Transcriptomic Profiling of Quinoa Reveals Distinct Defense Responses to Exogenous Methyl Jasmonate and Salicylic Acid
Abstract
1. Introduction
2. Results
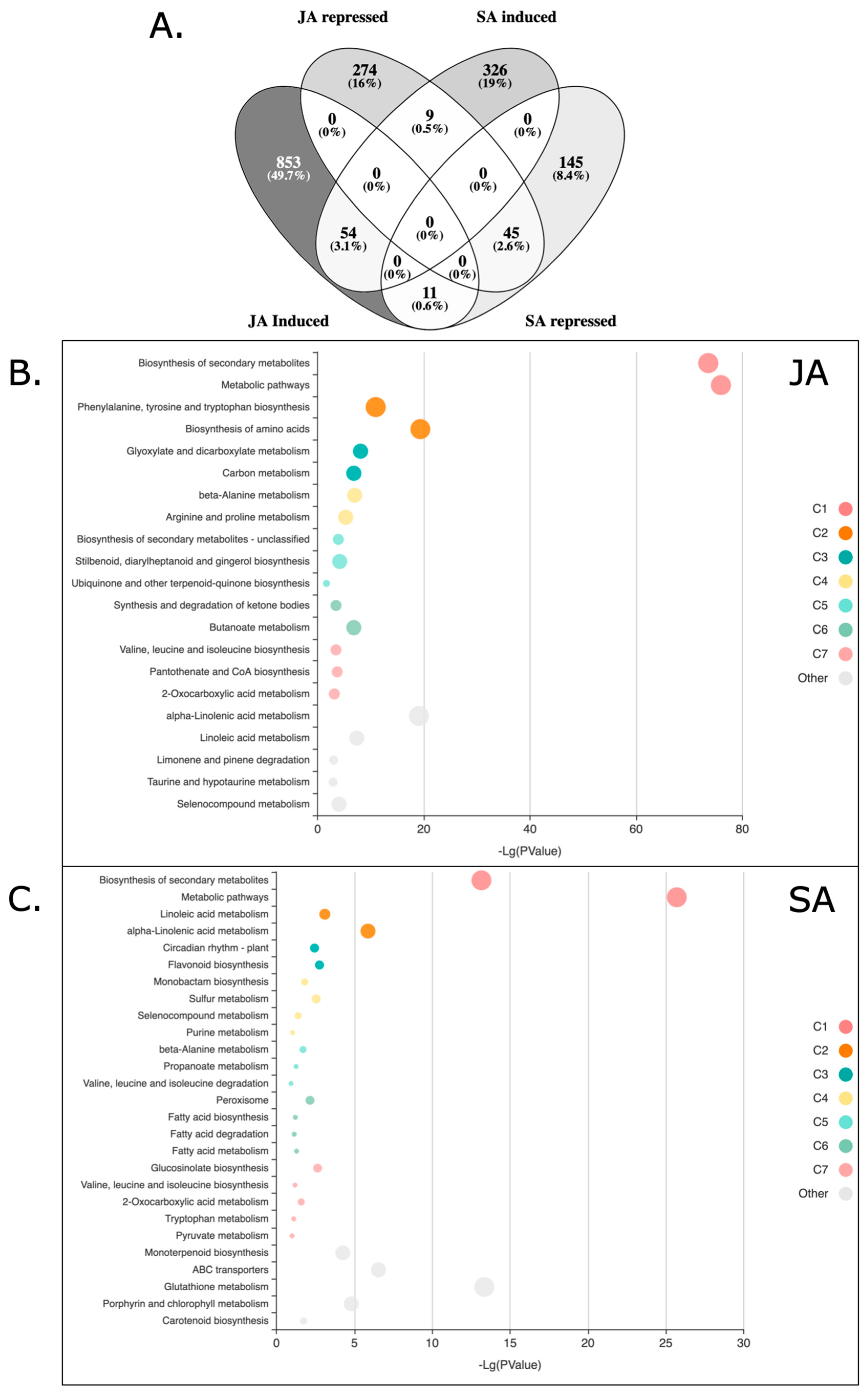
2.1. Transcriptomic Analysis of Quinoa Seedlings Treated with JA or SA
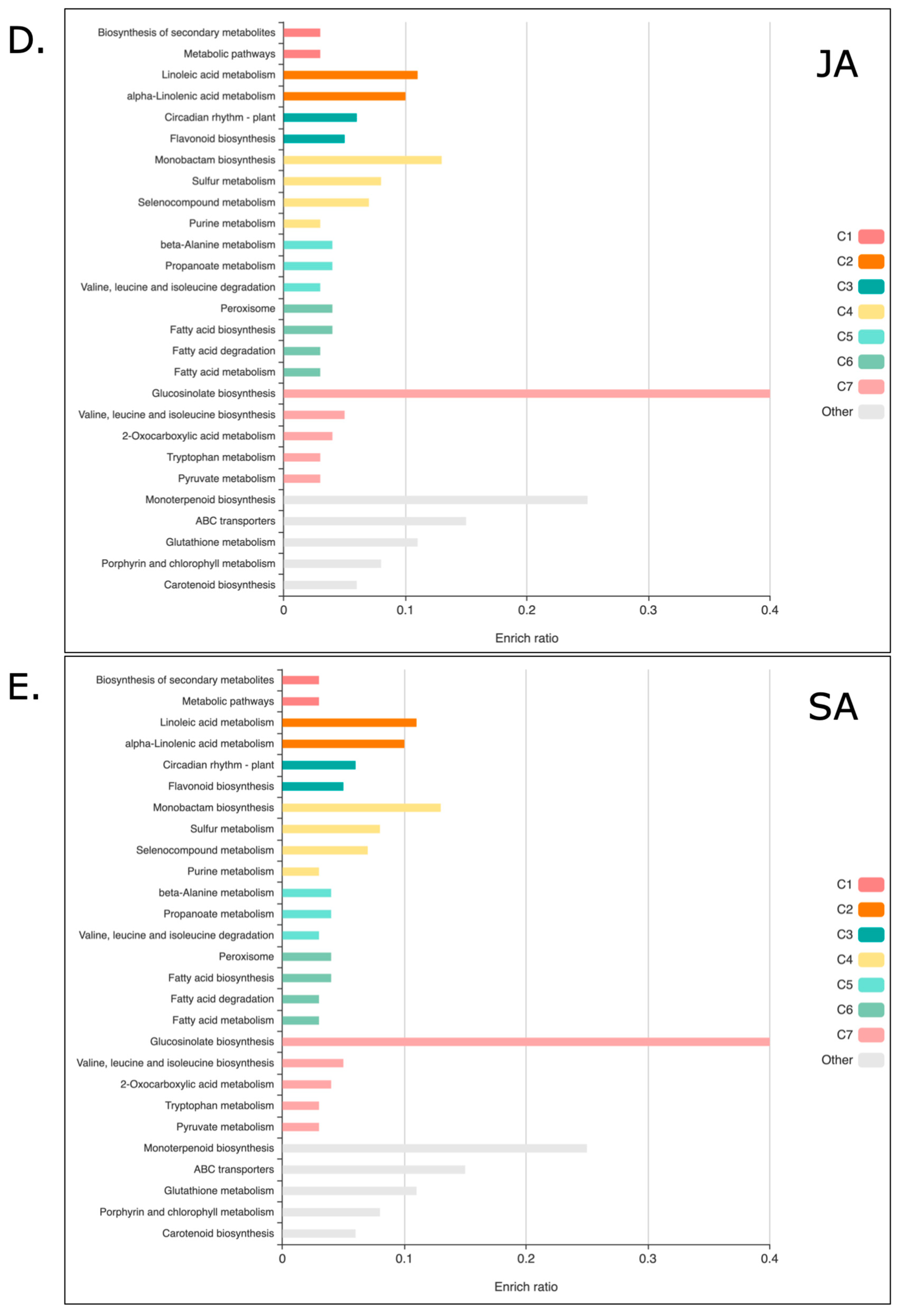
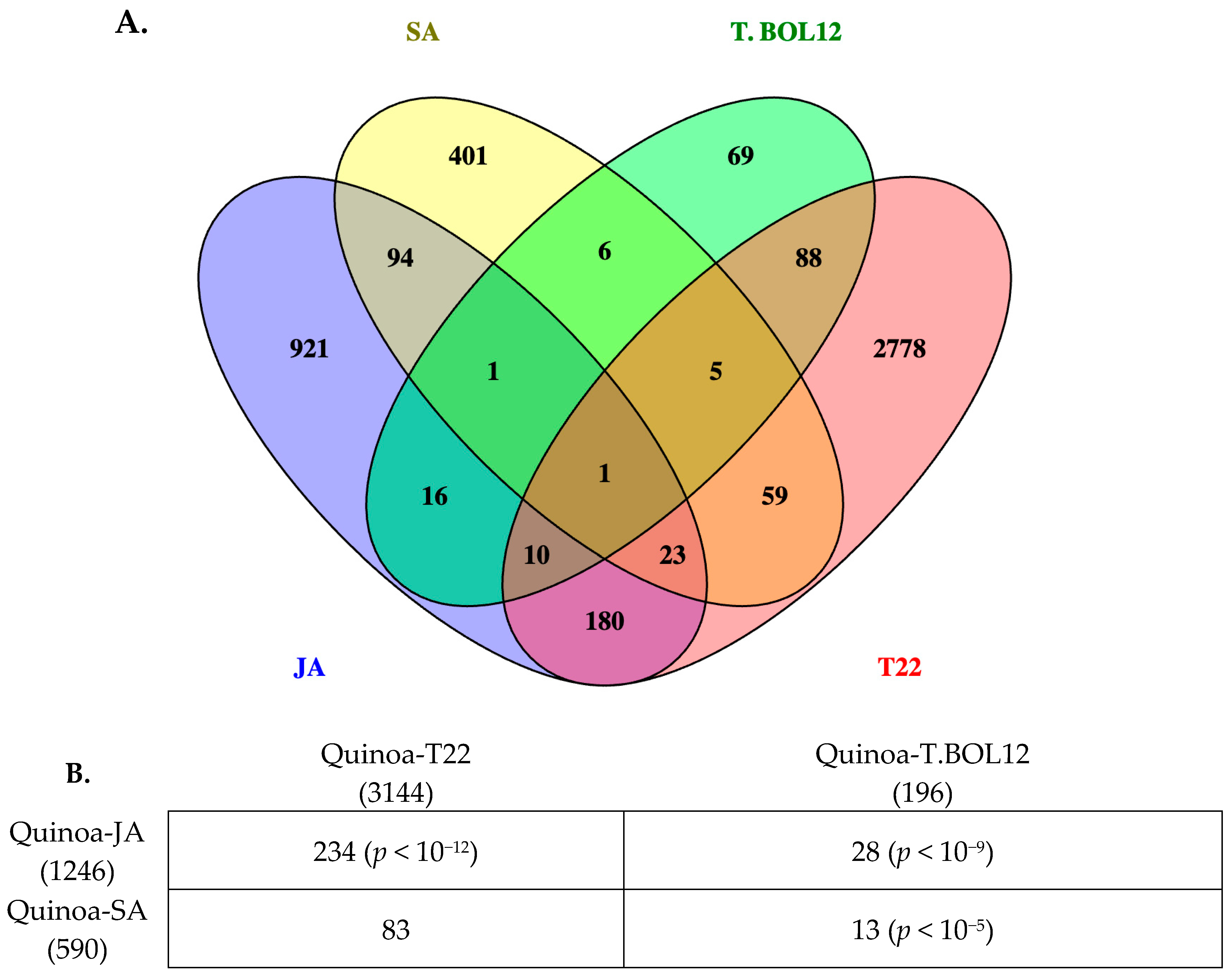
2.2. KOBAS-i Gene Set Enrichment Analysis of Quinoa Treated with JA and SA
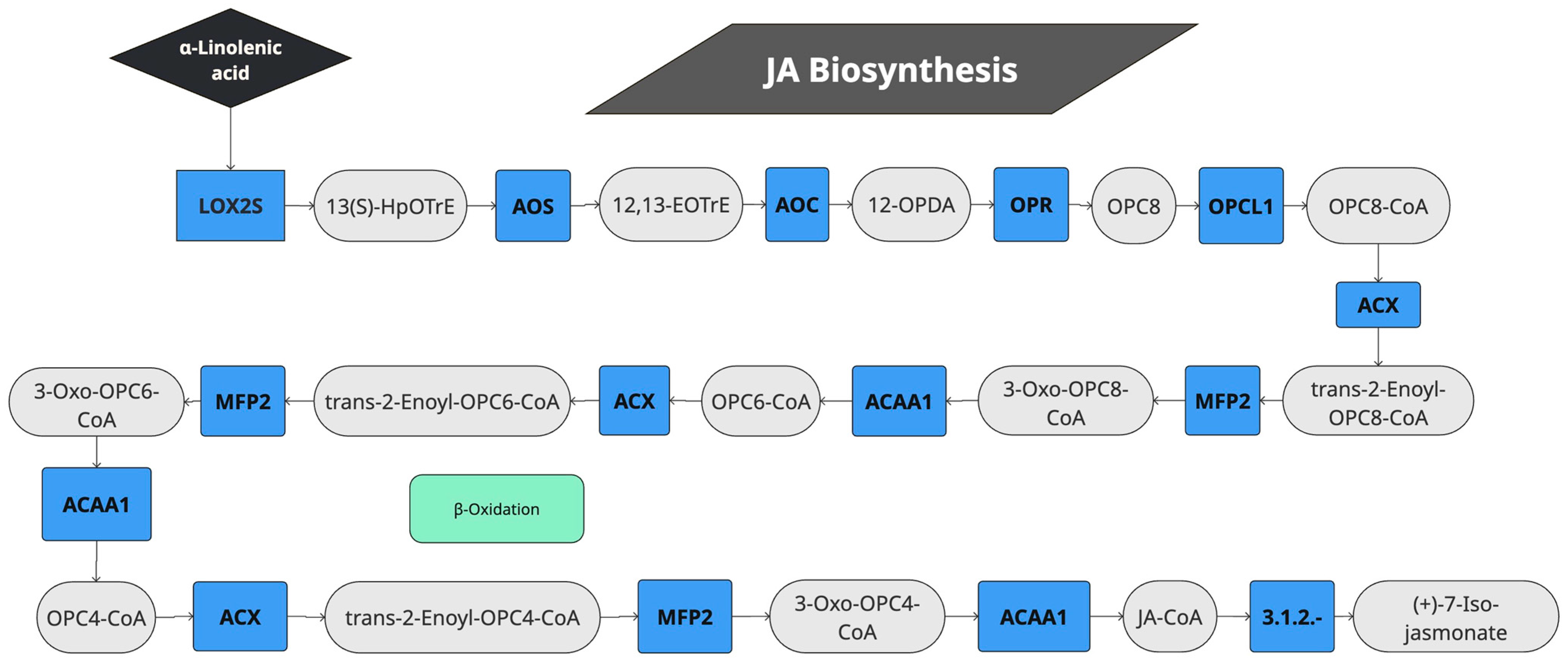
2.3. KOALA Annotation and KEGG Mapping Reveals That JA Application Induces Genes for Biosynthesis of Jasmonates
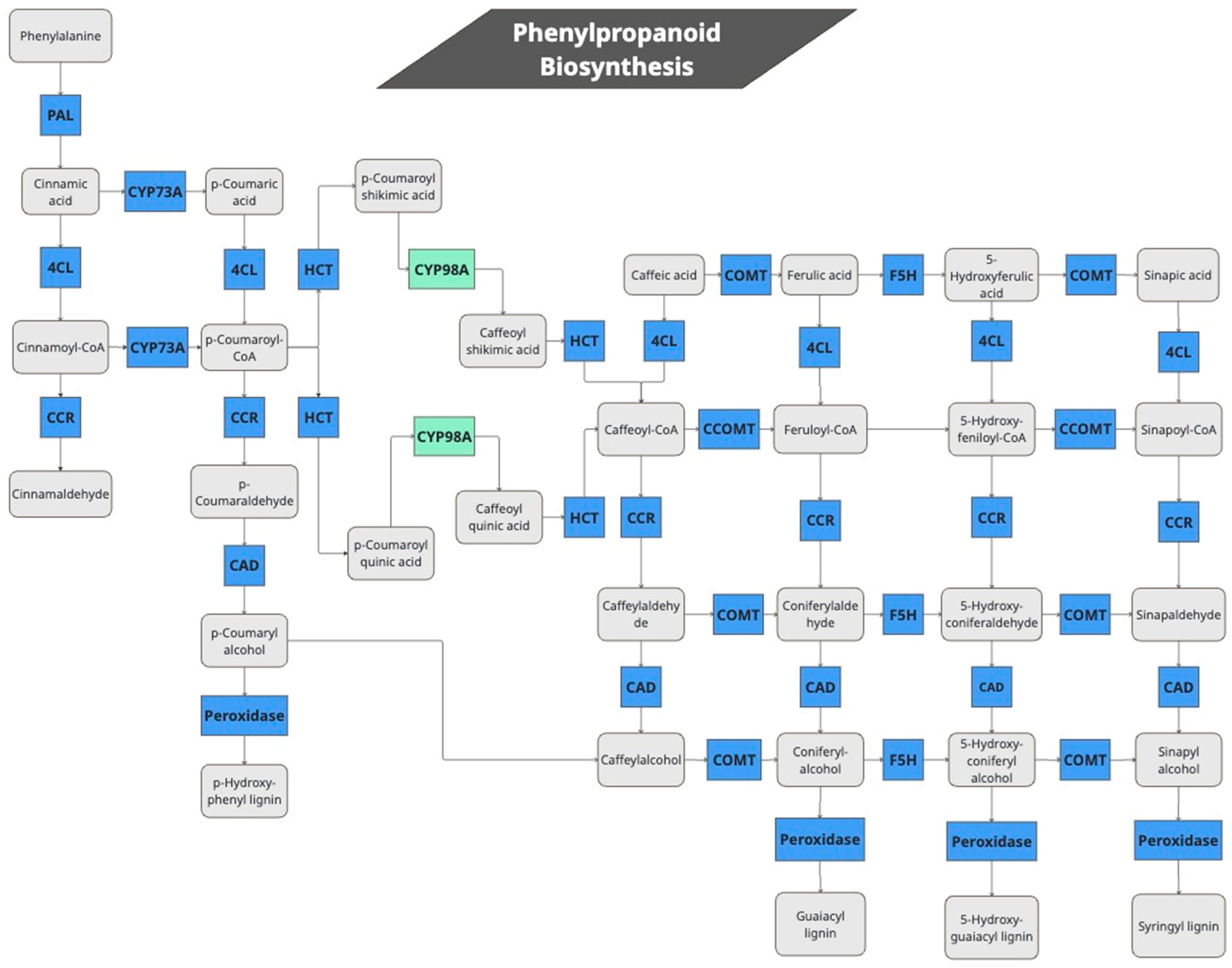
2.4. Quinoa Treatment with JA Induces Genes Involved in Lignin Biosynthesis
2.5. JA May Induce the Biosynthesis of Saponins
2.6. Other Pathways Differentially Expressed by JA Treatment in Quinoa
2.7. Quinoa Differential Gene Expression in Response to SA Treatment
3. Discussion
4. Materials and Methods
4.1. Biological Materials
4.2. Hormone Treatments
4.3. Sample Collection and RNA Extraction
4.4. RNA-Seq Library Construction and Sequencing
4.5. Transcriptomic Analysis
4.6. Functional Annotation of Differentially Expressed Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| JA | Jasmonic acid |
| SA | Salicylic acid |
| DE | Differentially expressed |
| DGE | Differential gene expression |
| H | hours |
| CPM | Counts per million |
| NGS | Next-Generation sequencing |
| RNA | Ribonucleic Acid |
| DNA | Deoxyribonucleic Acid |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KO | KEGG Orthology |
| QC | Quality Control |
References
- Vega-Gálvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martínez, E.A. Nutrition Facts and Functional Potential of Quinoa (Chenopodium quinoa Willd.), an Ancient Andean Grain: A Review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef]
- Alandia, G.; Rodriguez, J.P.; Jacobsen, S.E.; Bazile, D.; Condori, B. Global Expansion of Quinoa and Challenges for the Andean Region. Glob. Food Secur. 2020, 26, 100429. [Google Scholar] [CrossRef]
- Pathan, S.; Siddiqui, R.A. Nutritional Composition and Bioactive Components in Quinoa (Chenopodium quinoa Willd.) Greens: A Review. Nutrients 2022, 14, 558. [Google Scholar] [CrossRef] [PubMed]
- Bazile, D.; Jacobsen, S.E.; Verniau, A. The Global Expansion of Quinoa: Trends and Limits. Front. Plant Sci. 2016, 7, 622. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The Genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Mujica, A.; Jensen, C.R. The Resistance of Quinoa (Chenopodium quinoa Willd.) to Adverse Abiotic Factors. Food Rev. Int. 2003, 19, 99–109. [Google Scholar] [CrossRef]
- Ruiz, K.B.; Biondi, S.; Oses, R.; Acuña-Rodríguez, I.S.; Antognoni, F.; Martinez-Mosqueira, E.A.; Coulibaly, A.; Canahua-Murillo, A.; Pinto, M.; Zurita-Silva, A.; et al. Quinoa Biodiversity and Sustainability for Food Security under Climate Change. A Review. Agron. Sustain. Dev. 2014, 34, 349–359. [Google Scholar] [CrossRef]
- Testen, A.L. Diseases of Quinoa. In Handbook of Vegetable and Herb Diseases; Springer Nature: Cham, Switzerland, 2025; pp. 1–27. [Google Scholar]
- Antonio, G.; Saravia, R.; Plata, G.; Quispe, R.; Ortiz-Romero, R.; Bazile, D.; Bertero, H.D.; Nieto, C. Principle Quinoa Pests and Diseases. In State of the Art Report on Quinoa; FAO: Rome, Italy, 2015; p. 192. [Google Scholar]
- Danielsen, S.; Jacobsen, S.E.; Echegaray, J.; Ames, T. Impact of Downy Mildew on the Yield of Quinoa. CIP Program Rep. 2000, 2000, 397–401. [Google Scholar]
- Böndel, K.B.; Schmid, K.J. Quinoa Diversity and Its Implications for Breeding. In Quinoa Genome; Springer Nature: Cham, Switzerland, 2021; pp. 107–118. [Google Scholar]
- Rollano-Peñaloza, O.M.; Palma-Encinas, V.; Widell, S.; Mollinedo, P.; Rasmusson, A.G. The Disease Progression and Molecular Defense Response in Chenopodium quinoa Infected with Peronospora variabilis, the Causal Agent of Quinoa Downy Mildew. Plants 2022, 11, 2946. [Google Scholar] [CrossRef]
- Julio, G.; Luna, N.; Vargas, A.; Jury, M.; Angulo, A.; La Torre, J.; Bonifacio, A. Quinoa from Valley (Chenopodium quinoa Willd.): Valuable Source of Genetic Resistance to Powdery Mildew (Peronospora farinosa Willd.). J. Selva Andin. Res. Soc. 2012, 3, 27–44. [Google Scholar]
- Colque-Little, C.; Abondano, M.C.; Lund, O.S.; Amby, D.B.; Piepho, H.-P.; Andreasen, C.; Schmöckel, S.; Schmid, K. Genetic Variation for Tolerance to the Downy Mildew Pathogen Peronospora variabilis in Genetic Resources of Quinoa (Chenopodium quinoa). BMC Plant Biol. 2021, 21, 41. [Google Scholar] [CrossRef] [PubMed]
- Rollano-Peñaloza, O.M.; Widell, S.; Mollinedo, P.; Rasmusson, A.G. Trichoderma Harzianum T-22 and Bol-12qd Inhibit Lateral Root Development of Chenopodium quinoa in Axenic Co-Culture. Cogent Biol. 2018, 4, 1530493. [Google Scholar] [CrossRef]
- Variety of Quinoa Kurmi. In Technical Sheet; PROINPA Foundation: Cochabamba, Bolivia, 2005; No. 12-2005.
- Pieterse, C.M.J.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C.M. Networking by Small-Molecule Hormones in Plant Immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef]
- Caarls, L.; Pieterse, C.M.; Van Wees, S.C. How Salicylic Acid Takes Transcriptional Control over Jasmonic Acid Signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jonathan, J.D.G. Role of Plant Hormones in Plant Defence Responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Guerreiro, A.; Figueiredo, J.; Silva, M.S.; Figueiredo, A. Linking Jasmonic Acid to Grapevine Resistance against the Biotrophic Oomycete Plasmopara Viticola. Front. Plant Sci. 2016, 7, 565. [Google Scholar] [CrossRef]
- Fugate, K.K.; Ferrareze, J.P.; Bolton, M.D.; Deckard, E.L.; Campbell, L.G. Postharvest Jasmonic Acid Treatment of Sugarbeet Roots Reduces Rot Due to Botrytis cinerea, Penicillium claviforme, and Phoma betae. Postharvest Biol. Technol. 2012, 65, 1–4. [Google Scholar] [CrossRef]
- Forouzandeh, M.; Parsa, S.; Mahmoodi, S.; Izanloo, A. Physiological, Biochemical, and Molecular Responses of Quinoa (Chenopodium quinoa Willd.) to Elicitors under Drought Stress. Plant Mol. Biol. Report. 2024, 42, 515–531. [Google Scholar] [CrossRef]
- Fiallos-Jurado, J.; Pollier, J.; Moses, T.; Arendt, P.; Barriga-Medina, N.; Morillo, E.; Arahana, V.; de Lourdes Torres, M.; Goossens, A.; Leon-Reyes, A. Saponin Determination, Expression Analysis and Functional Characterization of Saponin Biosynthetic Genes in Chenopodium quinoa Leaves. Plant Sci. 2016, 250, 188–197. [Google Scholar] [CrossRef]
- Rollano-Peñaloza, O.M.; Mollinedo, P.A.; Widell, S.; Rasmusson, A.G. Transcriptomic Analysis of Quinoa Reveals a Group of Germin-Like Proteins Induced by Trichoderma. Front. Fungal Biol. 2021, 2, 768648. [Google Scholar] [CrossRef]
- Payá-Milans, M.; Olmstead, J.W.; Nunez, G.; Rinehart, T.A.; Staton, M. Comprehensive Evaluation of RNA-Seq Analysis Pipelines in Diploid and Polyploid Species. GigaScience 2018, 7, giy132. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. Jasmonate-Insensitive1 Encodes a Myc Transcription Factor Essential to Discriminate between Different Jasmonate-Regulated Defense Responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhao, J.; Tzeng, D.T.W.; Liu, Y.; Deng, L.; Yang, T.; Zhai, Q.; Wu, F.; Huang, Z.; Zhou, M. Myc2 Orchestrates a Hierarchical Transcriptional Cascade That Regulates Jasmonate-Mediated Plant Immunity in Tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Koo, A.J.K.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and Function of Arabidopsis Jasmonate Zim-Domain Genes in Response to Wounding and Herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate Passes Muster: A Receptor and Targets for the Defense Hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, Metabolism, and Signaling by Proteins Activating and Repressing Transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Sasaki, Y.; Asamizu, E.; Shibata, D.; Nakamura, Y.; Kaneko, T.; Awai, K.; Amagai, M.; Kuwata, C.; Tsugane, T.; Masuda, T.; et al. Monitoring of Methyl Jasmonate-Responsive Genes in Arabidopsis by cDNA Macroarray: Self-Activation of Jasmonic Acid Biosynthesis and Crosstalk with Other Phytohormone Signaling Pathways. DNA Res. 2001, 8, 153–161. [Google Scholar] [CrossRef]
- Bhuiyan, N.H.; Selvaraj, G.; Wei, Y.; King, J. Role of Lignification in Plant Defense. Plant Signal. Behav. 2009, 4, 158–159. [Google Scholar] [CrossRef]
- Pauwels, L.; Morreel, K.; De Witte, E.; Lammertyn, F.; Van Montagu, M.; Boerjan, W.; Inzé, D.; Goossens, A. Mapping Methyl Jasmonate-Mediated Transcriptional Reprogramming of Metabolism and Cell Cycle Progression in Cultured Arabidopsis Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 1380–1385. [Google Scholar] [CrossRef]
- Liu, C.; Yang, N.; Teng, R.-M.; Li, J.-W.; Chen, Y.; Hu, Z.-H.; Li, T.; Zhuang, J. Exogenous Methyl Jasmonate and Cytokinin Antagonistically Regulate Lignin Biosynthesis by Mediating Cshct Expression in Camellia sinensis. Protoplasma 2023, 260, 869–884. [Google Scholar] [CrossRef]
- Zhao, X.; Jiang, X.; Li, Z.; Song, Q.; Xu, C.; Luo, K. Jasmonic Acid Regulates Lignin Deposition in Poplar through Jaz5-Myb/Nac Interaction. Front. Plant Sci. 2023, 14, 1232880. [Google Scholar] [CrossRef] [PubMed]
- Hyde, L.S.; Pellny, T.K.; Freeman, J.; Michaelson, L.V.; Simister, R.; McQueen-Mason, S.J.; Mitchell, R.A.C. Response of Cell-Wall Composition and Rna-Seq Transcriptome to Methyl-Jasmonate in Brachypodium distachyon Callus. Planta 2018, 248, 1213–1229. [Google Scholar] [CrossRef]
- Hudgins, J.W.; Christiansen, E.; Franceschi, V.R. Induction of Anatomically Based Defense Responses in Stems of Diverse Conifers by Methyl Jasmonate: A Phylogenetic Perspective. Tree Physiol. 2004, 24, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.-N.; Son, S.; Jordan, M.C.; Levin, D.B.; Ayele, B.T. Lignin Biosynthesis in Wheat (Triticum aestivum L.): Its Response to Waterlogging and Association with Hormonal Levels. BMC Plant Biol. 2016, 16, 28. [Google Scholar] [CrossRef]
- Bonawitz, N.D.; Chapple, C. The Genetics of Lignin Biosynthesis: Connecting Genotype to Phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef]
- Khalifa, W.; Khalil, H.B.; Thabet, M. Unraveling Quinoa (Chenopodium quinoa Willd.) Defense against Downy Mildew (Peronospora variabilis): Comparative Molecular Analysis of Resistant “Hualhuas” and Susceptible “Real” Cultivars. Plants 2024, 13, 3344. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Wu, X.; Liu, D.; Li, J.; Li, J.; Liu, S.; Gao, W. Jasmonic Acid and Methyl Dihydrojasmonate Enhance Saponin Biosynthesis as Well as Expression of Functional Genes in Adventitious Roots of Panax notoginseng, F.H. Chen. Biotechnol. Appl. Biochem. 2017, 64, 225–238. [Google Scholar] [CrossRef]
- Hou, L.; Li, S.; Zhang, F.; Gu, Y.; Li, J. Effect of Exogenous Jasmonic Acid on Physiology and Steroidal Saponin Accumulation in Dioscorea zingiberensis. Plant Physiol. Biochem. 2022, 186, 1–10. [Google Scholar] [CrossRef]
- Yadav, D.; Boyidi, P.; Ahmed, I.; Kirti, P.B. Plant Annexins and Their Involvement in Stress Responses. Environ. Exp. Bot. 2018, 155, 293–306. [Google Scholar] [CrossRef]
- Saad, R.B.; Romdhane, W.B.; Hsouna, A.B.; Mihoubi, W.; Harbaoui, M.; Brini, F. Insights into Plant Annexins Function in Abiotic and Biotic Stress Tolerance. Plant Signal. Behav. 2020, 15, 1699264. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The Wrky70 Transcription Factor: A Node of Convergence for Jasmonate-Mediated and Salicylate-Mediated Signals in Plant Defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Somssich, I.E. Networks of Wrky Transcription Factors in Defense Signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef] [PubMed]
- Soltani, N.; Staton, M.; Gwinn, K.D. Response of Bitter and Sweet Chenopodium quinoa Varieties to Cucumber Mosaic Virus: Transcriptome and Small Rna-Seq Perspective. PLoS ONE 2021, 16, e0244364. [Google Scholar] [CrossRef]
- Wenig, M.; Ghirardo, A.; Sales, J.H.; Pabst, E.S.; Breitenbach, H.H.; Antritter, F.; Weber, B.; Lange, B.; Lenk, M.; Cameron, R.K. Systemic Acquired Resistance Networks Amplify Airborne Defense Cues. Nat. Commun. 2019, 10, 3813. [Google Scholar] [CrossRef]
- Hayat, Q.; Hayat, S.; Irfan, M.; Ahmad, A. Effect of Exogenous Salicylic Acid under Changing Environment: A Review. Environ. Exp. Bot. 2010, 68, 14–25. [Google Scholar] [CrossRef]
- Halkier, B.A.; Du, L. The Biosynthesis of Glucosinolates. Trends Plant Sci. 1997, 2, 425–431. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.-J.; Zhang, K. S5h/Dmr6 Encodes a Salicylic Acid 5-Hydroxylase That Fine-Tunes Salicylic Acid Homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef]
- Liang, B.; Bai, Y.; Zang, C.; Pei, X.; Xie, J.; Lin, Y.; Liu, X.; Ahsan, T.; Liang, C. Overexpression of the First Peanut-Susceptible Gene, Ahs5h1 or Ahs5h2, Enhanced Susceptibility to Pst Dc3000 in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 14210. [Google Scholar] [CrossRef]
- Thomazella, D.P.D.T.; Seong, K.; Mackelprang, R.; Dahlbeck, D.; Geng, Y.; Gill, U.S.; Qi, T.; Pham, J.; Giuseppe, P.; Lee, C.Y.; et al. Loss of Function of a Dmr6 Ortholog in Tomato Confers Broad-Spectrum Disease Resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2026152118. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, Q.; Gao, S.; Yu, N.; Zhao, L.; Wang, J.; Zhao, J.; Huang, P.; Yao, L.; Wang, M.; et al. Disruption of the Primary Salicylic Acid Hydroxylases in Rice Enhances Broad-Spectrum Resistance against Pathogens. Plant Cell Environ. 2022, 45, 2211–2225. [Google Scholar] [CrossRef]
- Silverman, P.; Seskar, M.; Kanter, D.; Schweizer, P.; Metraux, J.P.; Raskin, I. Salicylic Acid in Rice (Biosynthesis, Conjugation, and Possible Role). Plant Physiol. 1995, 108, 633–639. [Google Scholar] [CrossRef]
- Shoresh, M.; Yedidia, I.; Chet, I. Involvement of Jasmonic Acid/Ethylene Signaling Pathway in the Systemic Resistance Induced in Cucumber by Trichoderma asperellum T203. Phytopathology 2005, 95, 76–84. [Google Scholar] [CrossRef]
- Salas-Marina, M.A.; Silva-Flores, M.A.; Uresti-Rivera, E.E.; Castro-Longoria, E.; Herrera-Estrella, A.; Casas-Flores, S. Colonization of Arabidopsis Roots by Trichoderma atroviride Promotes Growth and Enhances Systemic Disease Resistance through Jasmonic Acid/Ethylene and Salicylic Acid Pathways. Eur. J. Plant Pathol. 2011, 131, 15–26. [Google Scholar] [CrossRef]
- Mathys, J.; De Cremer, K.; Timmermans, P.; Van Kerkhove, S.; Lievens, B.; Vanhaecke, M.; Cammue, B.; De Coninck, B. Genome-Wide Characterization of Isr Induced in Arabidopsis thaliana by Trichoderma hamatum T382 against Botrytis cinerea Infection. Front. Plant Sci. 2012, 3, 108. [Google Scholar] [CrossRef]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-Based Differential Expression Analysis of Rna Sequencing Data Using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Dillies, M.A.; Rau, A.; Aubert, J.; Hennequet-Antier, C.; Jeanmougin, M.; Servant, N.; Keime, C.; Marot, G.; Castel, D.; Estelle, J.; et al. A Comprehensive Evaluation of Normalization Methods for Illumina High-Throughput RNA Sequencing Data Analysis. Brief. Bioinform. 2013, 14, 671–683. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J. Kobas-I: Intelligent Prioritization and Exploratory Visualization of Biological Functions for Gene Enrichment Analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. Blastkoala and Ghostkoala: Kegg Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. 2007–2015. 2016. Available online: https://bioinfogp.cnb.csic.es/tools/venny/ (accessed on 22 April 2025).
| Sample | Treatment | Total Reads 1 | Mapped Reads | % | Unique Reads 2 | % | Non-Unique Reads 3 | % | Noncoding RNA Reads 4 | % | Ambiguous |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Control 1 | 4,030,580 | 3,983,129 | 96.1 | 497,482 | 12.5 | 1,328,848 | 33.4 | 2,033,141 | 51.0 | 622 |
| 2 | Control 2 | 4,171,906 | 4,171,831 | 94.3 | 478,990 | 11.5 | 1,391,787 | 33.4 | 2,142,277 | 51.4 | 608 |
| 3 | Control 3 | 4,534,183 | 4,486,832 | 96.1 | 561,575 | 12.5 | 1,527,014 | 34.0 | 2,256,616 | 50.3 | 691 |
| 4 | Control 4 | 4,297,813 | 4,240,505 | 95.7 | 595,001 | 14.0 | 1,349,547 | 31.8 | 2,149,134 | 50.7 | 750 |
| 5 | SA 1 | 4,221,902 | 4,176,269 | 96.0 | 468,141 | 11.2 | 1,452,097 | 34.8 | 2,119,621 | 50.8 | 610 |
| 6 | SA 2 | 4,100,965 | 4,059,417 | 96.4 | 477,640 | 11.8 | 1,633,839 | 40.2 | 1,828,580 | 45.0 | 612 |
| 7 | SA 3 | 3,984,027 | 3,935,189 | 95.1 | 564,365 | 14.3 | 1,307,064 | 33.2 | 1,906,205 | 48.4 | 785 |
| 8 | SA 4 | 4,111,569 | 4,070,911 | 95.7 | 614,441 | 15.1 | 1,418,030 | 34.8 | 1,891,153 | 46.5 | 846 |
| 9 | JA 1 | 4,032,015 | 3,975,797 | 95.1 | 656,029 | 16.5 | 1,263,491 | 31.8 | 1,897,901 | 47.7 | 745 |
| 10 | JA 2 | 4,376,388 | 4,327,057 | 94.9 | 596,965 | 13.8 | 1,495,474 | 34.6 | 2,045,298 | 47.3 | 664 |
| 11 | JA 3 | 3,872,463 | 3,826,879 | 95.2 | 590,513 | 15.4 | 1,265,202 | 33.1 | 1,818,469 | 47.5 | 598 |
| 12 | JA 4 | 4,171,328 | 4,128,924 | 94.5 | 685,783 | 16.6 | 1,367,084 | 33.1 | 1,878,286 | 45.5 | 772 |
| KEGG Code | Number of Quinoa Genes Induced/Genome Annotated | Enzyme Description | Enzyme Code Figure 2 | Enzyme Code | |
|---|---|---|---|---|---|
| 1 | K00454 | 8/14 | Lipoxygenase | LOX2S | 1.13.11.12 |
| 2 | K01723 | 2/7 | Hydroperoxide dehydratase | AOS | 4.2.1.92 |
| 3 | K10525 | 3/6 | Allene oxide cyclase | AOC | 5.3.99.6 |
| 4 | K05894 | 4/10 | 12-oxophytodienoic acid reductase | OPR | 1.3.1.42 |
| 5 | K10526 | 2/2 | OPC-8:0 CoA ligase 1 | OPCL1 | 6.2.1. |
| 6 | K00232 | 2/7 | Acyl-CoA oxidase | ACOX1 | 1.3.3.6 |
| 7 | K10527 | 2/6 | Enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase | MFP2 | 4.2.1.17 |
| 8 | K07513 | 1/4 | Acetyl-CoA acyltransferase 1 | ACAA1 | 2.3.1.16 |
| KEGG Code | Number of Genes Responsive/Total | Enzyme Description | Enzyme Code Figure 2 | Enzyme Code | |
|---|---|---|---|---|---|
| 1 | K10775 | 2/2 | Phenylalanine ammonia-lyase | PAL | 4.3.1.24 |
| 2 | K00487 | 2/4 | Trans-cinnamate 4-monooxygenase | CYP73A | 1.14.14.91 |
| 3 | K01904 | 2/11 | 4-coumarate-CoA 1 ligase | 4CL | 6.2.1.12 |
| 4 | K09754 | 1/6 2 | 5-O-(4-coumaroyl)-D-quinate 3′-monooxygenase | CYP98A | 1.14.14.96 |
| 5 | K13065 | 1/22 | Shikimate O-hydroxycinnamoyltransferase | HCT | 2.3.1.133 |
| 6 | K00588 | 2/15 | Caffeoyl-CoA O-methyltransferase | CCOMT | 2.1.1.104 |
| 7 | K13066 | 6/18 | Caffeic acid 3-O-methyltransferase | COMT | 2.1.1.68 |
| 8 | K09755 | 1/1 | Ferulate-5-hydroxylase | F5H | 1.14.13.- |
| 9 | K09753 | 1/2 | Cinnamoyl-CoA 1 reductase | CCR | 1.2.1.44 |
| 10 | K00083 | 1/15 | Cinnamyl-alcohol dehydrogenase | CAD | 1.1.1.195 |
| 11 | K00430 | 15/199 | Peroxidases that produce 4 types of lignin subunits. | Peroxidase | 1.11.1.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rollano-Peñaloza, O.M.; Neyrot, S.; Bravo Barrera, J.A.; Mollinedo, P.; Rasmusson, A.G. Transcriptomic Profiling of Quinoa Reveals Distinct Defense Responses to Exogenous Methyl Jasmonate and Salicylic Acid. Plants 2025, 14, 1708. https://doi.org/10.3390/plants14111708
Rollano-Peñaloza OM, Neyrot S, Bravo Barrera JA, Mollinedo P, Rasmusson AG. Transcriptomic Profiling of Quinoa Reveals Distinct Defense Responses to Exogenous Methyl Jasmonate and Salicylic Acid. Plants. 2025; 14(11):1708. https://doi.org/10.3390/plants14111708
Chicago/Turabian StyleRollano-Peñaloza, Oscar M., Sara Neyrot, Jose A. Bravo Barrera, Patricia Mollinedo, and Allan G. Rasmusson. 2025. "Transcriptomic Profiling of Quinoa Reveals Distinct Defense Responses to Exogenous Methyl Jasmonate and Salicylic Acid" Plants 14, no. 11: 1708. https://doi.org/10.3390/plants14111708
APA StyleRollano-Peñaloza, O. M., Neyrot, S., Bravo Barrera, J. A., Mollinedo, P., & Rasmusson, A. G. (2025). Transcriptomic Profiling of Quinoa Reveals Distinct Defense Responses to Exogenous Methyl Jasmonate and Salicylic Acid. Plants, 14(11), 1708. https://doi.org/10.3390/plants14111708






