Functional Roles of the Seagrass (Zostera marina) Holobiont Change with Plant Development
Abstract
1. Introduction
2. Materials and Methods
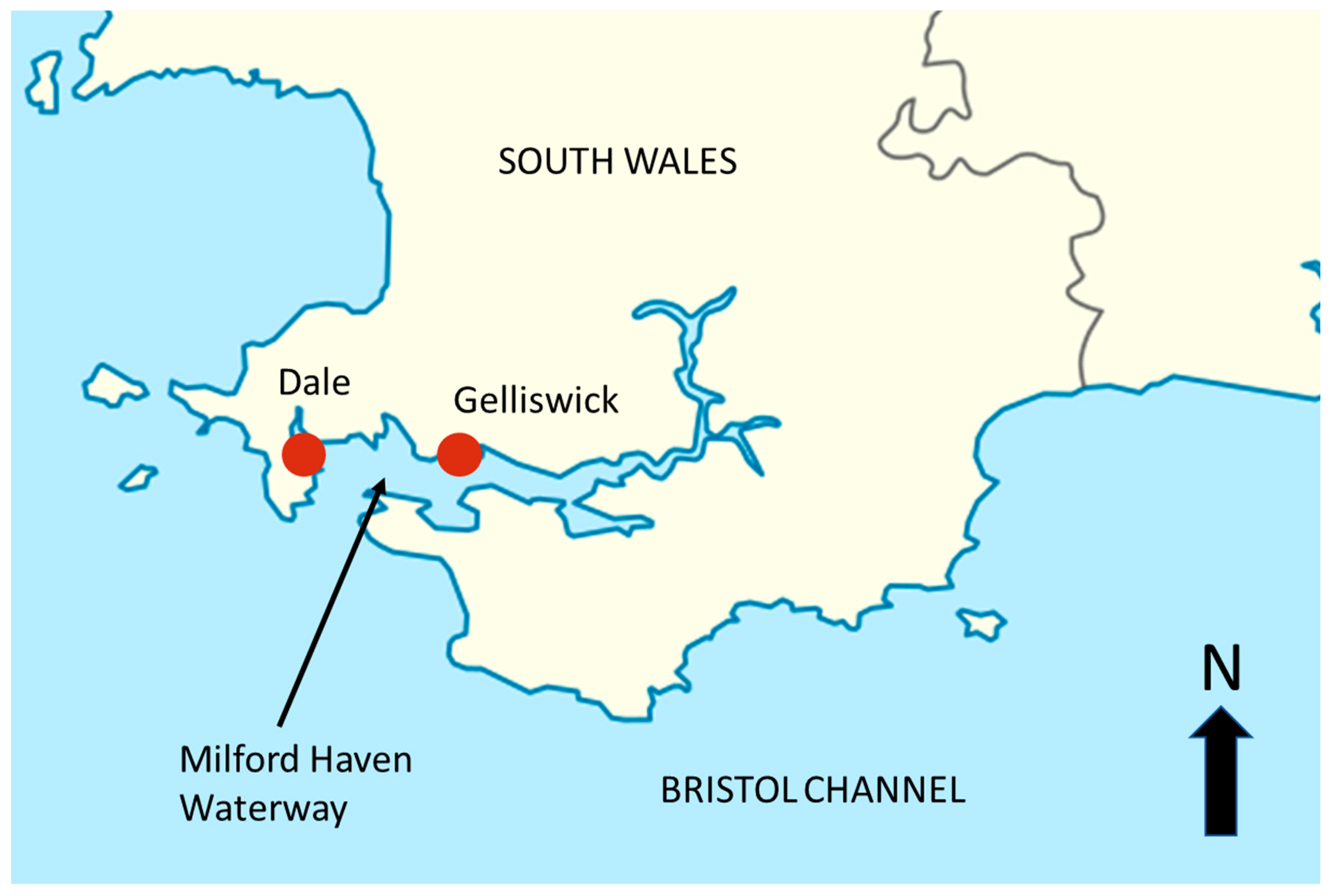
2.1. Study Sites
2.2. Sample Collection
2.3. DNA Extraction, PCR Amplification and High-Throughput Sequencing
2.4. Sequence Processing
2.5. Data Analysis
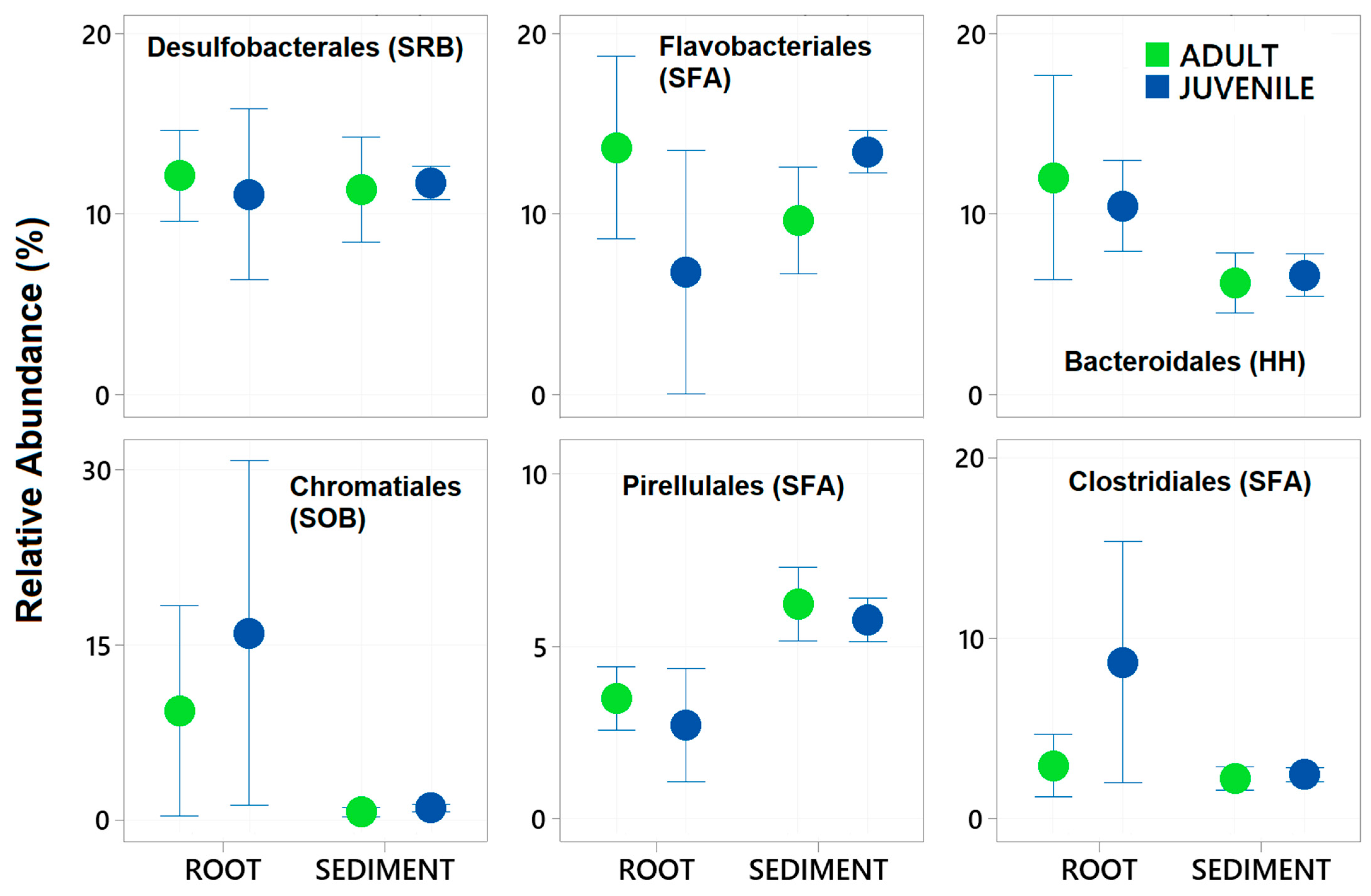
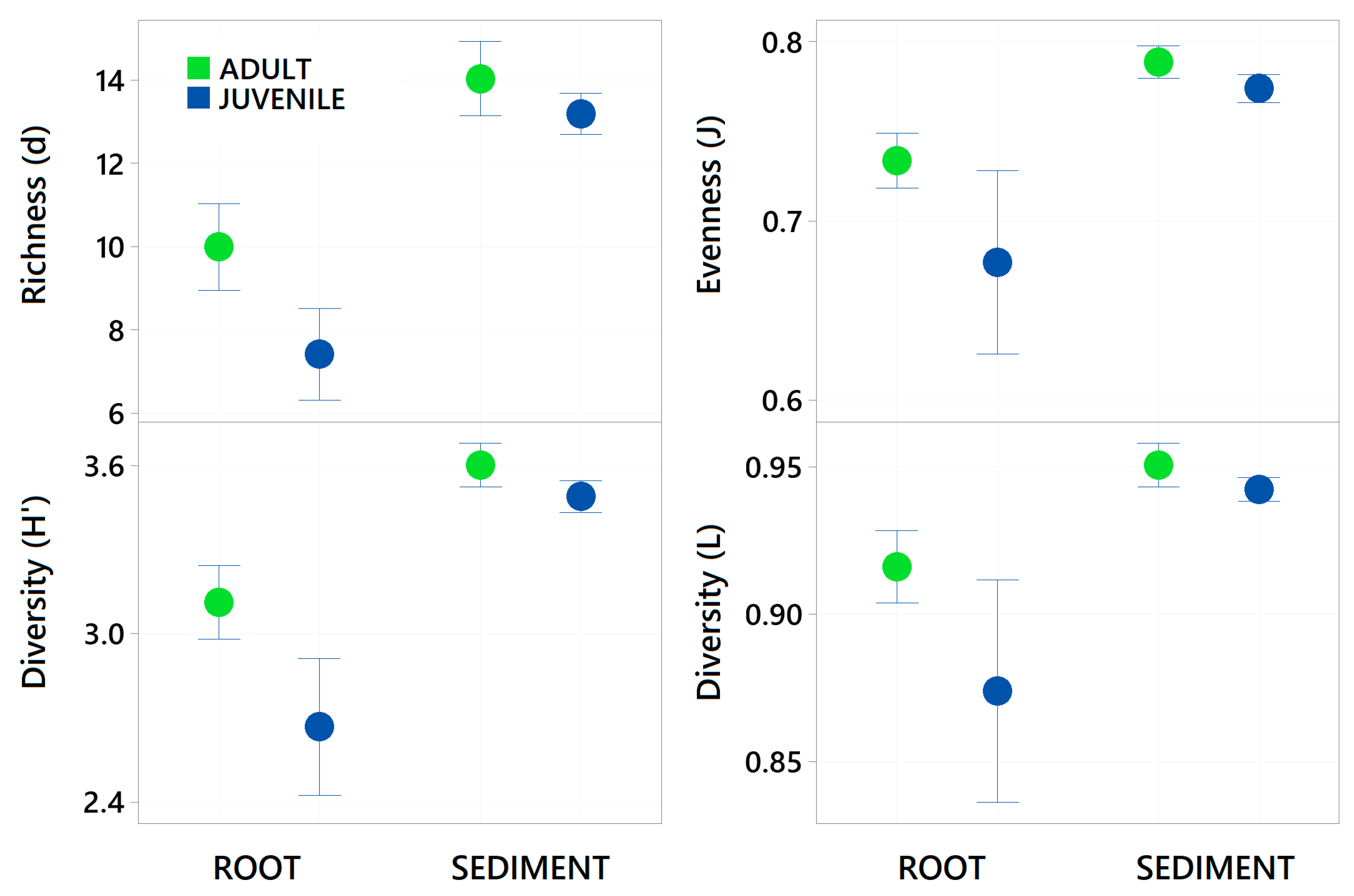
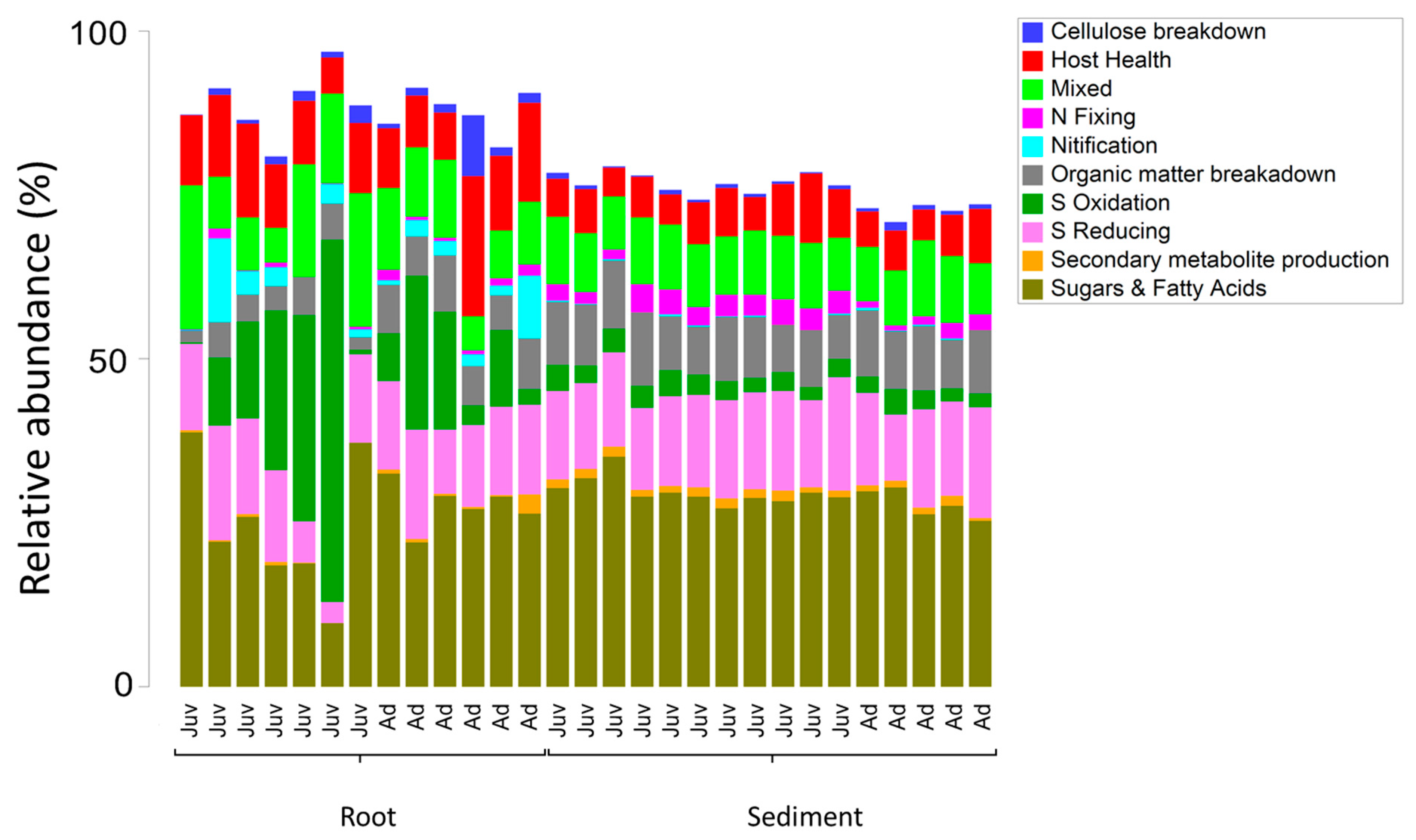
3. Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Microbial Taxa | Ecological Function |
|---|---|
| Desulfobacterales | S Reducing |
| Flavobacteriales | Sugars & Fatty Acids |
| Bacteroidales | Host Health |
| Chromatiales | S Oxidation |
| Pirellulales | Sugars & Fatty Acids |
| Clostridiales | Sugars & Fatty Acids |
| Chitinophagales | Organic matter breakadown |
| Gammaproteobacteria | Mixed |
| Cytophagales | Organic matter breakadown |
| Campylobacterales | Mixed |
| Spirochaetales | Sugars & Fatty Acids |
| Gammaproteobacteria Incertae Sedis | Mixed |
| Rhizobiales | Nitrogen Fixing |
| Cellvibrionales | Mixed |
| Steroidobacterales | Sugars & Fatty Acids |
| Nitrosococcales | Nitification |
| B2M28 | Unclear |
| Rhodobacterales | Mixed |
| Thiomicrospirales | S Oxidation |
| Thermoanaerobaculales | Sugars & Fatty Acids |
| Ectothiorhodospirales | S Reducing |
| Thiotrichales | S Oxidation |
| Myxobacteria | Secondary metabolite production |
| MSBL9 | Sugars & Fatty Acids |
| Alteromonadales | Organic matter breakadown |
| Oceanospirillales | Organic matter breakadown |
| Sphingobacteriales | Sugars & Fatty Acids |
| Microtrichales | Organic matter breakadown |
| Ignavibacteriales | Cellulose breakdown |
| Moduliflexales | Sugars & Fatty Acids |
| Betaproteobacteriales | Mixed |
| Desulfuromonadales | S Reducing |
| Kiritimatiellales | Sugars & Fatty Acids |
| Fibrobacterales | Cellulose breakdown |
References
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Blackall, L.L.; Dungan, A.M.; Hartman, L.M.; Van Oppen, M.J. Probiotics for corals. Microbiol. Aust. 2020, 41, 100–104. [Google Scholar] [CrossRef]
- Bressan, M.; Roncato, M.-A.; Bellvert, F.; Comte, G.; Haichar, F.E.Z.; Achouak, W.; Berge, O. Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 2009, 3, 1243–1257. [Google Scholar] [CrossRef]
- Brodersen, K.E.; Kühl, M. Photosynthetic capacity in seagrass seeds and early-stage seedlings of Zostera marina. New Phytol. 2023, 239, 1300–1314. [Google Scholar] [CrossRef]
- Brodersen, K.E.; Mosshammer, M.; Bittner, M.J.; Hallstrøm, S.; Santner, J.; Riemann, L.; Kühl, M. Seagrass-mediated rhizosphere redox gradients are linked with ammonium accumulation driven by diazotrophs. Microbiol. Spectr. 2024, 12, e03335-23. [Google Scholar] [CrossRef]
- Burkholder, J.M.; Tomasko, D.A.; Touchette, B.W. Seagrasses and eutrophication. J. Exp. Mar. Biol. Ecol. 2007, 350, 46–72. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Carey, D.A.; Hayn, M.; Germano, J.D.; Little, D.I.; Bullimore, B. Marine habitat mapping of the Milford Haven Waterway, Wales, UK: Comparison of facies mapping and EUNIS classification for monitoring sediment habitats in an industrialized estuary. J. Sea Res. 2015, 100, 99–119. [Google Scholar] [CrossRef]
- Clarke, K.R.; Gorley, R.N.; Somerfield, P.J.; Warwick, R.M. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation, 3rd ed.; PRIMER-E Ltd.: Plymouth, UK, 2014; 260p. [Google Scholar]
- Cúcio, C.; Overmars, L.; Engelen, A.H.; Muyzer, G. Metagenomic Analysis Shows the Presence of Bacteria Related to Free-Living Forms of Sulfur-Oxidizing Chemolithoautotrophic Symbionts in the Rhizosphere of the Seagrass Zostera marina. Front. Mar. Sci. 2018, 5, 171. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Voerman, S.E.; Lang, J.M.; Stachowicz, J.J.; Eisen, J.A. Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 2017, 5, e3246. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.W.; Gleeson, D.B.; Grierson, P.F.; Laverock, B.; Kendrick, G.A. Metagenomic Evidence of Microbial Community Responsiveness to Phosphorus and Salinity Gradients in Seagrass Sediments. Front. Microbiol. 2018, 9, 1703. [Google Scholar] [CrossRef]
- Garrity, G.M.; Staley, J.T.; Boone, D.R.; Brenner, D.J.; Devos, P.; Goodfellow, M.; Krieg, N.R.; Rainey, F.A.; Schlefer, K. Bergey’s Manual of Systematic Bacteriology; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Graham, O.J.; Adamczyk, E.M.; Schenk, S.; Dawkins, P.; Burke, S.; Chei, E.; Cisz, K.; Dayal, S.; Elstner, J.; Hausner, A.L.P.; et al. Manipulation of the seagrass-associated microbiome reduces disease severity. Environ. Microbiol. 2024, 26, e16582. [Google Scholar] [CrossRef]
- Han, S.; Van Treuren, W.; Fischer, C.R.; Merrill, B.D.; Defelice, B.C.; Sanchez, J.M.; Higginbottom, S.K.; Guthrie, L.; Fall, L.A.; Dodd, D.; et al. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature 2021, 595, 415–420. [Google Scholar] [CrossRef]
- Hemminga, M.A.; Duarte, C.M. Seagrass Ecology; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Herbert, R.A. Nitrogen cycling in coastal marine ecosystems. FEMS Microbiol. Rev. 1999, 23, 563–590. [Google Scholar] [CrossRef] [PubMed]
- Holmer, M. Chapter 13—Productivity and Biogeochemical Cycling in Seagrass Ecosystems. In Coastal Wetlands, 2nd ed.; Perillo, G.M.E., Wolanski, E., Cahoon, D.R., Hopkinson, C.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 443–477. [Google Scholar]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome—From metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Jensen, S.I.; Kühl, M.; Priemé, A. Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol. Ecol. 2007, 62, 108–117. [Google Scholar] [CrossRef]
- Jones, B.L.; Unsworth, R.K.F. The perilous state of seagrass in the British Isles. R. Soc. Open Sci. 2016, 3, e16582. [Google Scholar] [CrossRef]
- Kardish, M.R.; Stachowicz, J.J. Local environment drives rapid shifts in composition and phylogenetic clustering of seagrass microbiomes. Sci. Rep. 2023, 13, 3673. [Google Scholar] [CrossRef]
- Kay, Q.O.N. A Review of the Existing State of Knowledge of the Ecology and Distribution of Seagrass Beds Around the Coast of Wales; Science Report, 296; Countryside Council for Wales: Bangor, Wales, 1998. [Google Scholar]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Macreadie, P.I.; Atwood, T.B.; Seymour, J.R.; Fontes, M.L.S.; Sanderman, J.; Nielsen, D.A.; Connolly, R.M. Vulnerability of seagrass blue carbon to microbial attack following exposure to warming and oxygen. Sci. Total Environ. 2019, 686, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.C.; Bougoure, J.; Ryan, M.H.; Bennett, W.W.; Colmer, T.D.; Joyce, N.K.; Olsen, Y.S.; Kendrick, G.A. Oxygen loss from seagrass roots coincides with colonisation of sulphide-oxidising cable bacteria and reduces sulphide stress. ISME J. 2019, 13, 707–719. [Google Scholar] [CrossRef]
- Maúre, E.D.R.; Terauchi, G.; Ishizaka, J.; Clinton, N.; Dewitt, M. Globally consistent assessment of coastal eutrophication. Nat. Commun. 2021, 12, 6142. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.B. The seed microbiome: Origins, interactions, and impacts. Plant and Soil 2018, 422, 7–34. [Google Scholar] [CrossRef]
- Nrw. Access Our Data, Maps and Reports [Online]. 2024. Available online: https://naturalresources.wales/evidence-and-data/accessing-our-data/access-our-data-maps-and-reports/?lang=en (accessed on 19 May 2025).
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Rosenberg, E.; Delong, F.E.; Lory, S.; Stackebrandt, E.; Thompson, F. The Prokartyotes; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Schorn, S.; Ahmerkamp, S.; Bullock, E.; Weber, M.; Lott, C.; Liebeke, M.; Lavik, G.; Kuypers, M.M.M.; Graf, J.S.; Milucka, J. Diverse methylotrophic methanogenic archaea cause high methane emissions from seagrass meadows. Proc. Natl. Acad. Sci. USA 2022, 119, e2106628119. [Google Scholar] [CrossRef]
- Shade, A.; Jacques, M.-A.; Barret, M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 2017, 37, 15–22. [Google Scholar] [CrossRef]
- Sogin, E.M.; Michellod, D.; Gruber-Vodicka, H.R.; Bourceau, P.; Geier, B.; Meier, D.V.; Seidel, M.; Ahmerkamp, S.; Schorn, S.; D’angelo, G.; et al. Sugars dominate the seagrass rhizosphere. Nat. Ecol. Evol. 2022, 6, 866–877. [Google Scholar] [CrossRef]
- Tarquinio, F.; Hyndes, G.A.; Laverock, B.; Koenders, A.; Säwström, C. The seagrass holobiont: Understanding seagrass-bacteria interactions and their role in seagrass ecosystem functioning. FEMS Microbiol. Lett. 2019, 366, fnz057. [Google Scholar] [CrossRef]
- Thatcher, C.; Høj, L.; Bourne, D.G. Probiotics for coral aquaculture: Challenges and considerations. Curr. Opin. Biotechnol. 2022, 73, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [PubMed]
- Ugarelli, K.; Chakrabarti, S.; Laas, P.; Stingl, U. The Seagrass Holobiont and Its Microbiome. Microorganisms 2017, 5, 81. [Google Scholar] [CrossRef] [PubMed]
- Unsworth, R.K.F.; Bertelli, C.M.; Coals, L.; Cullen-Unsworth, L.C.; Den Haan, S.; Jones, B.L.H.; Rees, S.R.; Thomsen, E.; Wookey, A.; Walter, B. Bottlenecks to seed-based seagrass restoration reveal opportunities for improvement. Glob. Ecol. Conserv. 2023, 48, e02736. [Google Scholar] [CrossRef]
- Unsworth, R.K.F.; Bertelli, C.M.; Cullen-Unsworth, L.C.; Esteban, N.; Jones, B.L.; Lilley, R.; Lowe, C.; Nuuttila, H.K.; Rees, S.C. Sowing the Seeds of Seagrass Recovery Using Hessian Bags. Front. Ecol. Evol. 2019, 7, 311. [Google Scholar] [CrossRef]
- Unsworth, R.K.F.; Rees, S.C.; Bertelli, C.M.; Esteban, N.E.; Furness, E.J.; Walter, B. Nutrient additions to seagrass seed planting improve seedling emergence and growth. Front. Plant Sci. 2022, 13, 1013222. [Google Scholar] [CrossRef]
- Vieira, V.M.N.C.S.; Lobo-Arteaga, J.; Santos, R.; Leitão-Silva, D.; Veronez, A.; Neves, J.M.; Nogueira, M.; Creed, J.C.; Bertelli, C.M.; Samper-Villarreal, J.; et al. Seagrasses benefit from mild anthropogenic nutrient additions. Front. Mar. Sci. 2022, 9, 960249. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; Del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef]
- Wilkins, L.G.E.; Leray, M.; O’dea, A.; Yuen, B.; Peixoto, R.S.; Pereira, T.J.; Bik, H.M.; Coil, D.A.; Duffy, J.E.; Herre, E.A.; et al. Host-associated microbiomes drive structure and function of marine ecosystems. PLOS Biol. 2019, 17, e3000533. [Google Scholar] [CrossRef]
- Pan, X.; Raaijmakers, J.M.; Carrión, V.J. Importance of Bacteroidetes in host microbe interactions and ecosystem functioning. Trends Microbiol. 2023, 31, 959–971. [Google Scholar] [CrossRef]
- Mohr, W.; Lehnen, N.; Ahmerkamp, S.; Marchant, H.K.; Graf, J.S.; Tschitschko, B.; Yilmaz, P.; Littmann, S.; Gruber-Vodicka, H.; Leisch, N.; et al. Terrestrial-type nitrogen-fixing symbiosis between seagrass and a marine bacterium. Nature 2021, 600, 105–109. [Google Scholar] [CrossRef] [PubMed]

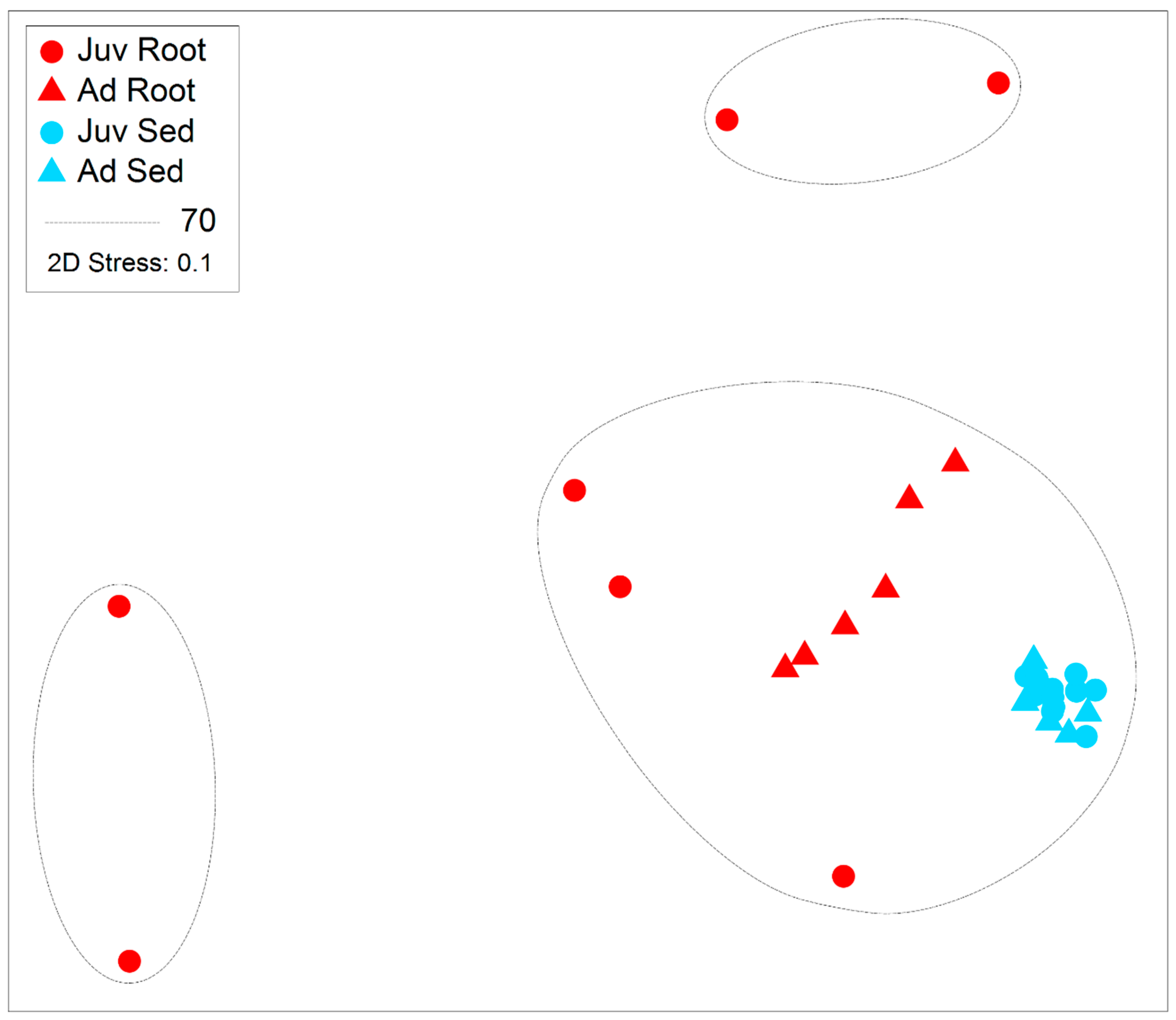

| DoF | Pseudo-F | Age | Pseudo-F | Structure | Pseudo-F | Interaction | |
|---|---|---|---|---|---|---|---|
| Assemblage | 1, 1, 25 | 3.524 | 0.001 | 11.14 | 0.001 | 2.689 | 0.001 |
| Richness (d) | 1, 1, 25 | 22.444 | 0.001 | 163.05 | 0.001 | 7.768 | 0.011 |
| Evenness (J) | 1, 1, 25 | 10.001 | 0.004 | 43.485 | 0.001 | 3.703 | 0.067 |
| Shannon (H’) | 1, 1, 25 | 20.976 | 0.001 | 110.22 | 0.001 | 8.350 | 0.01 |
| Simpsons (L) | 1, 1, 25 | 9.286 | 0.002 | 37.857 | 0.001 | 4.337 | 0.033 |
| Nitrogen Fixing | 1, 1, 25 | 2.5285 | 0.033 | 10.23 | 0.001 | 4.186 | 0.003 |
| Nitrification | 1, 1, 25 | 0.88636 | 0.413 | 18.025 | 0.001 | 0.244 | 0.813 |
| S Oxidation (SOB) | 1, 1, 25 | 0.789 | 0.414 | 10.08 | 0.004 | 1.059 | 0.314 |
| S Reducing (SRB) | 1, 1, 25 | 0.742 | 0.419 | 2.168 | 0.147 | 1.087 | 0.333 |
| Host Health (HH) | 1, 1, 25 | 0.052012 | 0.913 | 20.441 | 0.001 | 0.40844 | 0.577 |
| Secondary Metabolites | 1, 1, 25 | 2.2073 | 0.124 | 16.467 | 0.002 | 5.3014 | 0.015 |
| Cellulose | 1, 1, 25 | 3.258 | 0.05 | 6.625 | 0.005 | 0.182 | 0.869 |
| Mixed | 1, 1, 25 | 1.458 | 0.245 | 1.193 | 0.281 | 1.046 | 0.318 |
| Organic | 1, 1, 25 | 8.317 | 0.006 | 27.473 | 0.001 | 6.603 | 0.01 |
| Sugars (SFA) | 1, 1, 25 | 1.013 | 0.421 | 13.306 | 0.001 | 1.786 | 0.153 |
| Desulfobacterales (SRB) | 1, 1, 25 | 1.751 | 0.184 | 14.508 | 0.001 | 3.287 | 0.029 |
| Flavobacteriales (SFA) | 1, 1, 25 | 3.993 | 0.045 | 5.009 | 0.03 | 17.203 | 0.002 |
| Bacteroidales (HH) | 1, 1, 25 | 0.052 | 0.885 | 20.441 | 0.001 | 0.408 | 0.549 |
| Chromatiales (SOB) | 1, 1, 25 | 0.70781 | 0.55 | 14.28 | 0.001 | 1.298 | 0.261 |
| Pirellulales (SFA) | 1, 1, 25 | 3.0534 | 0.104 | 30.65 | 0.001 | 1.6787 | 0.225 |
| Clostridiales (SFA) | 1, 1, 25 | 6.8404 | 0.01 | 10.278 | 0.003 | 4.7801 | 0.031 |
| Parameter | Value |
|---|---|
| Site depth | 0.1 m (below chart datum) |
| Tidal range | 7.7 m |
| Salinity range | 33.5‰ |
| Temperature range | 8–17 °C |
| Porewater nitrogen | 280 ± 15 µmol.L−1 TON |
| Seagrass leaf nitrogen | 3.5% gDW−1 |
| Seagrass phosphorus | 0.43% gDW−1 |
| Porewater Phosphate | 30 ± 0.1 µmol.L−1 |
| Porewater Ammonium | 25 ± 0.2 µmol.L−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorvel, S.; Walter, B.; Taylor, J.D.; Unsworth, R.K.F. Functional Roles of the Seagrass (Zostera marina) Holobiont Change with Plant Development. Plants 2025, 14, 1584. https://doi.org/10.3390/plants14111584
Gorvel S, Walter B, Taylor JD, Unsworth RKF. Functional Roles of the Seagrass (Zostera marina) Holobiont Change with Plant Development. Plants. 2025; 14(11):1584. https://doi.org/10.3390/plants14111584
Chicago/Turabian StyleGorvel, Sam, Bettina Walter, Joe D. Taylor, and Richard K. F. Unsworth. 2025. "Functional Roles of the Seagrass (Zostera marina) Holobiont Change with Plant Development" Plants 14, no. 11: 1584. https://doi.org/10.3390/plants14111584
APA StyleGorvel, S., Walter, B., Taylor, J. D., & Unsworth, R. K. F. (2025). Functional Roles of the Seagrass (Zostera marina) Holobiont Change with Plant Development. Plants, 14(11), 1584. https://doi.org/10.3390/plants14111584







