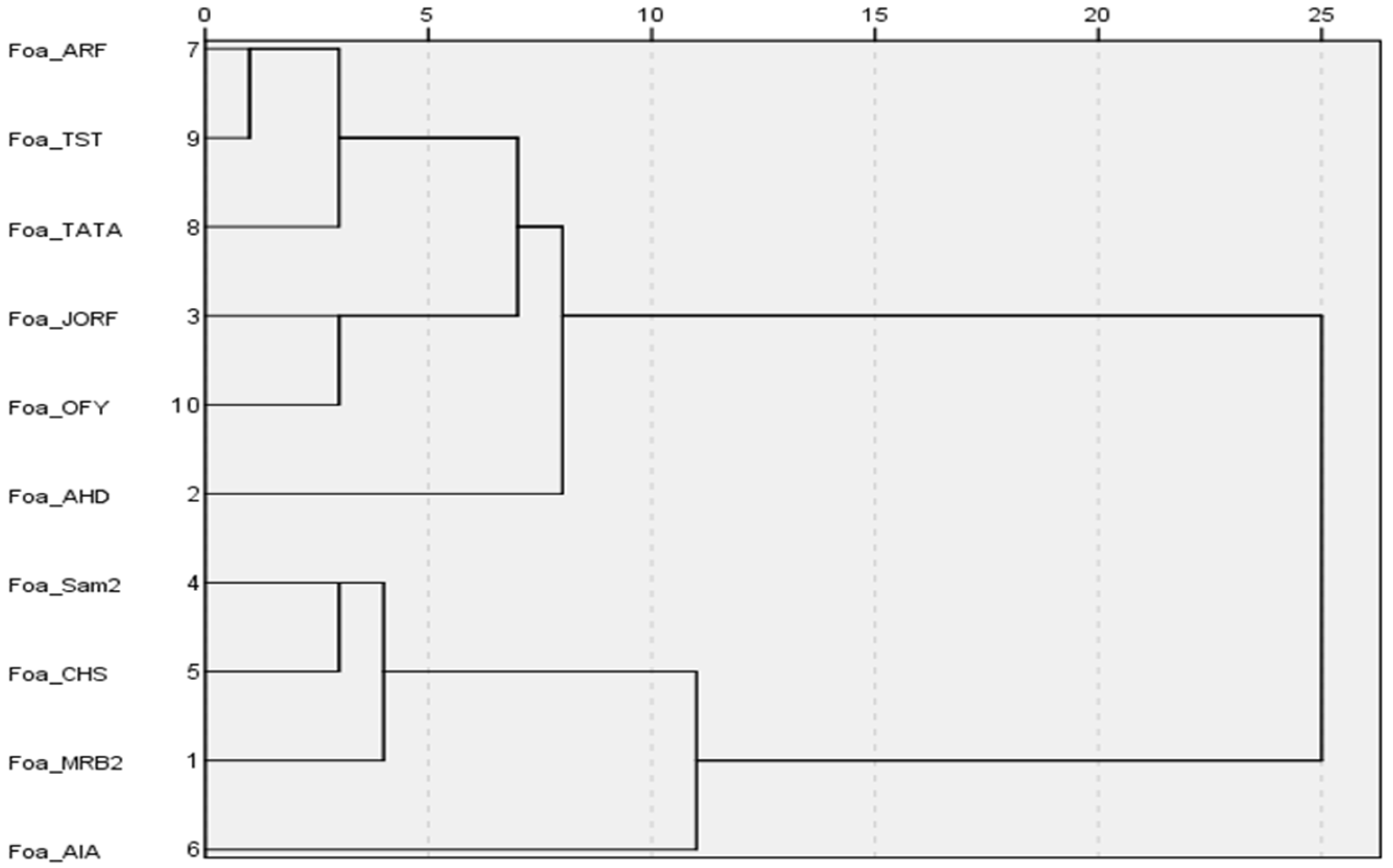
3.3. Relationship Between Aggressiveness and SIX Genes in Foa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sedra, M.H. Date Palm Status and Perspective in Morocco. In Date Palm Genetic Resources and Utilization; AlKhayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 257–323. [Google Scholar] [CrossRef]
- Saaidi, M. Comportement au champ de 32 cultivars de palmier dattier vis-à vis du bayoud: 25 années d’observations. Agronomie 1992, 12, 359–370. [Google Scholar] [CrossRef]
- El Modafar, C. Mechanisms of date palm resistance to Bayoud disease: Current state of knowledge and research prospects. Physiol. Mol. Plant Pathol. 2010, 74, 287–294. [Google Scholar] [CrossRef]
- Djerbi, M. Bayoud disease in North Africa: History, distribution, diagnostics and control. Date Palm J. 1982, 2, 153–198. [Google Scholar]
- Dihazi, A.; Serghini, M.A.; Jaiti, F.; Daayf, F.; Driouich, A.; Dihazi, H.; El Hadrami, I. Structural and biochemical changes in salicylic acid-treated date palm roots challenged with Fusarium oxysporum f. sp. albedinis. J. Pathog. 2011, 9, 280481. [Google Scholar] [CrossRef]
- Dihazi, A.; Naamani, K.; Nabgui, A.; El Meziane, A.; Elmodafar, C.; Dihazi, H. Proteome analysis of an aggressive and a hypoaggressive isolates of Fusarium oxysporum f. sp. albedinis showing several differently-expressed proteins related to the aggressiveness. Physiol. Mol. Plant Pathol. 2021, 116, 101738. [Google Scholar] [CrossRef]
- El Hassni, M.; J’Aiti, F.; Dihazi, A.; Ait Barka, S.; Daayf, F.; El Hadrami, I. Enhancement of defense responses against bayoud disease by treatment of date palm seedling with a hypoaggressive Fusarium oxysporum isolate. J. Phytopathol. 2004, 152, 182–189. [Google Scholar] [CrossRef]
- Ma, L.J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef]
- Jiming, L.; Fokkens, L.; Conneely, L.J.; Rep, M. Partial pathogenicity chromosomes in Fusarium oxysporum are sufficient to cause disease and can be horizontally transferred. Environ. Microbiol. 2020, 22, 4985–5004. [Google Scholar] [CrossRef]
- van Dam, P.; Fokkens, L.; Ayukawa, Y.; van der Gragt, M.; Ter Horst, A.; Brankovics, B. A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci. Rep. 2017, 7, 9042. [Google Scholar] [CrossRef]
- Schmidt, S.M.; Houterman, P.M.; Schreiver, I.; Ma, L.; Amyotte, S.; Chellappan, B.; Boeren, S.; Takken, F.L.; Rep, M. Mites in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genom. 2013, 4, 119. [Google Scholar] [CrossRef]
- Williams, A.H.; Sharma, M.; Thatcher, L.F.; Azam, S.; Hane, J.K.; Sperschneider, J.; Kidd, B.N.; Anderson, J.P.; Ghosh, R.; Garg, G.; et al. Comparative genomics and prediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genom. 2016, 17, 191. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Van Der Does, H.C.; Meijer, M.; Van Wijk, R.; Houterman, P.M.; Dekker, H.L.; de Koster, C.G.; Cornelissen, B.J.C. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 2004, 53, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Meijer, M.; Houterman, P.M.; van der Does, H.C.; Cornelissen, B.J.C. Fusarium oxysporum evades I-3-mediated resistance without altering the matching Avirulence gene. Mol. Plant-Microbe Interact. 2005, 18, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Arie, T. Fusarium diseases of cultivated plants, control, diagnosis, and molecular and genetic studies. J. Pestic. Sci. 2019, 44, 275–281. [Google Scholar] [CrossRef]
- Houterman, P.M.; Ma, L.; van Ooijen, G.; de Vroomen, M.J.; Cornelissen, B.J.; Takken, F.L.; Rep, M. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 2009, 58, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Gawehns, F.; Houterman, P.M.; Ait Ichou, F.; Michielse, C.B.; Hijdra, M.; Cornelissen, B.J.C.; Rep, M.; Takken, F.L.W. The Fusarium oxysporum Effector Six6 Contributes to Virulence and Suppresses I-2-Mediated Cell Death. Mol. Plant-Microbe Interact. 2014, 27, 336–348. [Google Scholar] [CrossRef]
- Ma, L.; Houterman, P.M.; Gawehns, F.; Cao, L.; Sillo, F.; Richter, H.; Clavijo-Ortiz, M.J.; Schmidt, S.M.; Boeren, S.; Vervoort, J.; et al. The AVR2-SIX5 gene pair is required to activate I-2-mediated immunity in tomato. New Phytol. 2015, 208, 507–518. [Google Scholar] [CrossRef]
- Lievens, B.; Houterman, P.M.; Rep, M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 2009, 300, 201–215. [Google Scholar] [CrossRef]
- Laurence, M.H.; Summerell, B.A.; Liew, E.C.Y. Fusarium oxysporum f. sp. canariensis: Evidence for horizontal gene transfer of putative pathogenicity genes. Plant Pathol. 2015, 64, 1068–1075. [Google Scholar] [CrossRef]
- Taylor, A.; Vagany, V.; Jackson, A.C.; Harrison, R.J.; Rainoni, A.; Clarkson, J.P. Identification of pathogenicity-related genes in Fusarium oxysporum f. sp. cepae. Mol. Plant Pathol. 2016, 17, 1032–1047. [Google Scholar] [CrossRef]
- van Dam, P.; Fokkens, L.; Schmidt, S.M.; Linmans, J.H.; Kistler, H.C.; Ma, L.J.; Rep, M. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 2016, 18, 4087–4102. [Google Scholar] [CrossRef] [PubMed]
- Czislowski, E.; Fraser-Smith, S.; Zander, M.; O’Neill, W.T.; Meldrum, R.A.; Tran-Nguyen, L.T.T.; Batley, J.; Aitken, E.A.B. Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol. Plant Pathol. 2018, 19, 1155–1171. [Google Scholar] [CrossRef] [PubMed]
- Simbaqueba, J.; Catanzariti, M.A.; Gonzalez, C.; Jones, D.A. Evidence for horizontal gene transfer and separation of effector recognition from effector function revealed by analysis of effector genes shared between cape goose berry- and tomato-infecting formae speciales of Fusarium oxysporum. Mol. Plant Pathol. 2018, 19, 2302–2318. [Google Scholar] [CrossRef]
- Ponukumati, S.V.; Elliott, M.L.; Desjardin, E.A. Comparison of Secreted in Xylem (SIX) genes in two Fusarium wilt pathogens of ornamental palms. Plant Pathol. 2019, 68, 1663–1681. [Google Scholar] [CrossRef]
- Jenkins, S.; Taylor, A.; Jackson, A.C.; Armitage, A.D.; Bates, H.J.; Mead, A.; Harrison, R.J.; Clarkson, J.P. Identification and expression of secreted in xylem pathogenicity genes in Fusarium oxysporum f. sp. pisi. Front. Microbiol. 2021, 12, 593140. [Google Scholar] [CrossRef] [PubMed]
- Widinugraheni, S.; Niño-Sánchez, J.; van der Does, H.C.; van Dam, P.; Garcia-Bastidas, F.A.; Subandiyah, S.; Meijer, H.J.G.; Kistler, H.C.; Kema, G.H.J.; Rep, M. A SIX1 homolog in Fusarium oxysporum f. sp. cubense tropical race 4 contributes to virulence towards Cavendish banana. PLoS ONE 2018, 13, e0205896. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Gardiner, D.M.; Kazan, K.; Manners, J.M. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol. Plant-Microbe Interact. 2012, 25, 180–190. [Google Scholar] [CrossRef]
- Rafiqi, M.; Jelonek, L.; Diouf, A.M.; Mbaye, A.; Rep, M.; Diarra, A. Profile of the in silico secretome of the palm dieback pathogen, Fusarium oxysporum f. sp. albedinis, a fungus that puts natural oases at risk. PLoS ONE 2022, 17, e0260830. [Google Scholar] [CrossRef]
- Gontia-Mishra, I.; Tripathi, N.; Tiwari, S. A simple and rapid DNA extraction protocol for filamentous fungi efficient for molecular studies. Ind. J. Biotechnol. 2014, 13, 536–539. [Google Scholar]
- Fernandez, D.; Ouinten, M.; Tantaoui, A.; Geiger, J.-P.; Daboussi, M.-J.; Langin, T. Fot1 insertions in the Fusarium oxysporum f. sp. albedinis genome provide diagnostic PCR targets for detection of the date palm pathogen. Appl. Environ. Microbiol. 1998, 64, 633–636. [Google Scholar] [CrossRef]
- Freeman, S.; Maymon, M. Reliable detection of the fungal pathogen Fusarium oxysporum f. sp. albedinis, causal agent of bayoud disease of date palm, using molecular techniques. Phytoparasitica 2000, 28, 341–348. [Google Scholar] [CrossRef]
- Meldrum, R.A.; Fraser-Smith, S.; Tran-Nguyen, L.T.T.; Daly, A.M.; Aitken, E.A.B. Presence of putative pathogenicity genes in isolates of Fusarium oxysporum f. sp. cubense from Australia. Austral. Plant Pathol. 2012, 41, 551–557. [Google Scholar]
- Van Der Does, H.C.; Lievens, B.; Claes, L.; Houterman, P.M.; Cornelissen, B.J.; Rep, M. The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ. Microbiol. 2008, 10, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Ploetz, R.C. Management of Fusarium wilt of banana: A review with special reference to tropical race 4. Crop Prot. 2015, 73, 7–15. [Google Scholar] [CrossRef]
- Maldonado-Bonilla, L.D.; Villarruel, O.J.L.; Calderón, O.M.A.; Sánchez, E.A.C. Secreted in Xylem (SIX) Genes in Fusarium oxysporum f. sp. cubense and Their Potential Acquisition by Horizontal Transfer. Adv. Biotechnol. Microbiol. 2018, 10, 555779. [Google Scholar] [CrossRef]
- Kotera, S.; Hishiike, M.; Saito, H.; Komatsu, K.; Arie, T. Differentiation of the Pea Wilt Pathogen Fusarium oxysporum f. sp. pisi from Other Isolates of Fusarium Species by PCR. Microbes Environ. 2022, 37, ME21061. [Google Scholar] [CrossRef]
- Sasaki, K.; Nakahara, K.; Tanaka, S.; Shigyo, M.; Ito, S.-I. Genetic and pathogenic variability of Fusarium oxysporum f. sp cepae isolated from onion and Welsh onion in Japan. Phytopathology 2015, 105, 525–532. [Google Scholar] [CrossRef]
- Khayi, S.; Khoulassa, S.; Gaboun, F.; Abdelwahd, R.; Diria, G.; Labhilili, M.; Iraqi, D.; El Guilli, M.; Fokar, M.; Mentag, R. Draft genome sequence of Fusarium oxysporum f. sp. albedinis strain Foa 133, the causal agent of Bayoud disease on date palm. Microbiol. Resour. Announc. 2020, 9, 10–1128. [Google Scholar] [CrossRef]
- Khayi, S.; Armitage, A.D.; Gaboun, F.; Meftah-kadmiri, I.; Lahlali, R.; Fokar, M.; Mentag, R. Chromosome-scale assembly uncovers genomic compartmentation of Fusarium oxysporum f. sp. albedinis, the causal agent of Bayoud disease in date palm. Front. Microbiol. 2023, 14, 1268051. [Google Scholar] [CrossRef]
- Khoulassa, S.; Elmoualij, B.; Benlyas, M.; Meziani, R.; Bouhlali, E.D.T.; Houria, B.; El Hilali Alaoui, Y.; Haridas, S.; Guo, J.; Lipzen, A.; et al. High-Quality Draft Nuclear and Mitochondrial Genome Sequence of Fusarium oxysporum f. sp. albedinis strain 9, the Causal Agent of Bayoud Disease on Date Palm. Plant Dis. 2022, 106, 1974–1976. [Google Scholar] [CrossRef]

| Primer | Sequence (5′-3′) | Target | Expected Fragment Size (bp) | Annealing Temperature | Sources |
|---|---|---|---|---|---|
| P12-F1 | GTATCCCTCCGGATTTTGAGC | SIX1 | 992 | 60 °C | [34] |
| P12-R1 | AATAGAGCCTGCAAAGCATG | [13] | |||
| SIX2-F2 | CAACGCCGTTTGAATAAGCA | SIX2 | 749 | 55.6 °C | [34] |
| SIX2-R2 | TCTATCCGCTTTCTTCTCTC | ||||
| SIX3-F1 | CCAGCCAGAAGGCCAGTTT | SIX3 | 608 | 55.5 °C | [34] |
| SIX3-R2 | GGCAATTAACCACTCTGCC | ||||
| SIX4-F1 | TCAGGCTTCACTTAGCATAC | SIX4 | 967 | 55.5 °C | [19] |
| SIX4-R2 | GCCGACCGAAAAACCCTAA | ||||
| SIX5-F1 | ACACGCTCTACTACTCTTCA | SIX5 | 667 | 59 °C | [19] |
| SIX5-R2 | GAAAACCTCAACGCGGCAAA | ||||
| SIX6-F1 | CTCTCCTGAACCATCAACTT | SIX6 | 793 | 56 °C | [19] |
| SIX6-R2 | CAAGACCAGGTGTAGGCATT | ||||
| SIX7-F1 | CATCTTTTCGCCGACTTGGT | SIX7 | 862 | 58 °C | [19] |
| SIX7-R2 | CTTAGCACCCTTGAGTAACT | ||||
| SIX8-F1 | TCGCCTGCATAACAGGTGCCG | SIX8 | 250 | 59 °C | [33] |
| SIX8-R2 | TTGTGTAGAAACTGGACAGTCGATGC | ||||
| SIX9-F1 | GGCCCAGCCCTAGTCTAACTCC | SIX9 | 458 | 69 °C | [21] |
| SIX9-R2 | AACTTAACATGCTGGCCGTCAATCG | ||||
| SIX10-F1 | GTTAGCAACTGCGAGACACTAGAA | SIX10 | 636 | 65 °C | [21] |
| SIX10-R2 | AGCAACTTCCTTCCTCTTACTAGC | ||||
| SIX11-F1 | ATTCCGGCTTCGGGTCTCGTTTAC | SIX11 | 559 | 61 °C | [21] |
| SIX11-R2 | GAGAGCCTTTTTGGTTGATTGTAT | ||||
| SIX12-F1 | CTAACGAAGTGAAAAGAAGTCCTC | SIX12 | 449 | 61 °C | [21] |
| SIX12-R2 | GCCTCGCTGGCAAGTATTTGTT | ||||
| SIX13-F1 | CCTTCATCRTCGACAGTACAACG | SIX13 | 1027 | 61 °C | [21] |
| SIX13-R2 | ATCAAACCCGTAACTCAGCTCC | ||||
| SIX14-F1 | ATAAAGTGCGACTGGACTTCTGCC | SIX14 | 449 | 67 °C | [21] |
| SIX14-R2 | ACCCCCATCCACATTCCTAAGCGA |
| Region | Locality | Isolate |
|---|---|---|
| Ouarzazate | Ahardane | AHR |
| Errachidia | Oufous Youssef | OFY |
| Jorf | JRF | |
| Sam2 | Sam2 | |
| Cheikh Sifa | CHS | |
| Arfoud | AOD | |
| Taznakht-Tata Axis | Ait Aissa | AIA |
| Mrabet2 | MRB2 | |
| Tissn’t | TST | |
| Tata | TATA |
| Species | Isolate | Fot 1 | Aggressiveness * | SIX 1 | SIX 2 | SIX 3 | SIX 4 | SIX 5 | SIX 6 | SIX 7 | SIX 8 | SIX 9 | SIX 10 | SIX 11 | SIX 12 | SIX 13 | SIX 14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Foa | MRB2 | + | ++ | + | + | − | − | − | − | + | + | + | + | + | + | + | + |
| Foa | AHD | + | − | − | − | − | − | − | − | − | + | − | + | − | − | − | + |
| Foa | Jorf2 | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Foa | SAM22 | + | ++ | + | + | + | − | + | + | − | + | + | + | + | + | + | + |
| Foa | CHS | + | ++ | − | + | + | − | + | − | + | + | + | + | + | + | + | + |
| Foa | AIA | + | ++ | − | − | + | + | + | − | + | + | + | − | − | + | + | + |
| Foa | ARF | + | − | − | − | + | − | + | − | − | − | + | − | − | − | − | + |
| Foa | TATA | + | + | − | − | + | + | + | − | − | + | + | − | − | − | − | + |
| Foa | TST | + | + | − | − | + | − | − | − | − | − | + | − | − | − | − | + |
| Foa | OFY | + | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − |
| Fol | Fol race1 CBS646.78 | − | NT | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Fol | Fol race2 CBS645.78 | − | NT | + | + | + | − | + | + | + | + | + | + | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dihazi, A.; Jouihri, Y.; Tadlaoui-Ouafi, A.; Alfeddy, M.N.; El Modafar, C.; Dihazi, H.; El Meziane, A.; Sayari, M.; Daayf, F. “Secreted in Xylem” Genes (SIX Genes): Relationship to the Aggressiveness of Fusarium oxysporum f. sp. albedinis. Plants 2025, 14, 1721. https://doi.org/10.3390/plants14111721
Dihazi A, Jouihri Y, Tadlaoui-Ouafi A, Alfeddy MN, El Modafar C, Dihazi H, El Meziane A, Sayari M, Daayf F. “Secreted in Xylem” Genes (SIX Genes): Relationship to the Aggressiveness of Fusarium oxysporum f. sp. albedinis. Plants. 2025; 14(11):1721. https://doi.org/10.3390/plants14111721
Chicago/Turabian StyleDihazi, Abdelhi, Youness Jouihri, Ahmed Tadlaoui-Ouafi, Mohamed Najib Alfeddy, Cherkaoui El Modafar, Hassan Dihazi, Abdellatif El Meziane, Mohammad Sayari, and Fouad Daayf. 2025. "“Secreted in Xylem” Genes (SIX Genes): Relationship to the Aggressiveness of Fusarium oxysporum f. sp. albedinis" Plants 14, no. 11: 1721. https://doi.org/10.3390/plants14111721
APA StyleDihazi, A., Jouihri, Y., Tadlaoui-Ouafi, A., Alfeddy, M. N., El Modafar, C., Dihazi, H., El Meziane, A., Sayari, M., & Daayf, F. (2025). “Secreted in Xylem” Genes (SIX Genes): Relationship to the Aggressiveness of Fusarium oxysporum f. sp. albedinis. Plants, 14(11), 1721. https://doi.org/10.3390/plants14111721









