Genome-Wide Association Study and RNA-Seq Elucidate the Genetic Mechanisms Behind Aphid (Rhopalosiphum maidis F.) Resistance in Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Germplasm Materials and Experimental Design
2.2. Phenotypic Data Collection
2.3. Statistical Analyses for Phenotypic Data
2.4. Genome-Wide Association Study
2.5. Total RNA Extraction and Sequencing
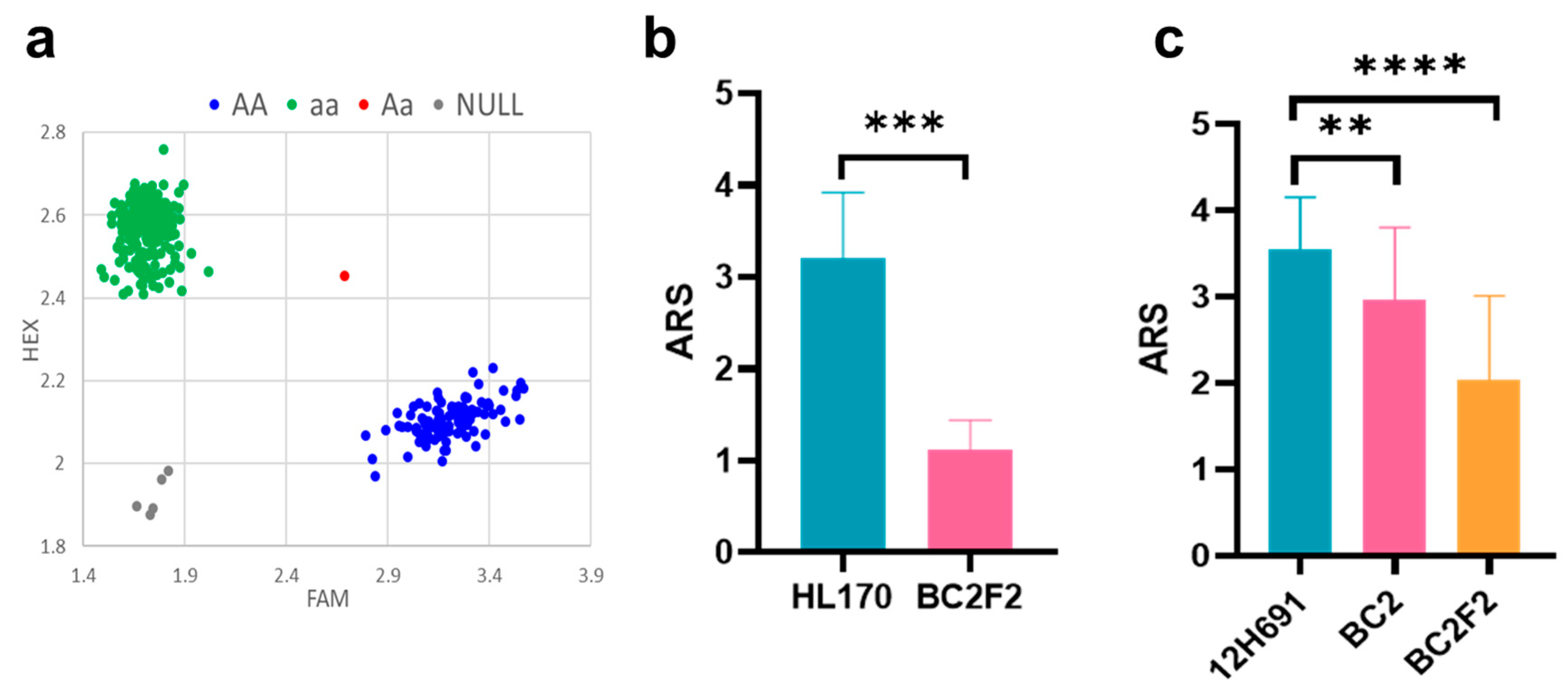
2.6. KASP Marker Development and Validation
3. Results
3.1. Phenotypic Data Analysis for Aphid Resistance
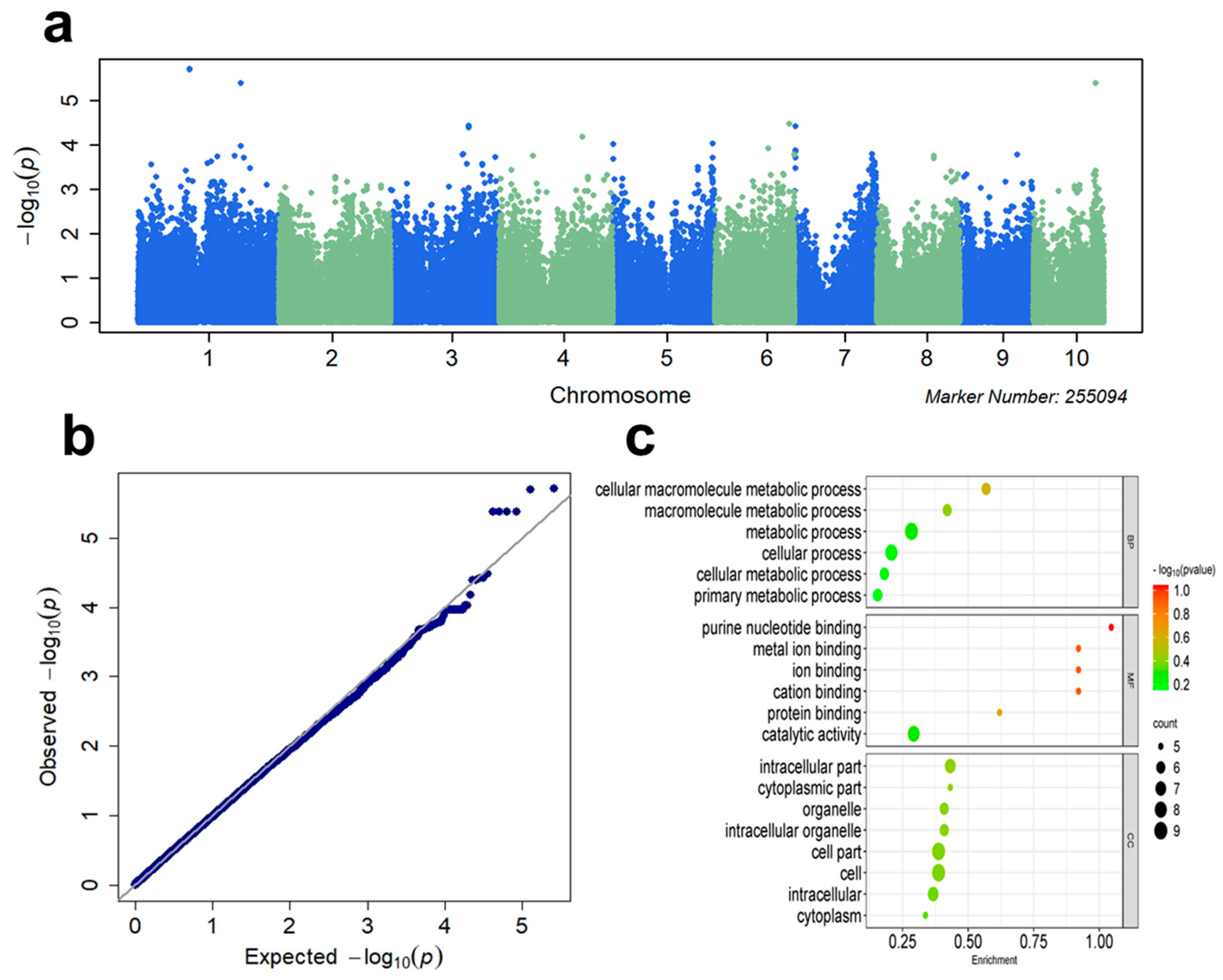
3.2. GWAS
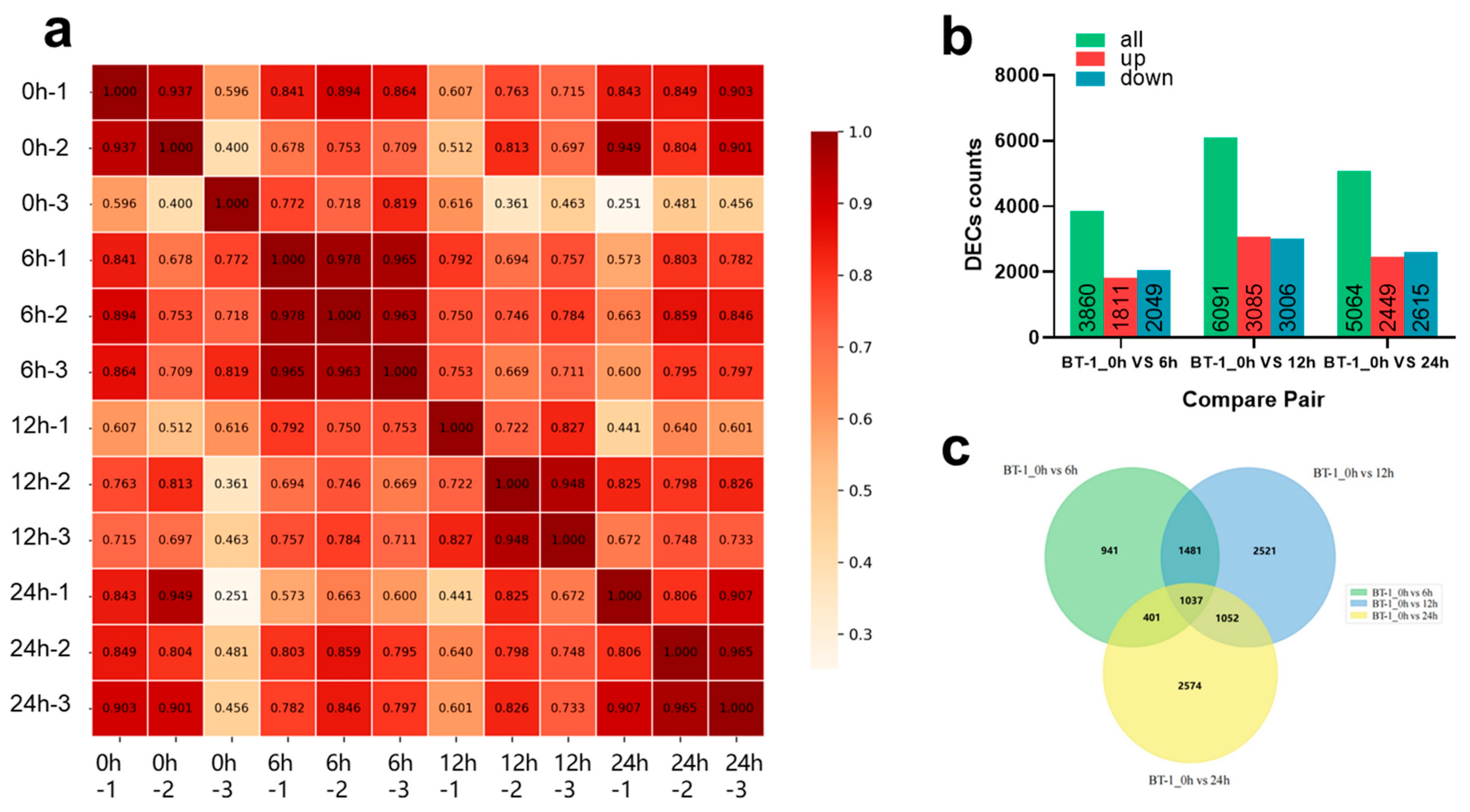
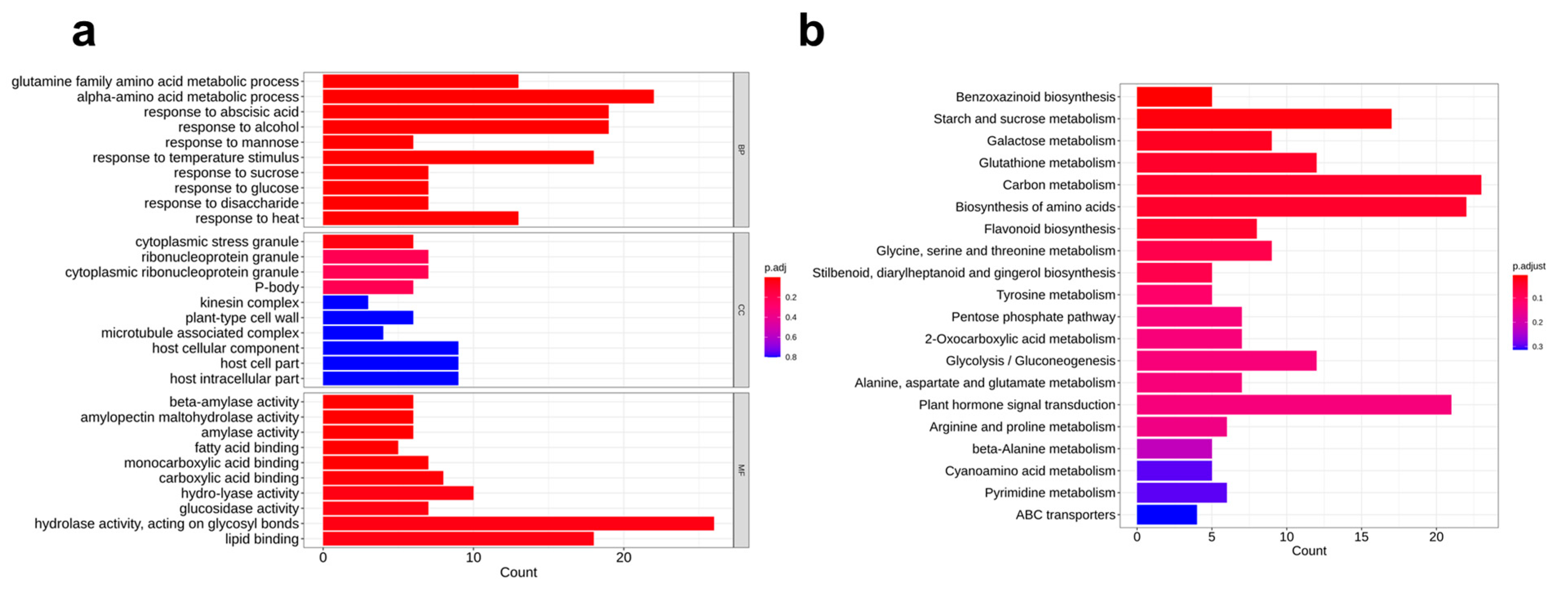
3.3. RNA-Seq
3.4. Functional Verification of the Gene Cluster on Chromosome 4S
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Meihls, L.N.; Handrick, V.; Glauser, G.; Barbier, H.; Kaur, H.; Haribal, M.M.; Lipka, A.E.; Gershenzon, J.; Buckler, E.S.; Erb, M.; et al. Natural Variation in Maize Aphid Resistance Is Associated with 2,4-Dihydroxy-7-Methoxy-1,4-Benzoxazin-3-One Glucoside Methyltransferase Activity. Plant Cell 2013, 25, 2341–2355. [Google Scholar] [CrossRef]
- Carena, M.J.; Glogoza, P. Resistance of Maize to the Corn Leaf Aphid: A Review. Maydica 2004, 49, 241–254. [Google Scholar]
- Regassa, B.; Abraham, A.; Wolde-Hawariat, Y.; Fininsa, C.; Wegary, D.; Atickem, A. Identification of Insect Vectors of Maize Lethal Necrosis Viruses and Their Virus-Transmission Ability in Ethiopia. Int. J. Trop. Insect Sci. 2024, 44, 843–854. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, J.; Qian, X.; Chao, Z.; Zong, X.; Jiang, Q.; An, S.; Shen, J.; Yan, S. Nanocarrier-Delivered Gene Silence in Juvenile Hormone Signaling Pathway: Conserved Dual Targets for Efficient Aphid Control. Entomol. Gen. 2024, 44, 1291–1299. [Google Scholar] [CrossRef]
- Yates, A.D.; Michel, A. Mechanisms of Aphid Adaptation to Host Plant Resistance. Curr. Opin. Insect Sci. 2018, 26, 41–49. [Google Scholar] [CrossRef]
- Talakayala, A.; Katta, S.; Garladinne, M. Genetic Engineering of Crops for Insect Resistance: An Overview. J. Biosci. 2020, 45, 114. [Google Scholar] [CrossRef]
- Ahmad, S.; Veyrat, N.; Gordon-Weeks, R.; Zhang, Y.; Martin, J.; Smart, L.; Glauser, G.; Erb, M.; Flors, V.; Frey, M.; et al. Benzoxazinoid Metabolites Regulate Innate Immunity against Aphids and Fungi in Maize. Plant Physiol. 2011, 157, 317–327. [Google Scholar] [CrossRef]
- Wang, T.; Wang, K.; Wang, C.; Zhao, Y.; Tao, Z.; Li, J.; Wang, L.; Shi, J.; Huang, S.; Xie, C.; et al. Combining Quantitative Trait Locus Mapping with Multiomics Profiling Reveals Genetic Control of Corn Leaf Aphid (Rhopalosiphum maidis) Resistance in Maize. J. Exp. Bot. 2023, 74, 3749–3764. [Google Scholar] [CrossRef]
- Cardona, J.B.; Grover, S.; Bowman, M.J.; Busta, L.; Kundu, P.; Koch, K.G.; Sarath, G.; Sattler, S.E.; Louis, J. Sugars and Cuticular Waxes Impact Sugarcane Aphid (Melanaphis sacchari) Colonization on Different Developmental Stages of Sorghum. Plant Sci. 2023, 330, 111646. [Google Scholar] [CrossRef]
- Wang, Z.; Yan, X.; Zhang, H.; Meng, Y.; Pan, Y.; Cui, H. NtCycB2 Negatively Regulates Tobacco Glandular Trichome Formation, Exudate Accumulation, and Aphid Resistance. Plant Mol. Biol. 2022, 108, 65–76. [Google Scholar] [CrossRef]
- Züst, T.; Agrawal, A.A. Mechanisms and Evolution of Plant Resistance to Aphids. Nat. Plants 2016, 2, 15206. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, J.; Mu, C.; Makumbi, D.; Xu, Y.; Mahuku, G. Combined Linkage and Association Mapping Reveal QTL for Host Plant Resistance to Common Rust (Puccinia sorghi) in Tropical Maize. BMC Plant Biol. 2018, 18, 310. [Google Scholar] [CrossRef]
- Chen, J.; Shrestha, R.; Ding, J.; Zheng, H.; Mu, C.; Wu, J.; Mahuku, G. Genome-Wide Association Study and QTL Mapping Reveal Genomic Loci Associated with Fusarium Ear Rot Resistance in Tropical Maize Germplasm. G3 Genes Genomes Genet. 2016, 6, 3803–3815. [Google Scholar] [CrossRef]
- Ju, M.; Zhou, Z.; Mu, C.; Zhang, X.; Gao, J.; Liang, Y.; Chen, J.; Wu, Y.; Li, X.; Wang, S.; et al. Dissecting the Genetic Architecture of Fusarium verticillioides Seed Rot Resistance in Maize by Combining QTL Mapping and Genome-Wide Association Analysis. Sci. Rep. 2017, 7, 46446. [Google Scholar] [CrossRef]
- Butrón, A.; Chen, Y.C.; Rottinghaus, G.E.; McMullen, M.D. Genetic Variation at Bx1 Controls DIMBOA Content in Maize. Theor. Appl. Genet. 2010, 120, 721–734. [Google Scholar] [CrossRef]
- Yan, J.; Lipka, A.E.; Schmelz, E.A.; Buckler, E.S.; Jander, G. Accumulation of 5-Hydroxynorvaline in Maize (Zea mays) Leaves Is Induced by Insect Feeding and Abiotic Stress. J. Exp. Bot. 2015, 66, 593–602. [Google Scholar] [CrossRef]
- Mijares, V.; Meihls, L.N.; Jander, G.; Tzin, V. Near-Isogenic Lines for Measuring Phenotypic Effects of DIMBOA-Glc Methyltransferase Activity in Maize. Plant Signal. Behav. 2013, 8, e26779. [Google Scholar] [CrossRef][Green Version]
- Betsiashvili, M.; Ahern, K.R.; Jander, G. Additive Effects of Two Quantitative Trait Loci That Confer Rhopalosiphum maidis (Corn Leaf Aphid) Resistance in Maize Inbred Line Mo17. J. Exp. Bot. 2015, 66, 571–578. [Google Scholar] [CrossRef]
- Niu, Y.; Zhao, X.; Chao, W.; Lu, P.; Bai, X.; Mao, T. Genetic Variation, DIMBOA Accumulation, and Candidate Gene Identification in Maize Multiple Insect-Resistance. Int. J. Mol. Sci. 2023, 24, 2138. [Google Scholar] [CrossRef]
- Chu, L.Y.; Liu, T.; Xia, P.L.; Li, J.P.; Tang, Z.R.; Zheng, Y.L.; Wang, X.P.; Zhang, J.M.; Xu, R.B. NtWRKY28 Orchestrates Flavonoid and Lignin Biosynthesis to Defense Aphid Attack in Tobacco Plants. Plant Physiol. Biochem. 2025, 221, 109673. [Google Scholar] [CrossRef]
- Buntjer, J.B.; Sørensen, A.P.; Peleman, J.D. Haplotype Diversity: The Link between Statistical and Biological Association. Trends Plant Sci. 2005, 10, 466–471. [Google Scholar] [CrossRef]
- Wu, X.; Li, Y.; Shi, Y.; Song, Y.; Zhang, D.; Li, C.; Buckler, E.S.; Li, Y.; Zhang, Z.; Wang, T. Joint-Linkage Mapping and GWAS Reveal Extensive Genetic Loci That Regulate Male Inflorescence Size in Maize. Plant Biotechnol. J. 2016, 14, 1551–1562. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, Z.; Dong, C.; Chen, J.; Ding, J.; Zhang, X.; Mu, C.; Chen, Y.; Li, X.; Li, H.; et al. Linkage Mapping and Genome-Wide Association Study Reveals Conservative QTL and Candidate Genes for Fusarium Rot Resistance in Maize. BMC Genom. 2020, 21, 357. [Google Scholar] [CrossRef]
- Zhou, B.; Zhou, Z.; Ding, J.; Zhang, X.; Mu, C.; Wu, Y.; Gao, J.; Song, Y.; Wang, S.; Ma, J.; et al. Combining Three Mapping Strategies to Reveal Quantitative Trait Loci and Candidate Genes for Maize Ear Length. Plant Genome 2018, 11, 170107. [Google Scholar] [CrossRef]
- Duan, H.; Li, J.; Sun, L.; Xiong, X.; Xu, S.; Sun, Y.; Ju, X.; Xue, Z.; Gao, J.; Wang, Y.; et al. Identification of Novel Loci Associated with Starch Content in Maize Kernels by a Genome-Wide Association Study Using an Enlarged SNP Panel. Mol. Breed. 2023, 43, 91. [Google Scholar] [CrossRef]
- Sun, G.; Zhang, X.; Duan, H.; Gao, J.; Li, N.; Su, P.; Xie, H.; Li, W.; Fu, Z.; Huang, Y.; et al. Dissection of the Genetic Architecture of Peduncle Vascular Bundle-Related Traits in Maize by a Genome-Wide Association Study. Plant Biotechnol. J. 2022, 20, 1042–1053. [Google Scholar] [CrossRef]
- Gao, J.; Wang, S.; Zhou, Z.; Wang, S.; Dong, C.; Mu, C.; Song, Y.; Ma, P.; Li, C.; Wang, Z.; et al. Linkage Mapping and Genome-Wide Association Reveal Candidate Genes Conferring Thermotolerance of Seed-Set in Maize. J. Exp. Bot. 2019, 70, 4849–4863. [Google Scholar] [CrossRef]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science (1979) 2008, 320, 1344–1349. [Google Scholar] [CrossRef]
- Koch, C.M.; Chiu, S.F.; Akbarpour, M.; Bharat, A.; Ridge, K.M.; Bartom, E.T.; Winter, D.R. A Beginner’s Guide to Analysis of RNA Sequencing Data. Am. J. Respir. Cell Mol. Biol. 2018, 59, 145–157. [Google Scholar] [CrossRef]
- Dou, H.; Sun, H.; Feng, X.; Wang, T.; Wang, Y.; Quan, J.; Yang, X. Full-Length Transcriptome Assembly of Platycladus Orientalis Root Integrated with RNA-Seq to Identify Genes in Response to Root Pruning. Forests 2024, 15, 1232. [Google Scholar] [CrossRef]
- Xu, F.; Chen, S.; Zhou, S.; Yue, C.; Yang, X.; Zhang, X.; Zhan, K.; He, D. Genome-Wide Association, RNA-Seq and ITRAQ Analyses Identify Candidate Genes Controlling Radicle Length of Wheat. Front. Plant Sci. 2022, 13, 939544. [Google Scholar] [CrossRef] [PubMed]
- Leng, D.; Zeng, B.; Wang, T.; Chen, B.-L.; Li, D.-Y.; Li, Z.-J.; Leng, D.; Zeng, B.; Wang, T.; Chen, B.-L.; et al. Single Nucleus/Cell RNA-Seq of the Chicken Hypothalamic-Pituitary-Ovarian Axis Offers New Insights into the Molecular Regulatory Mechanisms of Ovarian Development. Zool. Res. 2024, 45, 1088–1107. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, X.; Cui, R.; Wang, R.; Li, H.; Wang, J.; Chen, H.; Zhang, D. Identification of Soybean Phosphorous Efficiency QTLs and Genes Using Chlorophyll Fluorescence Parameters through GWAS and RNA-Seq. Planta 2021, 254, 110. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Feng, P.; Zhang, J.; Dong, C.; Wang, Z.; Chen, M.; Yu, Z.; Zhao, B.; Hou, X.; Wang, H.; et al. Enhancing Maize’s Nitrogen-Fixing Potential through ZmSBT3, a Gene Suppressing Mucilage Secretion. J. Integr. Plant Biol. 2023, 65, 2645–2659. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for Association Mapping of Complex Traits in Diverse Samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Altman, T.; Dale, J.M.; Dreher, K.; Fulcher, C.A.; Gilham, F.; Kaipa, P.; Karthikeyan, A.S.; Kothari, A.; Krummenacker, M.; et al. The MetaCyc Database of Metabolic Pathways and Enzymes and the BioCyc Collection of Pathway/Genome Databases. Nucleic Acids Res. 2010, 38, 473–479. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, L.; Huang, L.; Yang, T.; Ma, J.; Yu, T.; Zhu, W.; Zhang, Z.; Tang, J. Uncovering the Gene Regulatory Network of Maize Hybrid ZD309 under Heat Stress by Transcriptomic and Metabolomic Analysis. Plants 2022, 11, 677. [Google Scholar] [CrossRef]
- Horiguchi, G.; Van Lijsebettens, M.; Candela, H.; Micol, J.L.; Tsukaya, H. Ribosomes and Translation in Plant Developmental Control. Plant Sci. 2012, 191–192, 24–34. [Google Scholar] [CrossRef]
- Vaid, N.; Macovei, A.; Tuteja, N. Knights in Action: Lectin Receptor-like Kinases in Plant Development and Stress Responses. Mol. Plant 2013, 6, 1405–1418. [Google Scholar] [CrossRef]
- Xu, J.; Wang, X.Y.; Guo, W.Z. The Cytochrome P450 Superfamily: Key Players in Plant Development and Defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar] [CrossRef]
- Tian, P.; Liu, J.; Zhao, Y.; Huang, Y.; Lian, Y.; Wang, Y.; Ye, Y. Nitrogen Rates and Plant Density Interactions Enhance Radiation Interception, Yield, and Nitrogen Use Efficiencies of Maize. Front. Plant Sci. 2022, 13, 974714. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.; Powell, A.F.; Mirzaei, M.; Wang, L.J.; Movahed, N.; Miller, J.K.; Piñeros, M.A.; Jander, G. Indole-3-Glycerolphosphate Synthase, a Branchpoint for the Biosynthesis of Tryptophan, Indole, and Benzoxazinoids in Maize. Plant J. 2021, 106, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Glauser, G.; Marti, G.; Villard, N.; Doyen, G.A.; Wolfender, J.L.; Turlings, T.C.J.; Erb, M. Induction and Detoxification of Maize 1,4-Benzoxazin-3-Ones by Insect Herbivores. Plant J. 2011, 68, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, A.; Ishihara, A.; Hasegawa, M.; Kodama, O.; Iwamura, H. Induced Accumulation of 2-Hydroxy-4,7-Dimethoxy-1,4-Benzoxazin-3-One Glucoside (HDMBOA-Glc) in Maize Leaves. Phytochemistry 2001, 56, 669–675. [Google Scholar] [CrossRef]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single Nucleotide Polymorphism Genotyping Using Kompetitive Allele Specific PCR (KASP): Overview of the Technology and Its Application in Crop Improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Koch, K.G.; Chapman, K.; Louis, J.; Heng-Moss, T.; Sarath, G. Plant Tolerance: A Unique Approach to Control Hemipteran Pests. Front. Plant Sci. 2016, 7, 1363. [Google Scholar] [CrossRef]

| Aphid Resistant Score | Symptom |
|---|---|
| 0 | No aphid |
| 1 | Aphid cover area is less than 5% of the tassel or leaf. |
| 2 | Aphid cover area is more than 5% and less than 30%, and there are very few aphids in the lower leaves. |
| 3 | Aphid cover area is more than 30% and less than 50%, and there are very few aphids in the lower leaves. |
| 4 | The aphid covers the tassel more than 50%, and there are some aphids in the lower leaves. |
| 5 | The tassels and lower leaves are covered with dense aphids. |
| Environments | Mean ± SD | Range | σg2 | σge2 | σe2 | H2 |
|---|---|---|---|---|---|---|
| ARS_E1 | 2.83 ± 1.76 | 0.00–5.00 | 2.74 | 0.32 | 0.94 | |
| ARS_E2 | 2.98 ± 1.37 | 0.00–5.00 | 1.44 | 0.29 | 0.91 | |
| ARS_E3 | 2.92 ± 1.58 | 0.00–5.00 | 1.61 | 0.85 | 0.78 | |
| Combine | 2.91 ± 1.58 | 0.00–5.00 | 1.17 | 0.78 | 0.47 | 0.77 |
| Chr | Position | Gene | Function Annotation |
|---|---|---|---|
| 1 | 219548351 | GRMZM2G127361 | Oxidoreductase zinc-binding dehydrogenase family protein |
| 1 | 82154848 | GRMZM2G132690 | Heavy metal transport/detoxification superfamily protein |
| 2 | 196411877 | GRMZM2G114107 | winged-helix DNA-binding transcription factor family protein |
| 2 | 3387748 | GRMZM2G047448 | (PGA6, WUS, WUS1) Homeodomain-like superfamily protein |
| 3 | 196434589 | GRMZM2G352234 | Pentatricopeptide repeat (PPR) superfamily protein |
| 4 | 3261014 | GRMZM2G085661 | (CYP71B37) cytochrome P450 family 71 subfamily B polypeptide 37, benzoxazinone synthesis 2 |
| 4 | 240329225 | GRMZM2G054162 | (IBM1) Transcription factor jumonji (jmjC) domain-containing protein |
| 5 | 15323527 | GRMZM2G161411 | (ATWRKY23, WRKY23) WRKY DNA-binding protein 23 |
| 5 | 201461600 | GRMZM2G147430 | Phosphoribulokinase/Uridine kinase family |
| 5 | 147624523 | GRMZM2G026490 | NHL domain-containing protein |
| 6 | 145107284 | GRMZM2G479523 | (GL22) germin-like protein subfamily 2 member 2 precursor |
| 6 | 158441896 | GRMZM2G176133 | Ribosomal protein L33 family protein |
| 6 | 161283910 | GRMZM2G034225 | expressed protein |
| 7 | 8677546 | GRMZM2G107408 | expressed protein |
| 7 | 167601337 | GRMZM2G056407 | (ATMYB94, ATMYBCP70, MYB94) myb domain protein 94 |
| 7 | 156116120 | GRMZM2G465999 | S-locus lectin protein kinase family protein |
| 8 | 151270345 | GRMZM2G017305 | P-loop containing nucleoside triphosphate hydrolases superfamily protein |
| 8 | 163281806 | GRMZM2G065893 | (RGLG2) RING domain ligase2 |
| 9 | 100282128 | GRMZM2G423693 | Nucleotide-diphospho-sugar transferase family protein |
| 10 | 133276617 | GRMZM2G123719 | (ATSCO1, ATSCO1/CPEF-G, SCO1) Translation elongation factor EFG/EF2 protein |
| 10 | 133332879 | GRMZM2G054418 | ATP binding microtubule motor family protein |
| 10 | 137656350 | GRMZM2G155762 | basic helix–loop–helix (bHLH) DNA-binding superfamily protein |
| Pathway ID | KEGG Pathway | Gene Number | p_adj |
|---|---|---|---|
| ko00402 | Benzoxazinoid biosynthesis | 5 | 5.9 × 10−3 |
| ko00500 | Starch and sucrose metabolism | 17 | 1.6 × 10−2 |
| ko00052 | Galactose metabolism | 9 | 3.1 × 10−2 |
| ko00480 | Glutathione metabolism | 12 | 4.0 × 10−2 |
| ko01200 | Carbon metabolism | 23 | 4.2 × 10−2 |
| Allele | Number | Average Disease Grade | Increased Resistance | p-Value |
|---|---|---|---|---|
| Favorite allele | 145 | 4.75 | 18.50% | 1.29 × 10−5 |
| Normal allele | 313 | 5.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, D.; Wei, Y.; Han, C.; Li, X.; Zhang, Z.; Wang, S.; Zhou, Z.; Gao, J.; Chen, J.; Wu, J. Genome-Wide Association Study and RNA-Seq Elucidate the Genetic Mechanisms Behind Aphid (Rhopalosiphum maidis F.) Resistance in Maize. Plants 2025, 14, 1614. https://doi.org/10.3390/plants14111614
Sun D, Wei Y, Han C, Li X, Zhang Z, Wang S, Zhou Z, Gao J, Chen J, Wu J. Genome-Wide Association Study and RNA-Seq Elucidate the Genetic Mechanisms Behind Aphid (Rhopalosiphum maidis F.) Resistance in Maize. Plants. 2025; 14(11):1614. https://doi.org/10.3390/plants14111614
Chicago/Turabian StyleSun, Doudou, Yijun Wei, Chunyan Han, Xiaopeng Li, Zhen Zhang, Shiwei Wang, Zijian Zhou, Jingyang Gao, Jiafa Chen, and Jianyu Wu. 2025. "Genome-Wide Association Study and RNA-Seq Elucidate the Genetic Mechanisms Behind Aphid (Rhopalosiphum maidis F.) Resistance in Maize" Plants 14, no. 11: 1614. https://doi.org/10.3390/plants14111614
APA StyleSun, D., Wei, Y., Han, C., Li, X., Zhang, Z., Wang, S., Zhou, Z., Gao, J., Chen, J., & Wu, J. (2025). Genome-Wide Association Study and RNA-Seq Elucidate the Genetic Mechanisms Behind Aphid (Rhopalosiphum maidis F.) Resistance in Maize. Plants, 14(11), 1614. https://doi.org/10.3390/plants14111614






