Dual Regulatory Roles of SlGAMYB1 in Tomato Development: GA-Dependent and GA-Independent Mechanisms
Abstract
1. Introduction
2. Results
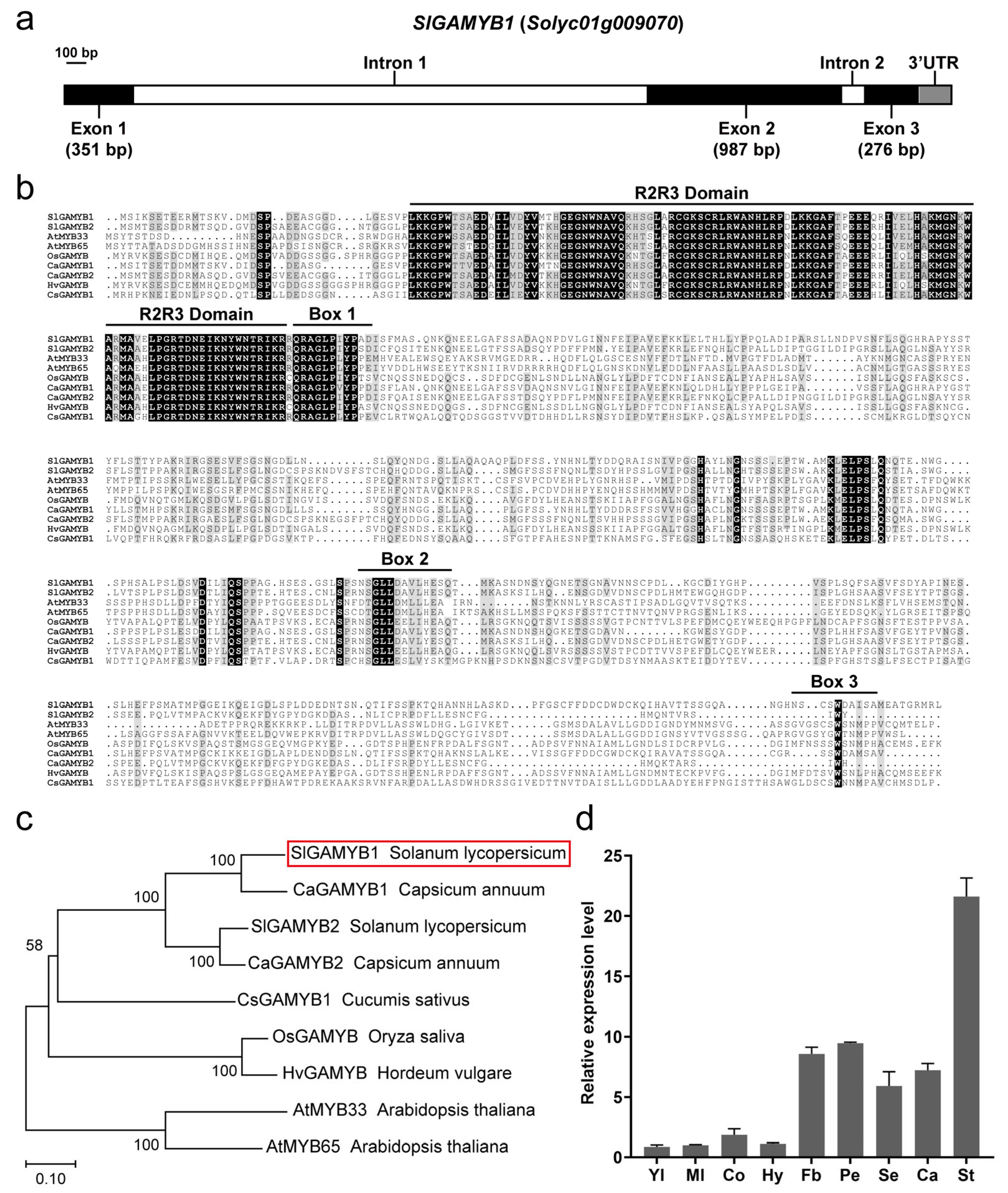
2.1. Characterization and Expression Pattern of SlGAMYB1
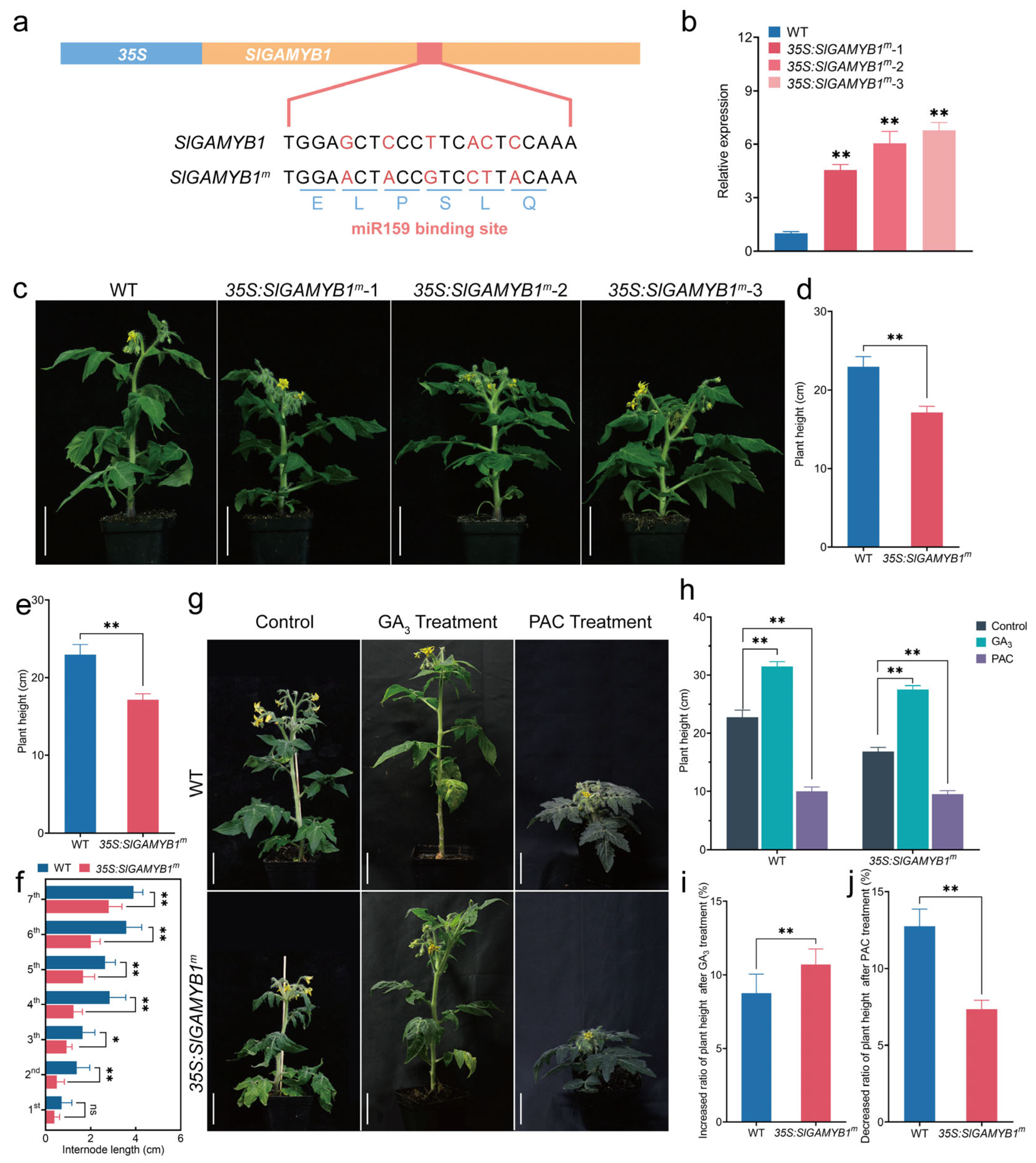
2.2. Overexpression of SlGAMYB1 Results in Plant Dwarfism via GA Deficiency in Tomato
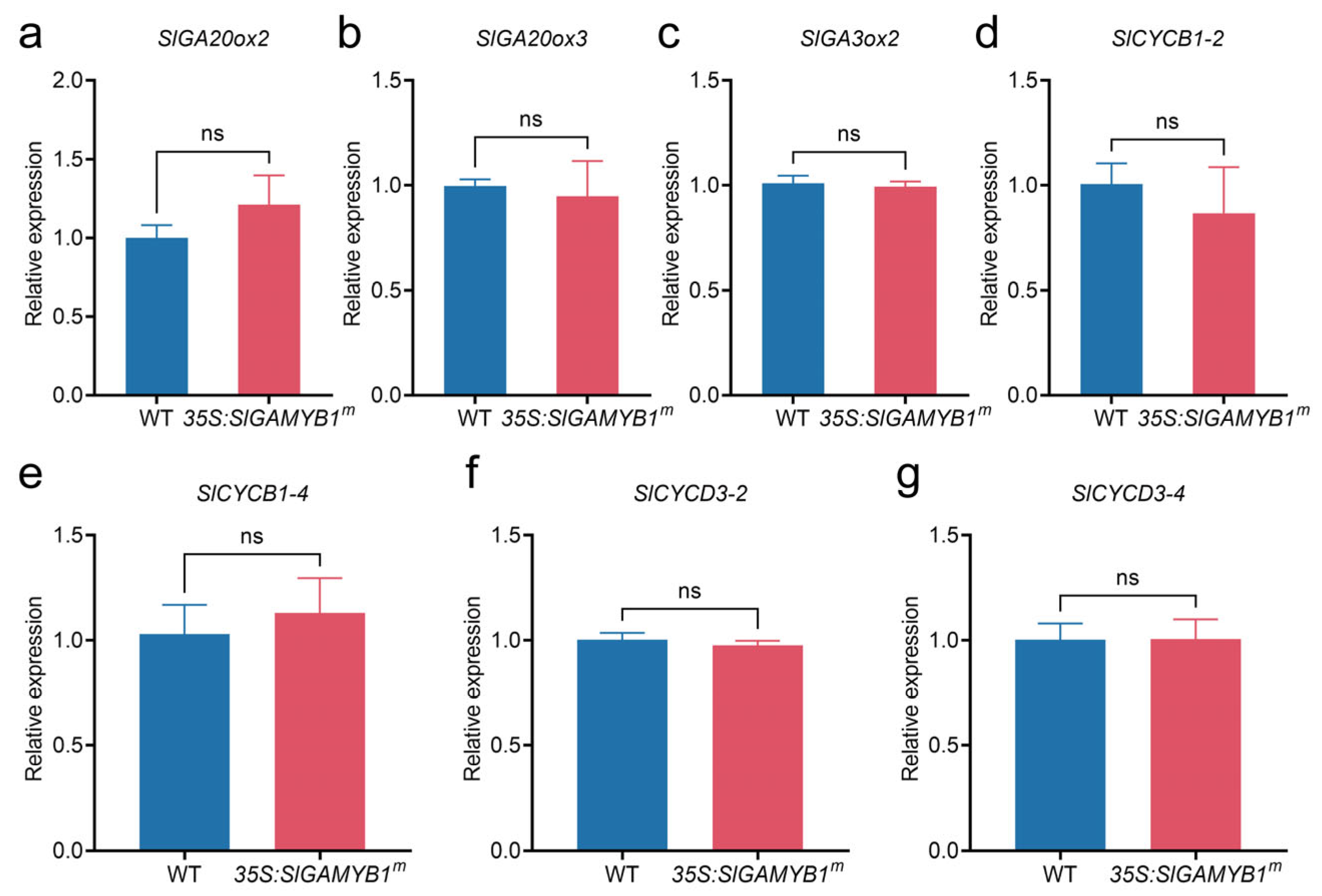
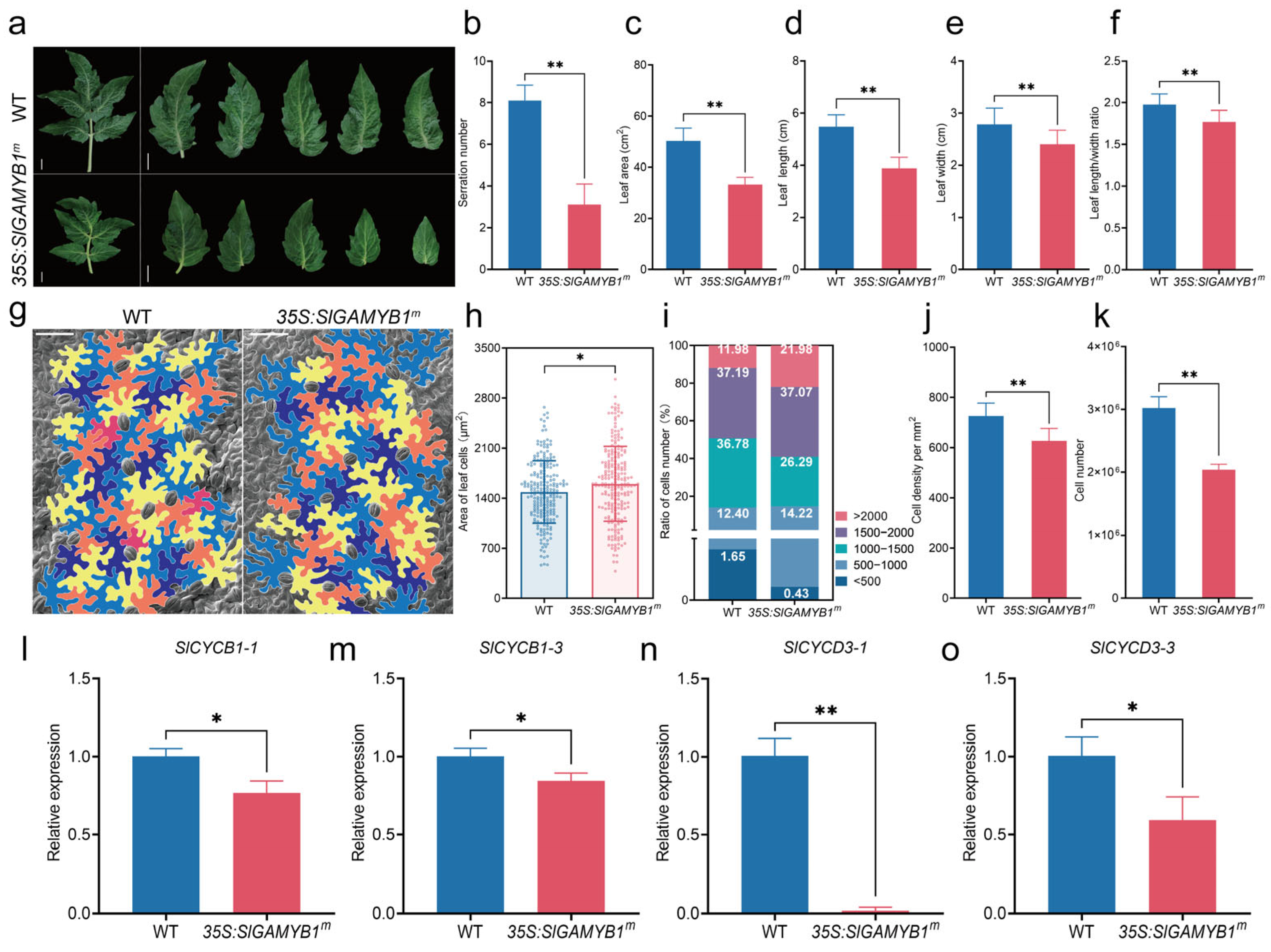
2.3. SlGAMYB1 Plays a Crucial Role in Controlling Leaf Morphology and Cell Characteristics in Tomato
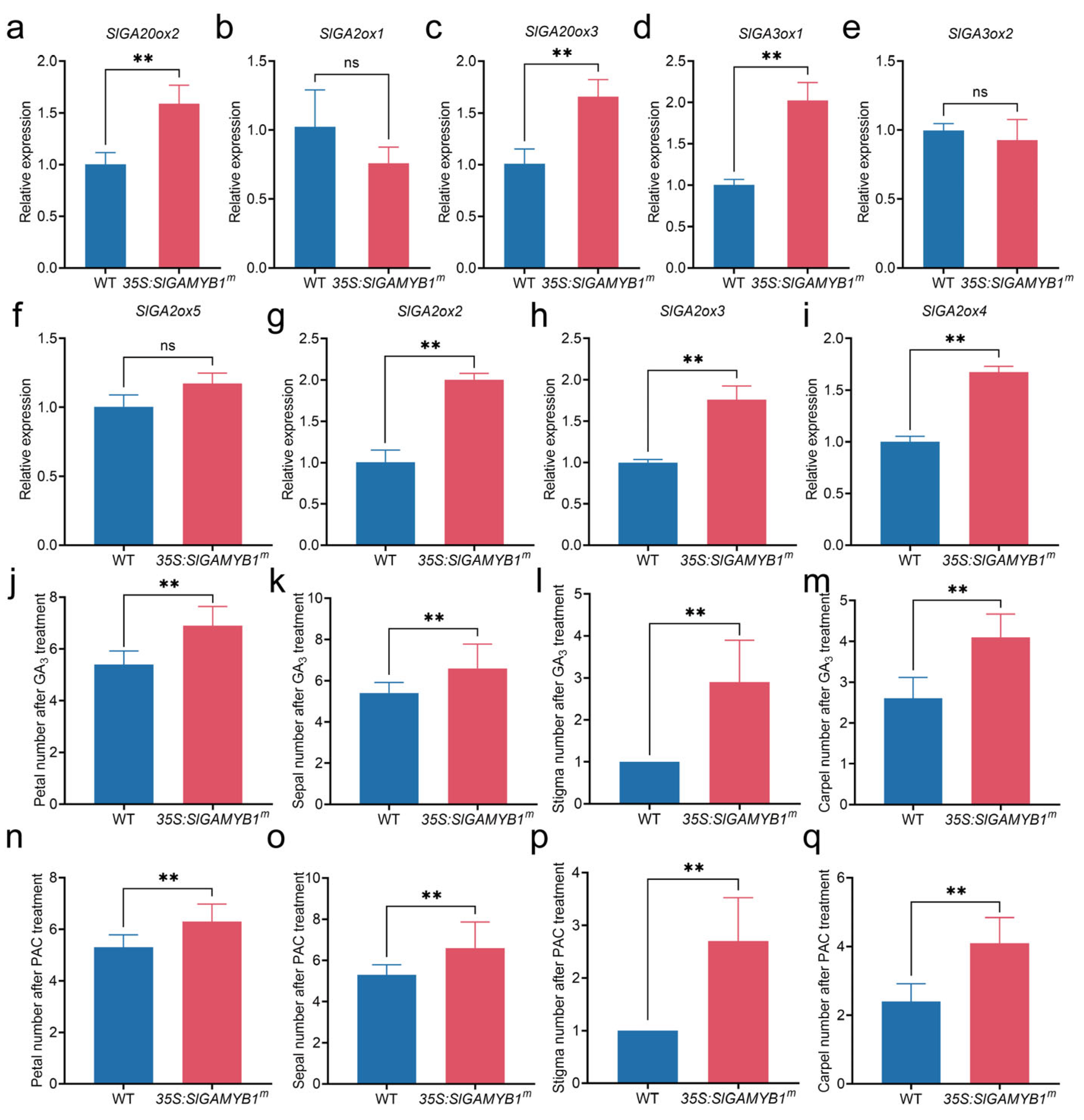
2.4. Overexpression of SlGAMYB1 in Tomato Promotes Flowering and Leads to an Increased Number of Floral Organs
3. Discussion
3.1. Evolutionary Conservation and Functional Diversification of SlGAMYB1 in Tomato
3.2. SlGAMYB1 Orchestrates GA Homeostasis to Fine-Tune Plant Stature in Tomato
3.3. SlGAMYB1 Modulates Leaf Development Through Differential Regulation of Cyclin-Dependent Cell Cycle Progression
3.4. SlGAMYB1 Promotes Floral Organogenesis Through WUS-Mediated Shoot Apical Meristem Regulation Independent of GA Signaling
4. Materials and Methods
4.1. Sequence Alignment and Phylogenetic Analysis
4.2. Plant Materials and Growth Conditions
4.3. Vector Construction and Plant Transformation
4.4. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
4.5. Plant Height Analyses
4.6. Gibberellin Quantification and GA3/Paclobutrazol (PAC) Treatment
4.7. Scanning Electron Microscopy (SEM)
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| GA | Gibberellin |
| PAC | Paclobutrazol |
| SEM | Scanning electron microscopy |
| qRT-PCR | Quantitative real-time PCR |
| GA1/3/4 | Gibberellic acid (bioactive GA form) |
| LC-MS | Liquid chromatography–mass spectrometry |
| ORF | Open reading frame |
| UTR | Untranslated region |
| NJ | Neighbor-joining |
| SAM | Shoot apical meristem |
| SEM | Standard error of the mean |
| ANOVA | Analysis of variance |
Appendix A

| Different Sizes of Cell | WT | 35S:SlGAMYB1m |
|---|---|---|
| <500 μm2 | 1.65% | 0.43% |
| 500–1000 μm2 | 12.40% | 14.22% |
| Gene | Primer Name | Sequence | Purpose |
|---|---|---|---|
| SlGAMYB1 | SlGAMYB1-qF | GAGATTAAGCAAGAGATTGGCG | qRT-PCR |
| SlGAMYB1 | SlGAMYB1-qR | AACCAAAAGGATCTTTCGAAGC | qRT-PCR |
| Actin | Actin-F | TGTTGCTATTCAGGCTGTGC | qRT-PCR |
| Actin | Actin-R | CTGCTCCTGGCAGTTTCAAT | qRT-PCR |
| SlGAMYB1 | PBI-SlGAMYB1m-F1 | ATGAGCATCAAAAGTGAAACC | Overexpression |
| SlGAMYB1 | PBI-SlGAMYB1m-R1 | TCATAATCTCATTCTTCCTGTTG | Overexpression |
| SlGAMYB1 | PBI-SlGAMYB1m-F2 | CAGAGCCCACATGGGCAATGAAGCTGGAACTACCGTCCTTACAAAACCAGACAGAGAAC | Overexpression |
| SlGAMYB1 | PBI-SlGAMYB1m-R2 | TAAGGACGGTAGTTCCAGCTTCATTGCCCATGTGGGCTCTGAAGAGGAGTTGCCATTTA | Overexpression |
| SlGAMYB1 | SlGAMYB1-qF | GAGATTAAGCAAGAGATTGGCG | qRT-PCR |
| TUB | TUB-F | TTGGTTTTGCACCACTGACTTC | qRT-PCR |
| TUB | TUB-R | AAGCTCTGGCACTGTCAAAGC | qRT-PCR |
| EF-1α | EF-1α-F | ATTGGAAATGGATATGCTCCA | qRT-PCR |
| EF-1α | EF-1α-R | TCCTTACCTGAACGCCTGTCA | qRT-PCR |
| UBI | UBI-F | TCGTAAGGAGTGCCCTAATGCTGA | qRT-PCR |
| UBI | UBI-R | CAATCGCCTCCAGCCTTGTTGTAA | qRT-PCR |
| SlCycB1-1 | SlCycB1-1-F | CTGGTTTCTCAGAGTCTCAAGT | qRT-PCR |
| SlCycB1-1 | SlCycB1-1-R | ACCTTAAGCTTGTGATTTGCAG | qRT-PCR |
| SlCycB1-2 | SlCycB1-2-F | GGACAGTTGGAGTGGTACTTAA | qRT-PCR |
| SlCycB1-2 | SlCycB1-2-R | GCATAATTCATCAGCCCCAATT | qRT-PCR |
| SlCycB1-3 | SlCycB1-3-F | GGCTGATGAACTACACTACTGT | qRT-PCR |
| SlCycB1-3 | SlCycB1-3-R | AACTTCCTATAAACCGCCTTCA | qRT-PCR |
| SlCycB1-4 | SlCycB1-4-F | GCTGCGGATGTTGATAATCATT | qRT-PCR |
| SlCycB1-4 | SlCycB1-4-R | TGTAGTCATTCACTCGACCTTC | qRT-PCR |
| SlCycD3-1 | SlCycD3-1-F | CTGTTTTTGAGAATCGAGTCCG | qRT-PCR |
| SlCycD3-1 | SlCycD3-1-R | TCATCCTCTAACAAATCACCCC | qRT-PCR |
| SlCycD3-2 | SlCycD3-2-F | CTCTGCTCAAACTGCAATTCTT | qRT-PCR |
| SlCycD3-2 | SlCycD3-2-R | TGGGTCTCTTCAATTTTTGCAG | qRT-PCR |
| SlCycD3-3 | SlCycD3-3-F | GGAAGAAGAAGAACTTACCTCTCT | qRT-PCR |
| SlCycD3-3 | SlCycD3-3-R | ACTGCAAGAAATCCAGTTTGAG | qRT-PCR |
| SlCycD3-4 | SlCycD3-4-F | TACTGCTACCACTGCTGTTTTA | qRT-PCR |
| SlCycD3-4 | SlCycD3-4-R | CTGACTCATCCAAGGCTTATCT | qRT-PCR |
| SlCycD3-5 | SlCycD3-5-F | ATGTGACATGTTCTGGGAAGAT | qRT-PCR |
| SlCycD3-5 | SlCycD3-5-R | CTAACACAGCAGTCAAAGCATT | qRT-PCR |
| SlCycD3-6 | SlCycD3-6-F | CCAAAGTATGTGTTTGAGGCAA | qRT-PCR |
| SlCycD3-6 | SlCycD3-6-R | GGTGTCACTGGATTCATTTTCC | qRT-PCR |
| SlCycD3-7 | SlCycD3-7-F | AGATGAGGGAGATTTGGGAGGA | qRT-PCR |
| SlCycD3-7 | SlCycD3-7-R | TCTTAACCCCAACAAAACCCCA | qRT-PCR |
| SlGA20ox2 | RT-SlGA20ox2-F | GTGATCCGATTGCAGCTAAGCG | qRT-PCR |
| SlGA20ox2 | RT-SlGA20ox2-R | ACCGATGTGCAAGTGAGATAAG | qRT-PCR |
| SlGA20ox3 | RT-SlGA20ox3-F | CTAGTGTTACTAGAGAACTACA | qRT-PCR |
| SlGA20ox3 | RT-SlGA20ox3-R | TGTCAACCCATGGTTAACCAC | qRT-PCR |
| SlGA3ox1 | RT-SlGA3ox1-F | GAATCCCATGCATGGAATCAT | qRT-PCR |
| SlGA3ox1 | RT-SlGA3ox1-R | TGTTATCGAGGTCGATCACTGG | qRT-PCR |
| SlGA3ox2 | RT-SlGA3ox2-F | ATTGGACGACGATGGATCGCG | qRT-PCR |
| SlGA3ox2 | RT-SlGA3ox2-R | GCATGCATGGCCAATTGTATCC | qRT-PCR |
| SlGA2ox1 | RT-SlGA2ox1-F | CATAGTGAAAGCCTCTGAAG | qRT-PCR |
| SlGA2ox1 | RT-SlGA2ox1-R | CAACTTCACCATTATCTCCA | qRT-PCR |
| SlGA2ox2 | RT-SlGA2ox2-F | CTCATCGTTAATGCCTGCGAAG | qRT-PCR |
| SlGA2ox2 | RT-SlGA2ox2-R | ACTTGATGGCTTCGGATTCGAG | qRT-PCR |
| SlGA2ox4 | RT-SlGA2ox4-F | CTCATCGTTAATGCCTGCGAAG | qRT-PCR |
| SlGA2ox4 | RT-SlGA2ox4-R | GATCAGCAGGCCCTGCCTTTAG | qRT-PCR |
| SlGA2ox5 | RT-SlGA2ox5-F | GAACCTCATCGTTGAGGCCTGC | qRT-PCR |
| SlGA2ox5 | RT-SlGA2ox5-R | GATGGCTTCGGATTCGAGTTTAC | qRT-PCR |
| DELLA | RT-DELLA-F | CGATGGTTACAGGGTGGAAGAA | qRT-PCR |
| DELLA | RT-DELLA-R | CAGGCGGAGGTAGCTATAAGTG | qRT-PCR |
| Protein | Species | Amino Acid Sequences |
|---|---|---|
| SlGAMYB1 | Solanum lycopersicum | MSIKSETEERMTSKVDMDSPDEASGGDLGESVPLKKGPWTSAEDVILVDYVMTHGEGNWNAVQRHSGLARCGKSCRLRWANHLRPDLKKGAFTPEEEQRIVELHAKMGNKWARMAVELPGRTDNEIKNYWNTRIKRRQRAGLPIYPADISFMASQNKQNEELGAFSSADAQNPDVLGINNFEIPAVEFKKLELTHLLYPPQLADIPARSLLNDPVSNFLSQGHRAPYSSTYFLSTTYPAKRIRGSESVFSGSNGDLLNSLQYQNDGSLLAQAQAQPLDFSSYNHNLTYDDQRAISNIVPGGHAYLNGNSSSEPTWAMKLELPSLQNQTENWGSPHSALPSLDSVDILIQSPPAGHSESGSLSPSNSGLLDAVLHESQTMKASNDNSYQGNETSGNAVNNSCPDLKGCDIYGHPVSPLSQFSASVFSDYAPINESSLHEFPSMATMPGGEIKQEIGDLSPLDDEDNTSNQTIFSSPKTQHANNHLASKDPFGSCFFDDCDWDCKQIHAVTTSSGQANGHNSCSWDAISAMEATGRMRL |
| SlGAMYB2 | Solanum lycopersicum | MSMTSESDDRMTSQDGVDSPSAEEACGGGNTGGGLPLKKGPWTSAEDAILVEYVTKHGEGNWNAVQKHSGLARCGKSCRLRWANHLRPDLKKGAFTPEEERHIIELHAKMGNKWARMAAELPGRTDNEIKNYWNTRIKRRQRAGLPIYPSDICFQSITENKQNEELGTFSSADSQYPDFFPMNYEIPAVEFKRLEFNQHLCPPALLDIPTGGILDIPGRSLLAQGLNSAYYSRSFLSTTP |
| AtMYB33 | Arabidopsis thaliana | PAKRIRGSESLFSGLNGDCSPSKNDVSFSTCHQHQDDGSLLAQSMGFSSSFNQNLTSDYHPSSLGVIPGSHALLNGHTSSSEPSWAKKLELPSLQSTIASWGLVTSPLPSLDSVDTLIQSPPTEHTESCNLSPRNSGLLDAVLHESQTMKASKSILHQENSGDVVDNSCPDLHMTEWGQHGDPISPLGHSAASVFSEYTPTSGSSSEEPQLVTMPACKVKQEKFDYGPYDGKDDASNLICPRPDFLLESNCFGHMQNTVRSIWY |
| AtMYB65 | Arabidopsis thaliana | MSYTSTDSDHNESPAADDNGSDCRSRWDGHALKKGPWSSAEDDILIDYVNKHGEGNWNAVQKHTSLFRCGKSCRLRWANHLRPNLKKGAFSQEEEQLIVELHAKMGNRWARMAAHLPGRTDNEIKNYWNTRIKRRQRAGLPLYPPEMHVEALEWSQEYAKSRVMGEDRRHQDFLQLGSCESNVFFDTLNFTDMVPGTFDLADMTAYKNMGNCASSPRYENFMTPTIPSSKRLWESELLYPGCSSTIKQEFSSPEQFRNTSPQTISKTCSFSVPCDVEHPLYGNRHSPVMIPDSHTPTDGIVPYSKPLYGAVKLELPSFQYSETTFDQWKKSSSPPHSDLLDPFDTYIQSPPPPTGGEESDLYSNFDTGLLDMLLLEAKIRNNSTKNNLYRSCASTIPSADLGQVTVSQTKSEEFDNSLKSFLVHSEMSTQNADETPPRQREKKRKPLLDITRPDVLLASSWLDHGLGIVKETGSMSDALAVLLGDDIGNDYMNMSVGASSGVGSCSWSNMPPVCQMTELP |
| OsGAMYB | Oryza saliva | MSYTTATADSDDGMHSSIHNESPAPDSISNGCRSRGKRSVLKKGPWTSTEDGILIDYVKKHGEGNWNAVQKHTSLARCGKSCRLRWANHLRPNLKKGAFSQEEEQLIVEMHAKMGNKWAQMAEHLPGRTDNEIKNYWNTRIKRRQRAGLPLYPPEIYVDDLHWSEEYTKSNIIRVDRRRRHQDFLQLGNSKDNVLFDDLNFAASLLPAASDLSDLVACNMLGTGASSSRYESYMPPILPSPKQIWESGSRFPMCSSNIKHEFQSPEHFQNTAVQKNPRSCSISPCDVDHHPYENQHSSHMMMVPDSHTVTYGMHPTSKPLFGAVKLELPSFQYSETSAFDQWKTTPSPPHSDLLDSVDAYIQSPPPSQVEESDCFSSCDTGLLDMLLHEAKIKTSAKHSLLMSSPQKSFSSTTCTTNVTQNVPRGSENLIKSGEYEDSQKYLGRSEITSPSQLSAGGFSSAFAGNVVKTEELDQVWEPKRVDITRPDVLLASSWLDQGCYGIVSDTSSMSDALALLGGDDIGNSYVTVGSSSGQAPRGVGSYGWTNMPPVWSL |
| CaGAMYB1 | Capsicum annuum | MYRVKSSDCDMIHMDSVADDGSSGGSHRGGGKKGWTSADAIVDYVKKHGGNWNAVKNTGRCGKSCRRWANHRNKKGATARIIHSKMGNKWARMAAHGRTDNIKNYWNTRIKRCRAGIYTSVCNSSNDCSSDDCGNSNDNANGYDTCDNIANSAYAHSAVSISNGSASKSCSMDVNTGMKSDGVGSDTINGVISSVDSNDSKKAVGDYHANSTSKIIAGGANGSHANGNSASRTSGKMSDTSDNSWKYTVAATVDYSAATSVKSCASRNSGIHATRSGKNTSVISSSSSVGTCNTTVSDMCYWHGNDCASGNSTSTVSAASDISKVSASTSMGSGVMGKYGDTSHNRDASGNTADSVNNAIAMGNDSIDCRVGDGIMNSSSWSNMHACMSK |
| CaGAMYB2 | Capsicum annuum | MSITSETDDMMTSKVDIDSPDEASGGESVPLKKGPWTTVEDAILVDYVMTNGEGNWNAVQRHSGLARCGKSCRLRWANHLRPDLKKGAFTPEEERRILELHAKMGNKWARMAAELPGRTDNEIKNYWNTRIKRRQRAGLPVYPPDISFLANQNKQNEELGAFSSVDAQNSNVLGINNFEIPAVEFKNLQLDHLLYPPPLGEIPAVSSFLAQGHRAPYGSTYLLSTMHPSKRIRGSESMFSGSNGDLLLSSSQYHNGGSLLAQPLGFSSYNHHLTYDDDRSFSSVVHGGHACLNGNSSSSEPTWAMKLELPSLQNQTANWGSPPSPLPSLESDDILIQSPPAGNSESGSLSPSNSGLLDAVLYESQTMKASNDNSHQGKETSGDAVNKGWESYGDPVSPLHHFSASVFGEYTPVNGSSLHEFPSVATMPGCKIKKEIGDLAPLDENDDSLNQTIFSSPKTQHAKNSLALKEVISSGFFDDCGWDCKQIRAVATSSGQACGRSSWDAMSAV |
| CsGAMYB1 | Cucumis sativus | MSMTSESDDRMASQDGVDSPSVEEACGGGITGGGLPLKKGPWTSAEDAILMDYVTKHGEGNWNAVQKHSGLARCGKSCRLRWANHLRPDLKKGAFTPEEERRIIELHAKMGNKWARMAAELPGRTDNEIKNYWNTRIKRRQRAGLPIYPPDISFQAISENKQNEELGAFSSTDSQYPDFLPMNNFEIPAVEFKRLEFNKQLCPPALLDIPNGGILDIPGRSLLAQGLNSAYYSRSFLSTMPPAKRIRGAESLFSGLNGDCSPSKNEGSFPTCHQYQDDGSLLAQSMGFSSSFNQNLTSVHHPSSSGVIPGSHAPLNGKTSSSEPLWAEKLELPSFQSQMASWGLSSSPLPSLESVDTVIQSPPTEHTESCNLSPRNSGLLDAVLYESQTMRASKSILHQENSGDVVDNSCPDLHETGWETYGDPISPLGHSAASVFSEYTPTSGSSPEEPQLVTMPGCKVKQEKFDFGPYDGKEDASDLIFSRPDYLLESNCFGHMQKTARSIWH |
| HvGAMYB | Hordeum vulgare | MRHPKNEIEDNLPSQDQTLSPLLDEDSGGNASGIILKKGPWTSAEDEILIEYVKKHGEGNWNAVQKHSGLSRCGKSCRLRWANHLRPNLKKGAFTAEEEHLIIELHAKMGNKWARMAGHLPGRTDNEIKNYWNTRIKRRQRAGLPLYPPEVCLRTWQALQQTQDSGGSTVVDTDHHDLLRSNSYDIPDVTFHSLKPQSALSYMPELPDISSCMLKRGLDTSQYCNLVQPTFHRQKRFRDSASLFPGPDGSVKTPFHQFEDNSYSQAAQSFGTPFAHESNPTTKNAMSFGSFEGSHSLTNGNSSASQHSKETEKLELPSLQYPETDLTSWDTTIQPAMFESVDPFIQSTPTFVLAPDRTSPCHSGLLESLVYSKTMGPKNHPSDKNSNSCSVTPGDVTDSYNMAASKTEIDDYTEVISPFGHSTSSLFSECTPISATGSSYEDPTLTEAFSGSHVKSEPFDHAWTPDREKAAKSRVNFARPDALLASDWHDRSSGIVEDTTNVTDAISLLLGDDLAADYEHFPNGISTTHSAWGLDSCSWNNMPAVCHMSDLP |
References
- Shi, B.H.; Felipo-Benavent, A.; Cerutti, G.; Galvan-Ampudia, C.; Jilli, L.; Brunoud, G.; Mutterer, J.; Vallet, E.; Sakvarelidze-Achard, L.; Davière, J.M.; et al. A quantitative gibberellin signaling biosensor reveals a role for gibberellins in internode specification at the shoot apical meristem. Nat. Commun. 2024, 15, 3895. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tian, Y.H.; Wu, K.; Ye, Y.F.; Yu, J.P.; Zhang, J.Q.; Liu, Q.; Hu, M.Y.; Li, H.; Tong, Y.P.; et al. Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 2018, 560, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, A.M.; Pan, Y.Q.; Chen, Z.L.; Qin, C.X.; Su, Z.L.; Lakshmanan, P.; Song, J.M.; Liao, F.; Huang, D.L. Gibberellin biosynthesis gene ScGA20 oxidase enhances sugarcane growth by modulating genes associated with phytohormone and growth processes. Plant Physiol. Bioch. 2025, 221, 109652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, B.; Yang, T.; Zhang, J.; Liu, B.; Zhan, X.; Liang, Y. The GAMYB-like gene SlMYB33 mediates flowering and pollen development in tomato. Hortic. Res. 2020, 7, 133. [Google Scholar] [CrossRef]
- Chai, Z.; Fang, J.L.; Yao, W.; Zhao, Y.; Cheng, G.Y.; Akbar, S.; Khan, M.T.; Chen, B.S.; Zhang, M.Q. ScGAIL, a sugarcane N-terminal truncated DELLA-like protein, participates in gibberellin signaling in Arabidopsis. J. Exp. Bot. 2022, 73, 3462–3476. [Google Scholar] [CrossRef]
- Wu, X.Y.; Xia, M.; Su, P.; Zhang, Y.F.; Tu, L.C.; Zhao, H.; Gao, W.; Huang, L.Q.; Hu, Y.T. MYB transcription factors in plants: A comprehensive review of their discovery, structure, classification, functional diversity and regulatory mechanism. Int. J. Biol. Macromol. 2024, 282, 136652. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef]
- Tang, Y.H.; Lu, L.L.; Sheng, Z.P.; Zhao, D.Q.; Tao, J. An R2R3-MYB network modulates stem strength by regulating lignin biosynthesis and secondary cell wall thickening in herbaceous peony. Plant J. 2023, 113, 1237–1258. [Google Scholar] [CrossRef]
- Tang, F.; Jiao, B.; Zhang, M.; He, M.H.; Su, R.Y.; Luo, K.M.; Lan, T. PtoMYB031, the R2R3 MYB transcription factor involved in secondary cell wall biosynthesis in poplar. Front. Plant Sci. 2024, 14, 1341245. [Google Scholar] [CrossRef]
- Ko, S.S.; Li, M.J.; Ho, Y.C.; Yu, C.P.; Yang, T.T.; Lin, Y.J.; Hsing, H.C.; Chen, T.K.; Jhong, C.M.; Li, W.H.; et al. Rice transcription factor GAMYB modulates bHLH142 and is homeostatically regulated by TDR during anther tapetal and pollen development. J. Exp. Bot. 2021, 72, 4888–4903. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Lohe, A.; Wong, G. Biology and Function of miR159 in Plants. Plants 2019, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Wu, Z.; Feng, J.X.; Yuan, G.Z.; He, L.; Zhang, D.H.; Teng, N.J. A Novel R2R3-MYB Gene LoMYB33 From Lily Is Specifically Expressed in Anthers and Plays a Role in Pollen Development. Front. Plant Sci. 2021, 12, 730007. [Google Scholar] [CrossRef]
- Gubler, F.; Kalla, R.; Roberts, J.K.; Jacobsen, J.V. Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 1995, 7, 1879–1891. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, F.L.; Cao, H.; Peng, H.R.; Ni, Z.F.; Sun, Q.X.; Yao, Y.Y. TamiR159 Directed Wheat TaGAMYB Cleavage and Its Involvement in Anther Development and Heat Response. PLoS ONE 2012, 7, e48445. [Google Scholar] [CrossRef]
- Abraham, Z.; Iglesias-Fernández, R.; Martínez, M.; Rubio-Somoza, I.; Díaz, I.; Carbonero, P.; Vicente-Carbajosa, J. A Developmental Switch of Gene Expression in the Barley Seed Mediated by HvVP1 (Viviparous-1) and HvGAMYB Interactions. Plant Physiol. 2016, 170, 2146–2158. [Google Scholar] [CrossRef] [PubMed]
- Ogrodowicz, P.; Kuczynska, A.; Krajewski, P.; Kempa, M. The effects of heading time on yield performance and HvGAMYB expression in spring barley subjected to drought. J. Appl. Genet. 2023, 64, 289–302. [Google Scholar] [CrossRef]
- Wang, R.N.; Yang, X.Y.; Guo, S.; Wang, Z.H.; Zhang, Z.H.; Fang, Z.M. MiR319-targeted OsTCP21 and OsGAmyb regulate tillering and grain yield in rice. J. Integr. Plant Biol. 2021, 63, 1260–1272. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.T.; Wang, K.; Ge, L.M.; Chen, X.N.; Zhang, L.L.; Song, X.Y. Transcription factor TaGAMYB from wheat (Triticum aestivum L.) regulates flowering time and fertility in transgenic Arabidopsis thaliana. Planta 2023, 257, 16. [Google Scholar] [CrossRef]
- Allen, R.S.; Li, J.; Alonso-Peral, M.M.; White, R.G.; Gubler, F.; Millar, A.A. MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects. Silence 2010, 1, 18. [Google Scholar] [CrossRef]
- Imran, M.; Liu, T.F.; Wang, Z.; Wang, M.; Liu, S.L.; Gao, X.Y.; Wang, A.N.; Liu, S.F.; Tian, Z.X.; Zhang, M. Evolutionary conservation of nested MIR159 structural microRNA genes and their promoter characterization in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 948751. [Google Scholar] [CrossRef] [PubMed]
- Csukasi, F.; Donaire, L.; Casañal, A.; Martínez-Priego, L.; Botella, M.A.; Medina-Escobar, N.; Llave, C.; Valpuesta, V. Two strawberry miR159 family members display developmental-specific expression patterns in the fruit receptacle and cooperatively regulate Fa-GAMYB. New Phytol. 2012, 195, 47–57. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Wan, J.; Dang, K.T.; Meng, S.J.; Hu, D.S.; Lin, Y.; Qiu, X.Q.; Guo, Z.Y.; Fu, Z.Y.; Ding, D.; et al. zma-miR159 targets ZmMYB74 and ZmMYB138 transcription factors to regulate grain size and weight in maize. Plant Physiol. 2023, 193, 2430–2441. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, F.; Deng, Y.; Zhong, F.; Tian, P.; Lin, D.; Deng, J.; Zhang, Y.; Huang, T. Sly-miR159 regulates fruit morphology by modulating GA biosynthesis in tomato. Plant Biotechnol. J. 2022, 20, 833–845. [Google Scholar] [CrossRef]
- da Silva, E.M.; Silva, G.; Bidoia, D.B.; da Silva Azevedo, M.; de Jesus, F.A.; Pino, L.E.; Peres, L.E.P.; Carrera, E.; López-Díaz, I.; Nogueira, F.T.S. microRNA159-targeted SlGAMYB transcription factors are required for fruit set in tomato. Plant J. 2017, 92, 95–109. [Google Scholar] [CrossRef]
- Zhao, P.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol. Genet. Genom. 2014, 289, 1183–1207. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.L.; Xiao, P.; Chen, D.L.; Xu, L.; Zhang, B.H. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Liu, X.; Yang, W.; Wang, J.; Yang, M.; Wei, K.; Liu, X.; Qiu, Z.; van Giang, T.; Wang, X.; Guo, Y.; et al. SlGID1a Is a Putative Candidate Gene for qtph1.1, a Major-Effect Quantitative Trait Locus Controlling Tomato Plant Height. Front. Genet. 2020, 11, 881. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Tang, Y.; Chu, Z.; Peng, Y.; Chen, J.; Yu, H.; Shi, C.; Jafar, J.; Chen, R.; Tang, Y.; et al. SlZF3 regulates tomato plant height by directly repressing SlGA20ox4 in the gibberellic acid biosynthesis pathway. Hortic. Res. 2023, 10, uhad025. [Google Scholar] [CrossRef]
- Chu, L.L.; Liu, D.H.; Li, C.L.; Liu, J.H. Dwarfing of fruit trees: From old cognitions to new insights. Hortic. Adv. 2025, 3, 7. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Debernardi, J.M.; Palatnik, J.F. Morphogenesis of simple leaves: Regulation of leaf size and shape. Wiley. Interdiscip. Rev. Dev. Biol. 2014, 3, 41–57. [Google Scholar] [CrossRef] [PubMed]
- Ding, A.M.; Xu, C.T.; Xie, Q.; Zhang, M.J.; Yan, N.; Dai, C.B.; Lv, J.; Cui, M.M.; Wang, W.F.; Sun, Y.H. ERF4 interacts with and antagonizes TCP15 in regulating endoreduplication and cell growth in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 1673–1689. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Jenke, H.; Strauss, S.; Bazakos, C.; Mosca, G.; Lymbouridou, R.; Kierzkowski, D.; Neumann, U.; Naik, P.; Huijser, P.; et al. Cell-cycle-linked growth reprogramming encodes developmental time into leaf morphogenesis. Curr. Biol. 2024, 34, 541–556. [Google Scholar] [CrossRef]
- Zhang, T.Y.; Wang, X.; Lu, Y.G.; Cai, X.F.; Ye, Z.B.; Zhang, J.H. Genome-Wide Analysis of the Cyclin Gene Family in Tomato. Int. J. Mol. Sci. 2014, 15, 120–140. [Google Scholar] [CrossRef]
- Chu, Y.H.; Jang, J.C.; Huang, Z.; van der Knaap, E. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct. 2019, 3, e00142. [Google Scholar] [CrossRef]
- Yuste-Lisbona, F.J.; Fernández-Lozano, A.; Pineda, B.; Bretones, S.; Ortíz-Atienza, A.; García-Sogo, B.; Müller, N.A.; Angosto, T.; Capel, J.; Moreno, V.; et al. ENO regulates tomato fruit size through the floral meristem development network. Proc. Natl. Acad. Sci. USA 2020, 117, 8187–8195. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Liu, X.; Zhou, Y. Transcriptional circuits in control of shoot stem cell homeostasis. Curr. Opin. Plant Biol. 2020, 53, 50–56. [Google Scholar] [CrossRef]
- Becker, A. A molecular update on the origin of the carpel. Curr. Opin. Plant Biol. 2020, 53, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.F.; Hu, C.; Cui, Y.W.; Zeng, L.; Li, S.J.N.; Zhu, M.S.; Meng, F.H.; Huang, S.T.; Long, L.; Yi, J.; et al. Conserved and differentiated functions of CIK receptor kinases in modulating stem cell signaling in Arabidopsis. Mol. Plant 2021, 14, 1119–1134. [Google Scholar] [CrossRef]
- Hong, L.; Fletcher, J.C. Stem Cells: Engines of Plant Growth and Development. Int. J. Mol. Sci. 2023, 24, 14889. [Google Scholar] [CrossRef]
- Shan, F.; Zhang, R.; Zhang, J.; Wang, C.; Lyu, X.; Xin, T.; Yan, C.; Dong, S.; Ma, C.; Gong, Z. Study on the Regulatory Effects of GA3 on Soybean Internode Elongation. Plants 2021, 10, 1737. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; Jiao, Z.H.; Zhang, Y.H.; Shi, Q.B.; Wang, Q.B.; Zhou, F.L.; Xu, D.; Wang, G.D.; Kong, F.Y.; Zhang, H.S.; et al. DBB2 regulates plant height and shade avoidance responses in maize. J. Integr. Plant Biol. 2025, 67, 1323–1338. [Google Scholar] [CrossRef] [PubMed]
- Shohat, H.; Eliaz, N.I.; Weiss, D. Gibberellin in tomato: Metabolism, signaling and role in drought responses. Mol. Hortic. 2021, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.Y.; Hill, C.B.; Shabala, S.; Li, C.D.; Zhou, M.X. Manipulating GA-Related Genes for Cereal Crop Improvement. Int. J. Mol. Sci. 2022, 23, 14046. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, H.J.; Ma, Z.X.; Huang, J.R. DELLA Proteins Promote Anthocyanin Biosynthesis via Sequestering MYBL2 and JAZ Suppressors of the MYB/bHLH/WD40 Complex in Arabidopsis thaliana. Mol. Plant 2016, 9, 711–721. [Google Scholar] [CrossRef]
- Nohales, M.A.; Kay, S.A. GIGANTEA gates gibberellin signaling through stabilization of the DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 21893–21899. [Google Scholar] [CrossRef]
- Li, T.; Wang, Y.Q.; Natran, A.; Zhang, Y.; Wang, H.; Du, K.X.; Qin, P.; Yuan, H.; Chen, W.L.; Tu, B.; et al. C-TERMINAL DOMAIN PHOSPHATASE-LIKE 3 contributes to GA-mediated growth and flowering by interaction with DELLA proteins. New Phytol. 2024, 242, 2555–2569. [Google Scholar] [CrossRef]
- Bertoni, G. Cell Cycle Regulation by Chlamydomonas Cyclin-Dependent Protein Kinases. Plant Cell 2018, 30, 271. [Google Scholar] [CrossRef]
- Heinzle, C.; Höfler, A.; Yu, J.; Heid, P.; Kremer, N.; Schunk, R.; Stengel, F.; Bange, T.; Boland, A.; Mayer, T.U. Positively charged specificity site in cyclin B1 is essential for mitotic fidelity. Nat. Commun. 2025, 16, 853. [Google Scholar] [CrossRef]
- Cerbantez-Bueno, V.E.; Serwatowska, J.; Rodríguez-Ramos, C.; Cruz-Valderrama, J.E.; de Folter, S. The role of D3-type cyclins is related to cytokinin and the bHLH transcription factor SPATULA in Arabidopsis gynoecium development. Planta 2024, 260, 48. [Google Scholar] [CrossRef]
- Zheng, T.C.; Dai, L.J.; Li, S.; Liu, Y.; Zhao, Z.N.; Yang, C.P.; Qu, G.Z. Populus D-type cyclin gene PsnCYCD1;1 accelerates cell division and participates in secondary growth of vascular bundles. J. Exp. Bot. 2023, 74, 4077–4092. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.J.; Liu, L.; She, L.L.; Zhang, H.R.; Zhang, N.N.; Wang, Y.Q.; Ni, Y.T.; Chen, F.pG.; Wan, F.Y.; Dai, Y.Q.; et al. DRN facilitates WUS transcriptional regulatory activity by chromatin remodeling to regulate shoot stem cell homeostasis in Arabidopsis. PLoS Biol. 2024, 22, e3002878. [Google Scholar] [CrossRef] [PubMed]
- Chetty, V.J.; Ceballos, N.; Garcia, D.; Narváez-Vásquez, J.; Lopez, W.; Orozco-Cárdenas, M.L. Evaluation of four strains for the genetic transformation of tomato (Solanum lycopersicum L.) cultivar Micro-Tom. Plant Cell Rep. 2013, 32, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.L.; Wang, W.S.; Zhang, H.Y.; Liu, X.Q.; Yu, S.B.; Xiong, L.Z.; Luo, J. A Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Li, D.M.; Guo, Z.P.; Liu, C.M.; Li, J.C.; Xu, W.Z.; Chen, Y. Quantification of near-attomole gibberellins in floral organs dissected from a single Arabidopsis thaliana flower. Plant J. 2017, 91, 547–557. [Google Scholar] [CrossRef]
- Galdon-Armero, J.; Arce-Rodriguez, L.; Downie, M.; Li, J.; Martin, C. A Scanning Electron Micrograph-based Resource for Identification of Loci Involved in Epidermal Development in Tomato: Elucidation of a New Function for the Mixta-like Transcription Factor in Leaves. Plant Cell 2020, 32, 1414–1433. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, F.; Wang, F.; Chen, Z.; Huang, T.; Zhao, P. Dual Regulatory Roles of SlGAMYB1 in Tomato Development: GA-Dependent and GA-Independent Mechanisms. Plants 2025, 14, 1613. https://doi.org/10.3390/plants14111613
Zhong F, Wang F, Chen Z, Huang T, Zhao P. Dual Regulatory Roles of SlGAMYB1 in Tomato Development: GA-Dependent and GA-Independent Mechanisms. Plants. 2025; 14(11):1613. https://doi.org/10.3390/plants14111613
Chicago/Turabian StyleZhong, Fanjia, Fengpan Wang, Zike Chen, Tengbo Huang, and Panpan Zhao. 2025. "Dual Regulatory Roles of SlGAMYB1 in Tomato Development: GA-Dependent and GA-Independent Mechanisms" Plants 14, no. 11: 1613. https://doi.org/10.3390/plants14111613
APA StyleZhong, F., Wang, F., Chen, Z., Huang, T., & Zhao, P. (2025). Dual Regulatory Roles of SlGAMYB1 in Tomato Development: GA-Dependent and GA-Independent Mechanisms. Plants, 14(11), 1613. https://doi.org/10.3390/plants14111613






