Genome-Wide Analysis of the Polygalacturonase Gene Family in Macadamia and Identification of Members Involved in Fruit Abscission
Abstract
1. Introduction
2. Results
2.1. Identification of PG Gene Family Members in Macadamia
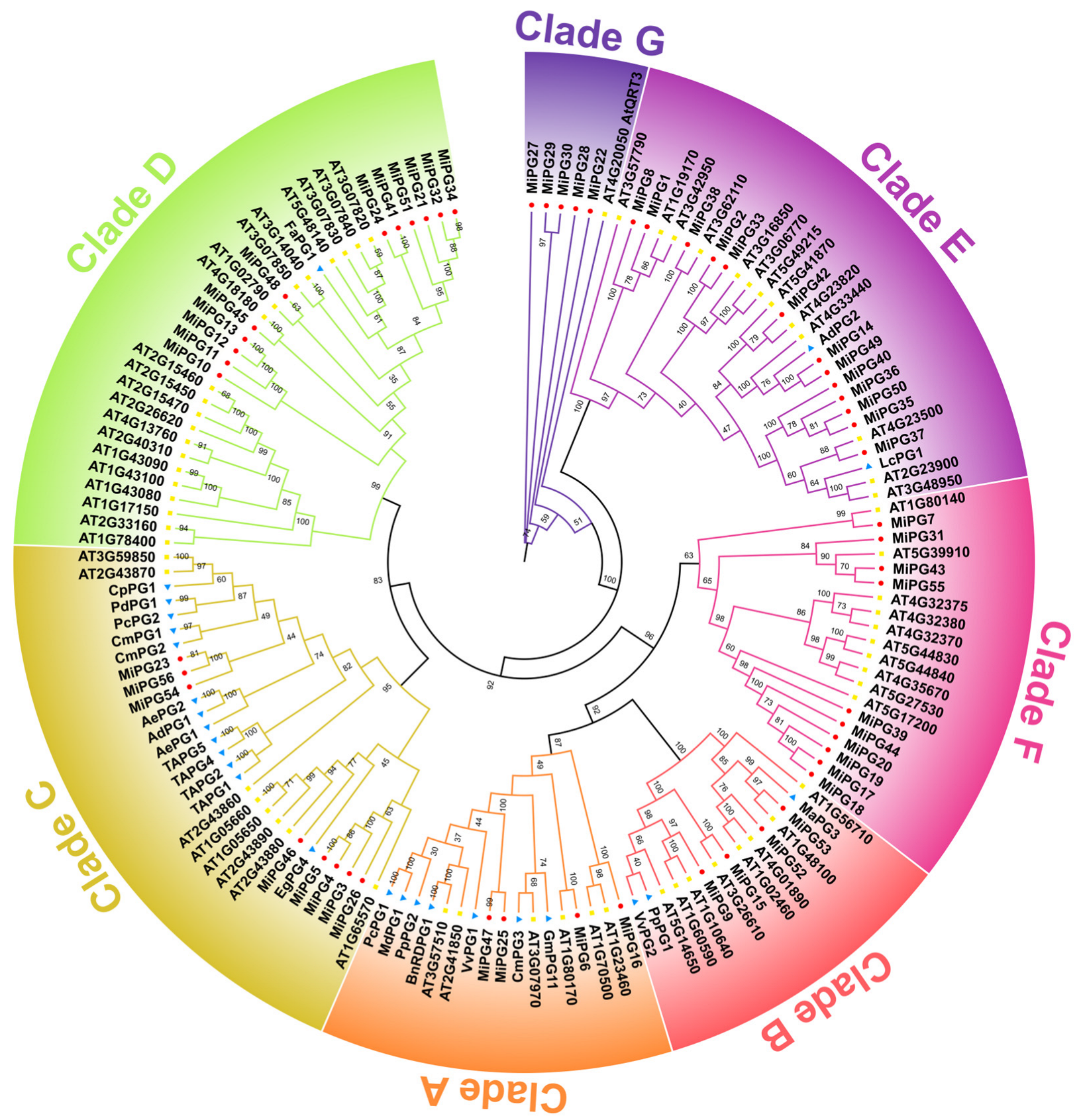
2.2. Phylogenetic Analysis of PG Gene Family Genes in Macadamia
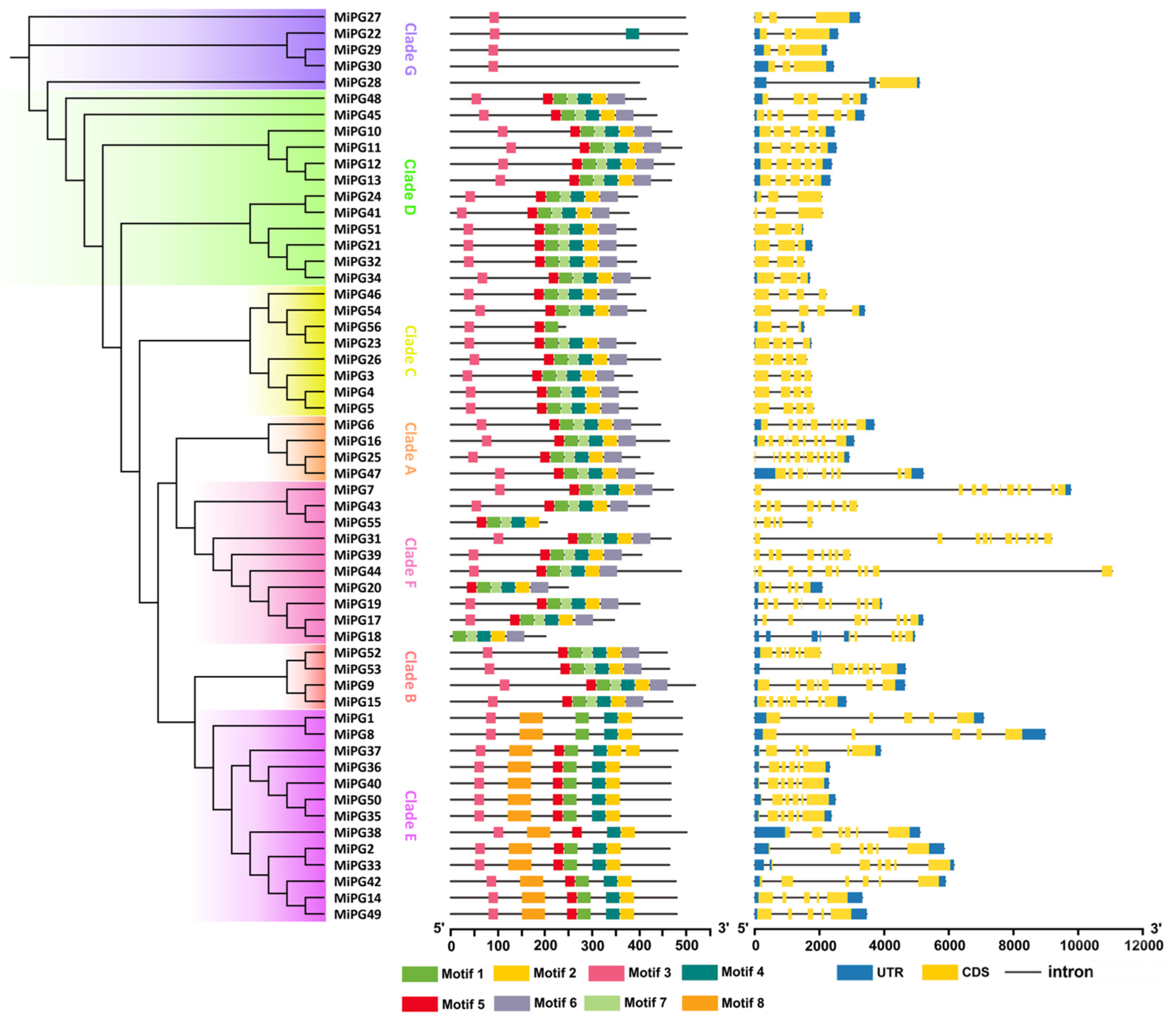
2.3. Gene Structure Analysis of PG Gene Family Genes in Macadamia
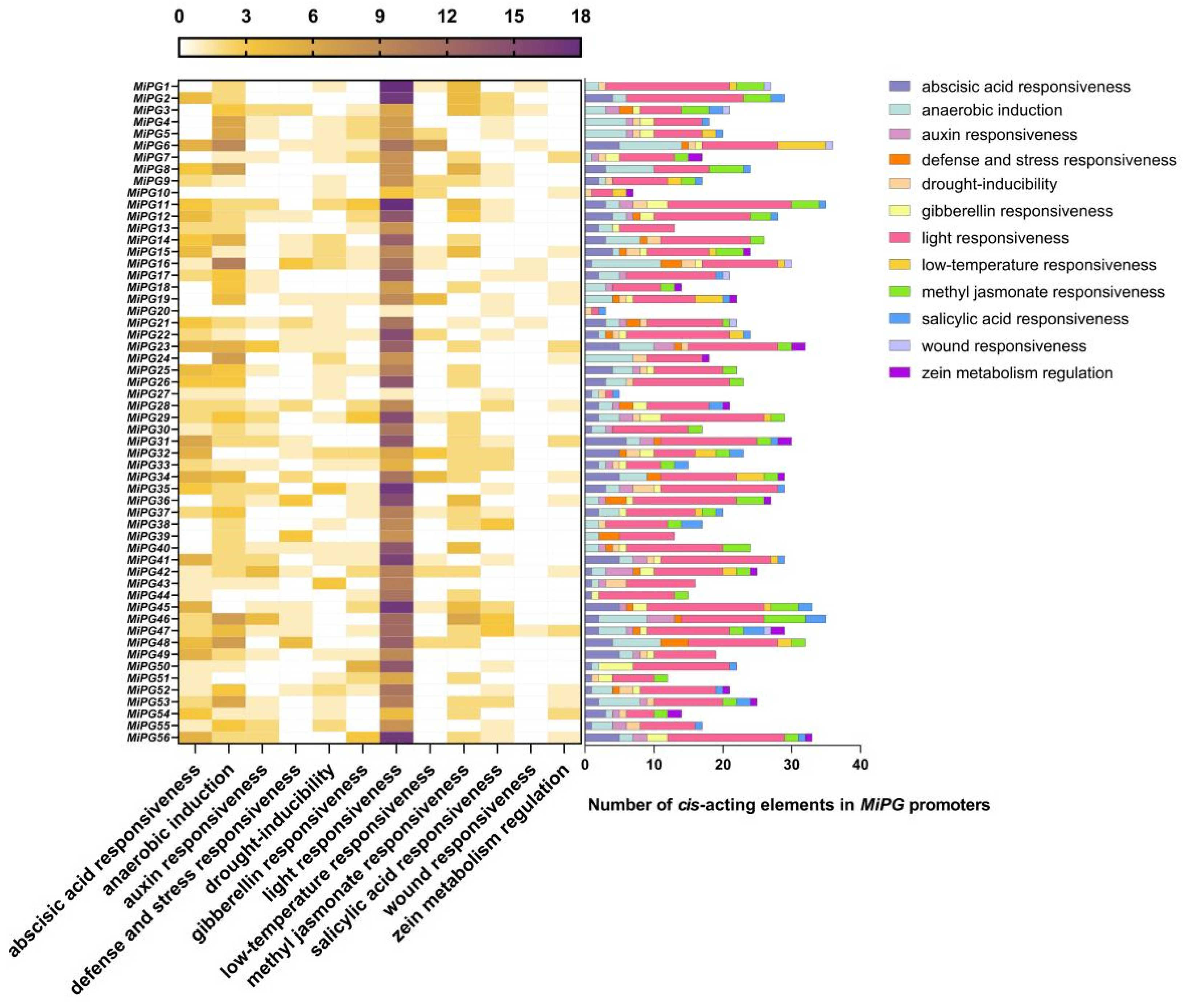
2.4. Cis-Element Analysis of the MiPG Genes
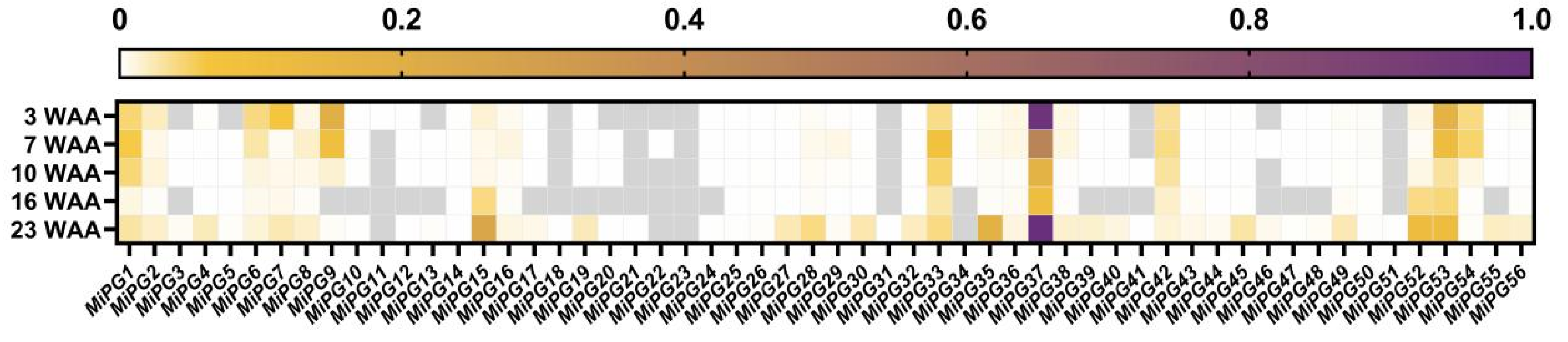
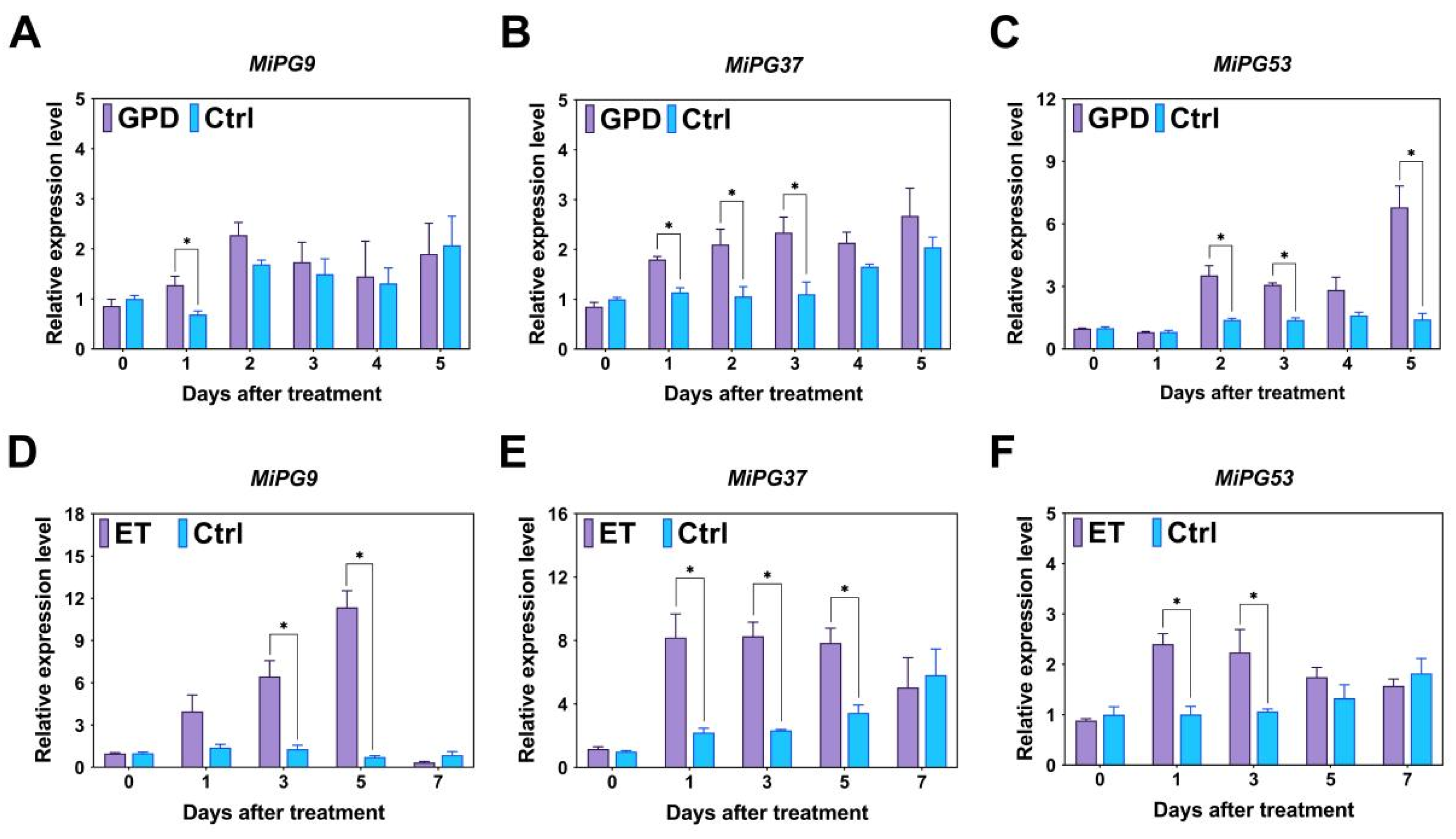
2.5. Expression Profiles of PG Gene Family Genes in Macadamia
2.6. Transient MiPG37 Overexpression Promoted Abscission in Lily Petals
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatment
4.2. Determination of Fruit Abscission
4.3. Identification of Macadamia PG Gene Family Members
4.4. Multiple Sequence Alignment, Phylogenetic Analysis, and Exon/Intron Structure
4.5. Cis-Element Analysis of Macadamia PG Gene Promoters
4.6. Quantitative Real-Time PCR Analysis
4.7. Transient MiPG37 Overexpression in Lily Petals
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| PG | polygalacturonase |
| AZ | abscission zone |
| QRT | QUARTET |
| WAA | weeks after anthesis |
| ABA | abscisic acid |
| qRT-PCR | quantitative real-time PCR |
| GPD | girdling with defoliation |
| ET | ethephon |
| CFAR | cumulative fruit abscission rate |
| Ctrl | Control |
References
- Xie, R.J.; Deng, L.; Jing, L.; He, S.L.; Ma, Y.T.; Yi, S.L.; Zheng, Y.Q.; Zheng, L. Recent advances in molecular events of fruit abscission. Biol. Plant. 2013, 57, 201–209. [Google Scholar] [CrossRef]
- Estornell, L.H.; Agustí, J.; Merelo, P.; Talón, M.; Tadeo, F.R. Elucidating mechanisms underlying organ abscission. Plant Sci. 2013, 199–200, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Chun, J.-P.; Tucker, M.L. Transcriptional regulation of abscission zones. Plants 2019, 8, 154. [Google Scholar] [CrossRef]
- Roberts, J.A.; Elliott, K.A.; Gonzalez-Carranza, Z.H. Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 2002, 53, 131–158. [Google Scholar] [CrossRef]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef]
- Yin, Y.; Chen, H.; Hahn, M.G.; Mohnen, D.; Xu, Y. Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant Physiol. 2010, 153, 1729–1746. [Google Scholar] [CrossRef]
- Jarvis, M.C.; Briggs, S.P.H.; Knox, J.P. Intercellular adhesion and cell separation in plants. Plant Cell Environ. 2003, 26, 977–989. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef]
- Kim, J.; Shiu, S.-H.; Thoma, S.; Li, W.-H.; Patterson, S.E. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006, 7, R87. [Google Scholar] [CrossRef]
- Bunya-atichart, K.; Ketsa, S.; van Doorn, W. Ethylene-sensitive and ethylene-insensitive abscission in Dendrobium: Correlation with polygalacturonase activity. Postharvest Biol. Technol. 2011, 60, 71–74. [Google Scholar] [CrossRef]
- Parra, R.; Paredes, M.A.; Labrador, J.; Nunes, C.; Coimbra, M.A.; Fernandez-Garcia, N.; Olmos, E.; Gallardo, M.; Gomez-Jimenez, M.C. Cell wall composition and ultrastructural immunolocalization of pectin and arabinogalactan protein during Olea europaea L. fruit abscission. Plant Cell Physiol. 2020, 61, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Roongsattham, P.; Morcillo, F.; Jantasuriyarat, C.; Pizot, M.; Moussu, S.; Jayaweera, D.; Collin, M.; Gonzalez-Carranza, Z.H.; Amblard, P.; Tregear, J.W.; et al. Temporal and spatial expression of polygalacturonase gene family members reveals divergent regulation during fleshy fruit ripening and abscission in the monocot species oil palm. BMC Plant Biol. 2012, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Riov, J. A polygalacturonase from citrus leaf explants: Role in abscission. Plant Physiol. 1974, 53, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Phetsirikoon, S.; Paull, R.E.; Chen, N.; Ketsa, S.; van Doorn, W.G. Increased hydrolase gene expression and hydrolase activity in the abscission zone involved in chilling-induced abscission of Dendrobium flowers. Postharvest Biol. Technol. 2016, 117, 217–229. [Google Scholar] [CrossRef]
- Xu, T.; Li, T.; Qi, M. Calcium effects on mediating polygalacturonan activity by mRNA expression and protein accumulation during tomato pedicel explant abscission. Plant Growth Regul. 2010, 60, 255–263. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Schröder, R.; Hallett, C.; Cohen, D.; MacRae, E.A. Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 2002, 129, 122–133. [Google Scholar] [CrossRef]
- Jiang, C.-Z.; Lu, F.; Imsabai, W.; Meir, S.; Reid, M.S. Silencing polygalacturonase expression inhibits tomato petiole abscission. J. Exp. Bot. 2008, 59, 973–979. [Google Scholar] [CrossRef]
- Chersicola, M.; Kladnik, A.; Žnidarič, M.T.; Mrak, T.; Gruden, K.; Dermastia, M. 1-aminocyclopropane-1-crboxylate oxidase induction in tomato flower pedicel phloem and abscission related processes are differentially sensitive to ethylene. Front. Plant Sci. 2017, 8, 464. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Somerville, C.R. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wal. Plant J. 1998, 15, 79–88. [Google Scholar] [CrossRef]
- Ogawa, M.; Kay, P.; Wilson, S.; Swaina, S.M. Arabidopsis dehiscence zone polygalacturonase1 (ADPG1), ADPG2, ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in arabidopsis. Plant Cell 2009, 21, 216–233. [Google Scholar] [CrossRef]
- Ge, T.; Huang, X.; Pan, X.; Zhang, J.; Xie, R. Genome-wide identifcation and expression analysis of citrus fruitlet abscission-related polygalacturonase genes. 3 Biotech. 2019, 9, 250. [Google Scholar] [CrossRef]
- Lu, L.; Hou, Q.; Wang, L.; Zhang, T.; Zhao, W.; Yan, T.; Zhao, L.; Li, J.; Wan, X. Genome-wide identification and characterization of polygalacturonase gene family in maize (Zea mays L.). Int. J. Mol. Sci. 2021, 222, 10722. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Zhang, Y.; Yan, X.; Han, M.; Li, J.; Li, F.; Li, F.; Zhang, D.; Zhao, C. Identification and expression analysis of polygalacturonase family members during peach fruit softening. Int. J. Mol. Sci. 2016, 17, 1933. [Google Scholar] [CrossRef]
- Ke, X.; Wang, H.; Li, Y.; Zhu, B.; Zang, Y.; He, Y.; Cao, J.; Zhu, Z.; Yu, Y. Genome-wide identification and analysis of polygalacturonase genes in Solanum lycopersicum. Int. J. Mol. Sci. 2018, 19, 2290. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.L.; Liu, H.J.; Wang, X.R.; Zeng, Q.Y. Molecular evolution and expression divergence of the Populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol. 2013, 197, 1353–1365. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, M.; Zhang, H.; Zhang, S.; Qian, M.; Zhang, Z.; Luo, W.; Fan, J.; Liu, Z.; Wang, L. Genome-wide analysis of polygalacturonase gene family from pear genome and identification of the member involved in pear softening. BMC Plant Biol. 2019, 19, 587. [Google Scholar] [CrossRef]
- Hadfield, K.A.; Bennett, A.B. Polygalacturonases: Many genes in search of a function. Plant Physiol. 1998, 117, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Markovič, O.; Janeček, Š. Pectin degrading glycoside hydrolases of family 28: Sequence-structural features, specificities and evolution. Protein Eng. Des. Sel. 2001, 14, 615–631. [Google Scholar] [CrossRef]
- González-Carranza, Z.H.; Elliott, K.A.; Roberts, J.A. Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thalian. J. Exp. Bot. 2007, 58, 3719–3730. [Google Scholar] [CrossRef]
- Peng, G.; Wua, J.; Lu, W.; Li, J. A polygalacturonase gene clustered into clade E involved in lychee fruitlet abscission. Sci. Hortic. 2013, 150, 244–250. [Google Scholar] [CrossRef]
- Nagao, M.A.; Hirae, H.H.; Stephenson, R.A. Macadamia: Cultivation and physiology. Crit. Rev. Plant Sci. 1992, 10, 441–470. [Google Scholar] [CrossRef]
- Hardner, C.M.; Wall, M.; Cho, A. Global macadamia science: Overview of the special section. HortScience 2019, 54, 592–595. [Google Scholar] [CrossRef]
- Trueman, S.J. The reproductive biology of macadamia. Sci. Hortic. 2013, 150, 354–359. [Google Scholar] [CrossRef]
- Zeng, H.; Yang, W.; Lu, C.; Lin, W.; Zou, M.; Zhang, H.; Wan, J.; Huang, X. Effect of CPPU on carbohydrate and endogenous hormone levels in young macadamia fruit. PLoS ONE 2016, 11, e0158705. [Google Scholar] [CrossRef]
- McFadyen, L.; Robertson, D.; Sedgley, M.; Kristiansen, P.; Olesen, T. Effects of the ethylene inhibitor aminoethoxyvinylglycine (AVG) on fruit abscission and yield on pruned and unpruned macadamia trees. Sci. Hortic. 2012, 137, 125–130. [Google Scholar] [CrossRef]
- Howlett, B.G.; Read, S.F.J.; Alavi, M.; Cutting, B.T.; Nelson, W.R.; Goodwin, R.M.; Cross, S.; Thorp, T.G.; Pattemore, D.E. Cross-pollination enhances macadamia yields, even with branch-level resource limitation. HortScience 2019, 54, 609–615. [Google Scholar] [CrossRef]
- Trueman, S.J.; Turnbull, C.G.N. Fruit set, abscission and dry matter accumulation on girdled branches of macadamia. Ann. Bot. 1994, 74, 667–674. [Google Scholar] [CrossRef]
- Yang, W.; Xiang, P. Changes of fruit abscission and carbohydrates, hormones, related gene expression in the fruit and pedicel of macadamia under starvation stress. Horticulturae 2022, 8, 398. [Google Scholar] [CrossRef]
- Trueman, S.J. Endogenous gibberellin levels during early fruit development of macadamia. Afr. J. Agric. Res. 2011, 60, 4785–4788. [Google Scholar] [CrossRef]
- Nock, C.J.; Baten, A.; Mauleon, R.; Langdon, K.S.; Topp, B.; Hardner, C.; Furtado, A.; Henry, R.J.; King, G.J. Chromosome-scale assembly and annotation of the macadamia genome (Macadamia integrifolia HAES 741). G3-Genes Genomes Genet. 2020, 10, 3497–3504. [Google Scholar] [CrossRef]
- Torki, M.; Mandaron, P.; Mache, R.; Falconet, D. Characterization of a ubiquitous expressed gene family encoding polygalacturonase in Arabidopsis thaliana. Gene 2000, 242, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Park, K.C.; Kwon, S.J.; Kim, N.S. Intron loss mediated structural dynamics and functional differentiation of the polygalacturonase gene family in land plants. Genes Genom. 2010, 32, 570–577. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Stratilová, E.; Mislovičová, D.; Kačuráková, M.; Machová, E.; Kolarová, N.; Markovič, O.; Jörnvall, H. The glycoprotein vharacter of multiple forms of aspergillus polygalacturonase. J. Protein Chem. 1998, 17, 173–179. [Google Scholar] [CrossRef]
- Wang, F.; Sun, X.; Shi, X.; Zhai, H.; Tian, C.; Kong, F.; Liu, B.; Yuan, X. A global analysis of the polygalacturonase gene family in soybean (Glycine max). PLoS ONE 2016, 11, e0163012. [Google Scholar] [CrossRef]
- Huang, W.; Chen, M.; Zhao, T.; Han, F.; Zhang, Q.; Liu, X.; Jiang, C.; Zhong, C. Genome-wide identification and expression analysis of polygalacturonase gene family in kiwifruit (Actinidia chinensis) during fruit softening. Plants 2020, 9, 327. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, Y.; Ying, L.; Anderson, C.T.; Cao, J. A profusionof molecular scissors for pectins: Classification, expression, and functions of plant polygalacturonases. Front. Plant Sci. 2018, 9, 1208. [Google Scholar] [CrossRef]
- Rao, M.N.; Kembhavi, A.A.; Pant, A. Implication of tryptophan and histidine in the active site of endo-polygalacturonase from Aspergillus ustus: Elucidation of the reaction mechanism. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 1996, 1296, 167–173. [Google Scholar] [CrossRef]
- Rhee, S.Y.; Osborne, E.; Poindexter, P.D.; Somerville, C.R. Microspore separation in the quartet 3 mutants of arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 2003, 133, 1170–1180. [Google Scholar] [CrossRef]
- Lyu, M.; Iftikhar, J.; Guo, R.; Wu, B.; Cao, J. Patterns of expansion and expression divergence of the polygalacturonase gene family in Brassica oleracea. Int. J. Mol. Sci. 2020, 21, 5706. [Google Scholar] [CrossRef]
- Pan, H.; Sun, Y.; Qiao, M.; Qi, H. Beta-galactosidase gene family genome-wide identification and expression analysis of members related to fruit softening in melon (Cucumis melo L.). BMC Genom. 2022, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xie, M.; Wang, X.; Wang, G.; Zhang, Y.; Li, Z.; Ma, Z. Identification of cell wall-associated kinases as important regulators involved in Gossypium hirsutum resistance to Verticillium dahliae. BMC Plant Biol. 2021, 21, 220. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Yang, H.; Yang, J.; Wang, Y.; Ye, T.; Xiang, L.; Chan, Z.; Wang, Y. Tulip transcription factor TgWRKY75 activates salicylic acid and abscisic acid biosynthesis to synergistically promote petal senescence. J. Exp. Bot. 2024, 75, 2435–2450. [Google Scholar] [CrossRef]
- Ma, X.; Li, C.; Huang, X.; Wang, H.; Wu, H.; Zhao, M.; Li, J.; Rennenberg, H. Involvement of HD-ZIP I transcription factors LcHB2 and LcHB3 in fruitlet abscission by promoting transcription of genes related to the biosynthesis of ethylene and ABA in litchi. Tree Physiol. 2019, 39, 1600–1613. [Google Scholar] [CrossRef]
- Einhorn, T.C.; Arrington, M. ABA and shading induce ‘Bartlett’ pear abscission and inhibit photosynthesis but are not additive. J. Plant Growth Regul. 2018, 37, 300–308. [Google Scholar] [CrossRef]
- Brummell, D.A.; Cin, V.D.; Crisosto, C.H.; Labavitch, J.M. Cell wall metabolism during maturation, ripening and senescence of peach fruit. J. Exp. Bot. 2004, 55, 2029–2039. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.W.; Wang, Y.; Ma, X.S.; Zhang, J.Q.; Zhao, M.L.; Huang, X.M.; Li, J.G.; Hu, G.-B.; Wang, H.C. LcERF2 modulates cell wall metabolism by directly targeting a UDP-glucose-4-epimerase gene to regulate pedicel development and fruit abscission of litchi. Plant J. 2021, 106, 801–816. [Google Scholar] [CrossRef]
- Zhai, Z.; Feng, C.; Wang, Y.; Sun, Y.; Peng, X.; Xiao, Y.; Zhang, X.; Zhou, X.; Jiao, J.; Wang, W.; et al. Genome-wide identification of the xyloglucan endotransglucosylase/hydrolase (XTH) and polygalacturonase (PG) genes and characterization of their role in fruit softening of sweet cherry. Int. J. Mol. Sci. 2021, 22, 12331. [Google Scholar] [CrossRef]
- Zhao, M.; Li, C.; Ma, X.; Xia, R.; Chen, J.; Liu, X.; Ying, P.; Peng, M.; Wang, J.; Shi, C.L.; et al. KNOX protein KNAT1 regulates fruitlet abscission in litchi by repressing ethylene biosynthetic genes. J. Exp. Bot. 2020, 71, 4069–4082. [Google Scholar] [CrossRef]
- Aruwajoye, N.N.; Olarewaju, O.O.; Oluwalana-Sanusi, A.E.; Mditshwa, A.; Magwaza, L.S.; Tesfay, S.Z. Accelerating abscission of macadamia nuts using ethephon: Are there implications for nut quality? J. Hortic. Sci. Biotechnol. 2024, 100, 153–163. [Google Scholar] [CrossRef]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis information resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2007, 36, D1009–D1014. [Google Scholar] [CrossRef]
- Wang, P.; Mo, Y.; Wang, Y.; Fei, Y.; Huang, J.; Ni, J.; Xu, Z.-F. Macadamia germplasm and genomic database (MacadamiaGGD): A comprehensive platform for germplasm innovation and functional genomics in Macadamia. Front. Plant Sci. 2022, 13, 100726. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Anderson, J.B.; Cherukuri, P.F.; DeWeese-Scott, C.; Geer, L.Y.; Gwadz, M.; He, S.; Hurwitz, D.I.; Jackson, J.D.; Ke, Z.; et al. CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 2005, 33, D192–D196. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; Heijne, G.v.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Yu, C.S.; Cheng, C.W.; Su, W.C.; Chang, K.C.; Huang, S.W.; Hwang, J.-K.; Lu, C.H. CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI′s conserved domain database and tools for protein domain analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, Z.; Zhou, Y.; Chen, D.; Heng, L. Screening of stable reference genes for qRT-PCR analysis in Macadamia integrifolia. Chin. J. Trop. Crops 2020, 40, 1505–1512. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fallahpour, M.; Ghanbari, A.; Koobaz, P.; Chamani, E.; Azadi, P.; Mii, M. Selection of suitable lily cultivars by using needle agroinfiltration for blue flower production. J. Hortic. Sci. Biotechnol. 2022, 98, 207–222. [Google Scholar] [CrossRef]
- Feng, Y.; Guo, Z.; Zhong, J.; Liang, Y.; Zhang, P.; Sun, M. The LibHLH22 and LibHLH63 from Lilium ‘Siberia’ can positively regulate volatile terpenoid biosynthesis. Horticulturae 2023, 9, 459. [Google Scholar] [CrossRef]
- Yin, X.; Zhang, Y.; Zhang, L.; Wang, B.; Zhao, Y.; Irfan, M.; Chen, L.; Feng, Y. Regulation of MYB transcription factors of anthocyanin synthesis in lily flowers. Front. Plant Sci. 2021, 12, 761668. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | Deduced Protein | Signal Peptide | Subcellular Localization | Domain | ||
|---|---|---|---|---|---|---|---|
| Length (aa) | Molecular Weight (kDa) | Isoelectric Points (pI) | |||||
| MiPG1 | LOC122081278 | 491 | 55.10 | 8.76 | − | CM | II IV |
| MiPG2 | LOC122081682 | 465 | 50.10 | 5.10 | + | CM | I II IV |
| MiPG3 | LOC122063767 | 385 | 41.23 | 9.25 | + | CM | I II III IV |
| MiPG4 | LOC122063777 | 396 | 42.08 | 7.93 | + | CM | I II III IV |
| MiPG5 | LOC122063785 | 396 | 42.09 | 8.66 | + | CM | I II III IV |
| MiPG6 | LOC122071976 | 445 | 48.72 | 8.34 | + | CM | I II III IV |
| MiPG7 | LOC122064844 | 472 | 50.83 | 8.94 | − | CM | I II III IV |
| MiPG8 | LOC122066229 | 491 | 54.99 | 8.59 | − | CM | II IV |
| MiPG9 | LOC122073447 | 519 | 56.38 | 7.52 | + | CM | I II III IV |
| MiPG10 | LOC122074598 | 469 | 48.01 | 7.47 | + | CM | I II III IV |
| MiPG11 | LOC122074599 | 490 | 49.83 | 8.14 | + | CM | I II III IV |
| MiPG12 | LOC122074600 | 474 | 48.98 | 8.28 | + | CM | I II III IV |
| MiPG13 | LOC122074601 | 468 | 48.34 | 7.47 | + | CM | I II III IV |
| MiPG14 | LOC122072816 | 480 | 52.49 | 6.79 | − | CM | I II IV |
| MiPG15 | LOC122075419 | 471 | 51.08 | 8.77 | + | CM | I II III IV |
| MiPG16 | LOC122076188 | 464 | 50.78 | 4.85 | + | CM | I II III IV |
| MiPG17 | LOC122077755 | 347 | 37.08 | 8.88 | + | CM | I II III IV |
| MiPG18 | LOC122078590 | 201 | 21.08 | 4.85 | − | CM | II III IV |
| MiPG19 | LOC122078589 | 401 | 42.42 | 4.94 | + | CM | I II III IV |
| MiPG20 | LOC122078252 | 249 | 26.32 | 6.14 | − | CM | I II III IV |
| MiPG21 | LOC122082848 | 393 | 42.78 | 8.91 | + | CM | I II III IV |
| MiPG22 * | LOC122081461 | 502 | 54.13 | 5.56 | + | CM Chl Cyt | |
| MiPG23 | LOC122083126 | 392 | 41.78 | 5.88 | + | CM | I II III IV |
| MiPG24 | LOC122084718 | 396 | 42.92 | 9.42 | + | CM | I II III IV |
| MiPG25 | LOC122089024 | 401 | 42.97 | 5.80 | − | CM | I II III IV |
| MiPG26 | LOC122092380 | 445 | 47.66 | 5.99 | + | CM | I II III IV |
| MiPG27 * | LOC122092659 | 498 | 54.13 | 9.45 | + | Chl | |
| MiPG28 * | LOC122093358 | 400 | 44.51 | 5.34 | − | Chl | |
| MiPG29 * | LOC122094310 | 484 | 51.71 | 8.26 | − | CM | |
| MiPG30 * | LOC122094116 | 482 | 51.89 | 8.55 | + | CM Chl | |
| MiPG31 | LOC122093962 | 467 | 50.92 | 5.33 | − | CM | I II III IV |
| MiPG32 | LOC122092887 | 394 | 42.77 | 8.86 | + | CM | I II III IV |
| MiPG33 | LOC122093810 | 464 | 49.69 | 5.23 | + | CM | I II IV |
| MiPG34 | LOC122057274 | 423 | 46.27 | 8.59 | − | CM | I II III IV |
| MiPG35 | LOC122058248 | 467 | 50.76 | 6.08 | + | CM | I II IV |
| MiPG36 | LOC122057404 | 467 | 50.67 | 6.70 | + | CM | I II IV |
| MiPG37 | LOC122058173 | 482 | 52.29 | 6.32 | + | CM | I II IV |
| MiPG38 | LOC122058047 | 501 | 55.45 | 6.46 | − | CM | II IV |
| MiPG39 | LOC122059778 | 405 | 43.35 | 6.06 | + | CM | I II III IV |
| MiPG40 | LOC122059401 | 467 | 50.60 | 6.31 | + | CM | I II IV |
| MiPG41 | LOC122059027 | 378 | 40.99 | 9.16 | − | CM | I II III IV |
| MiPG42 | LOC122061528 | 478 | 51.97 | 8.30 | − | CM | I II IV |
| MiPG43 | LOC122061700 | 421 | 46.05 | 5.41 | + | CM | I II III IV |
| MiPG44 | LOC122061732 | 489 | 51.74 | 5.61 | + | CM | I II III IV |
| MiPG45 | LOC122062845 | 437 | 45.85 | 8.82 | + | CM | I II III IV |
| MiPG46 | LOC122065217 | 392 | 41.54 | 9.43 | + | CM | I II III IV |
| MiPG47 | LOC122065260 | 430 | 47.16 | 5.18 | + | CM | I II III IV |
| MiPG48 | LOC122065662 | 414 | 44.01 | 8.75 | + | CM | I II III IV |
| MiPG49 | LOC122066012 | 480 | 52.51 | 7.10 | − | CM | I II IV |
| MiPG50 | LOC122066415 | 467 | 50.62 | 6.01 | + | CM | I II IV |
| MiPG51 | LOC122066705 | 393 | 42.81 | 5.54 | + | CM | I II III IV |
| MiPG52 | LOC122068767 | 459 | 50.30 | 5.98 | + | CM | I II III IV |
| MiPG53 | LOC122068773 | 464 | 49.70 | 4.99 | + | CM | I II III IV |
| MiPG54 | LOC122069182 | 414 | 43.73 | 8.45 | − | CM | I II III IV |
| MiPG55 | LOC122070809 | 204 | 21.71 | 6.21 | − | CM | I II III IV |
| MiPG56 | LOC122071794 | 243 | 26.05 | 9.08 | + | CM | I II |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fei, Y.-C.; Mo, Y.; Xu, J.; Lin, K.; Tao, L.; He, X.; Li, M.; Xu, Z.-F. Genome-Wide Analysis of the Polygalacturonase Gene Family in Macadamia and Identification of Members Involved in Fruit Abscission. Plants 2025, 14, 1610. https://doi.org/10.3390/plants14111610
Fei Y-C, Mo Y, Xu J, Lin K, Tao L, He X, Li M, Xu Z-F. Genome-Wide Analysis of the Polygalacturonase Gene Family in Macadamia and Identification of Members Involved in Fruit Abscission. Plants. 2025; 14(11):1610. https://doi.org/10.3390/plants14111610
Chicago/Turabian StyleFei, Yu-Chong, Yi Mo, Jiajing Xu, Kai Lin, Liang Tao, Xiyong He, Meng Li, and Zeng-Fu Xu. 2025. "Genome-Wide Analysis of the Polygalacturonase Gene Family in Macadamia and Identification of Members Involved in Fruit Abscission" Plants 14, no. 11: 1610. https://doi.org/10.3390/plants14111610
APA StyleFei, Y.-C., Mo, Y., Xu, J., Lin, K., Tao, L., He, X., Li, M., & Xu, Z.-F. (2025). Genome-Wide Analysis of the Polygalacturonase Gene Family in Macadamia and Identification of Members Involved in Fruit Abscission. Plants, 14(11), 1610. https://doi.org/10.3390/plants14111610







