Karyotype Variability in Wild Narcissus poeticus L. Populations from Different Environmental Conditions in the Dinaric Alps †
Abstract
1. Introduction
2. Results
2.1. General Karyotype Characteristics
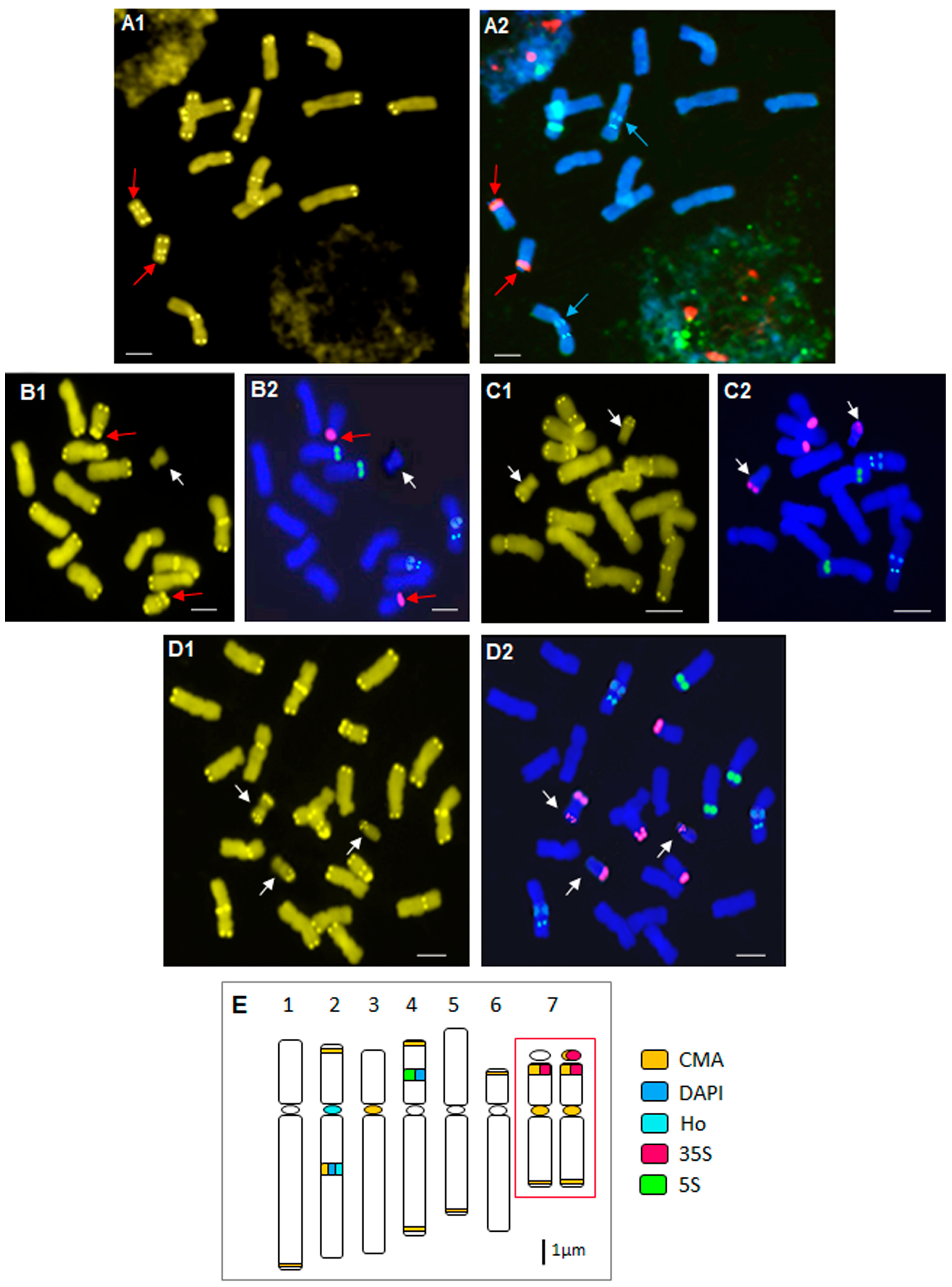
2.2. Heterochromatin Organization and Physical Mapping of rRNA Genes
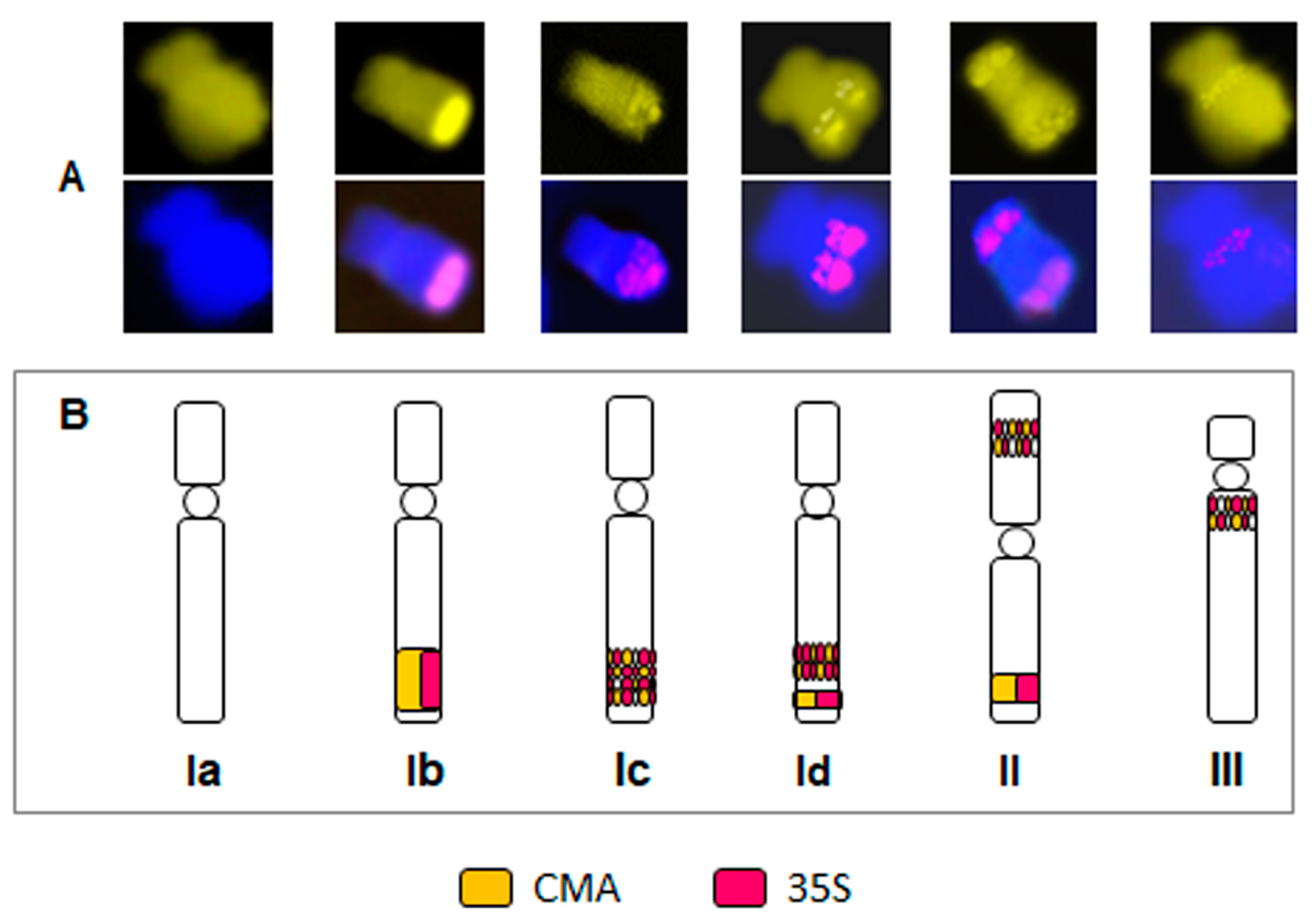
2.3. Particular B Chromosome Types in N. poeticus
2.4. Deviations from the Standard Karyotype Features
2.5. Genome Size Estimation
3. Discussion
3.1. Karyotype Characteristics and Heterochromatin Organization
3.2. The B Chromosomes in Narcissus poeticus
3.3. rDNA and Heterochromatin Organization on B Chromosomes
3.4. Natural Triploids of Narcissus poeticus
3.5. Deviant Forms of the Standard Karyotype
3.6. DNA Content Increases Due to the Presence of B Chromosomes and Triploidy
4. Materials and Methods
4.1. Plant Material
4.2. Genome Size Assessment
4.3. Chromosome Preparation
4.4. Feulgen Staining and Karyotype Features
4.5. Silver Staining
4.6. Slide Preparation for Fluorochrome Banding and FISH
4.7. Fluorochrome Banding
4.8. Fluorescence In Situ Hybridization (FISH)
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanks, G.R. Narcissus and Daffodil, The Genus Narcissus, 1st ed.; Taylor and Francis: London, UK, 2002; pp. 1–52. [Google Scholar]
- Fernandes, A. Sur la phylogenie des especes du genre Narcissus L. Bol. Soc. Brot. 1951, II 25, 113–190. [Google Scholar]
- Webb, D.A. Narcissus L. In Flora Europaea, Alismataceae to Orchidaceae (Monocotyledones); Tutin, T.G., Heywood, V.H., Burges, N.A., Moore, D.M., Valentine, D.H., Walters, S.M., Web, D.A., Chater, A.O., Richardson, I.B.K., Eds.; Cambridge University Press: Cambridge, UK, 1980; Volume 5, pp. 78–84. [Google Scholar]
- Graham, S.W.; Barrett, S.C.H. Phylogenetic reconstruction of the evolution of stylar polymorphism in Narcissus (Amaryllidaceae). Am. J. Bot. 2004, 91, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, B.J.M. The systematic value of nuclear DNA content for all species of Narcissus L. (Amaryllidaceae). Plant Syst. Evol. 2008, 275, 109–132. [Google Scholar] [CrossRef]
- RHS (Royal Horticultural Society). Narcissus Botanical Classification. 2016. Available online: https://www.rhs.org.uk/plants/pdfs/plant-registration-forms/daffbotanical (accessed on 10 May 2023).
- Kington, S. The International Daffodil Checklist; Royal Horticultural Society: London, UK, 1989. [Google Scholar]
- Wylie, A.P. The history of garden narcissi. Heredity 1952, 6, 137–156. [Google Scholar] [CrossRef]
- Brandham, P.E.; West, J.P. Correlation between nuclear DNA values and differing optimal ploidy levels in Narcissus, Hyacinthus and Tulipa cultivars. Genetica 1993, 90, 1–8. [Google Scholar] [CrossRef]
- Brandham, P.E. Chromosome numbers in narcissus cultivars and their significance to the plant breeder. Plantsman 1992, 14, 133–168. [Google Scholar]
- Marques, I.; Aguilar, J.F.; Martins-Louçao, M.A.; Moharrek, F.; Feliner, G.N. A three-genome five-gene comprehensive phylogeny of the bulbous genus Narcissus (Amaryllidaceae) challenges current classifications and reveals multiple hybridization events. Taxon 2017, 66, 832–854. [Google Scholar] [CrossRef]
- Leitch, I.J.; Johnston, E.; Pellicer, J.; Hidalgo, O.; Bennett, M.D. Plant DNA C-Values Database. 2019. Available online: https://cvalues.science.kew.org/ (accessed on 23 August 2023).
- Fernandes, A. L’evolution chez le genre Narcissus L. Anal. Inst. Bot. Cavanilles 1975, 32, 843–872. [Google Scholar]
- Tucci, G.F.; Winfield, M.O.; D’Amato, G.F.; Gregori, C.; Trombetta, B.; De Dominicis, R.I. Genetic diversity in Narcissus poëticus L. and N. radiiflorus Salisb. (Amaryllidaceae) in two different populations: AFLP and karyological studies. Caryologia 2004, 57, 405–411. [Google Scholar] [CrossRef]
- Fernandes, A. Sur le rôle probable des heterochromatinosomes dans l’évolution des nombres chromosomiques. Sci. Gen. 1952, 4, 168–181. [Google Scholar]
- Brandham, P.E.; Kirton, P.R. The chromosomes of species, hybrids and cultivars of Narcissus L. (Amaryllidaceae). Kew Bull. 1987, 42, 65–102. [Google Scholar] [CrossRef]
- Pogosian, A.I. Numeri Chromosomatum Magnoliophytorum Florae URSS, Aceraceae–Menyanthaceae; Takhtajan, A., Ed.; Nauka: Leningrad, Russia, 1990; p. 47. [Google Scholar]
- Jones, R.N. Tansley review no. 85: B chromosomes in plants. New Phytol. 1995, 131, 411–434. [Google Scholar] [CrossRef] [PubMed]
- Palestis, B.G.; Trivers, R.; Burt, A.; Jones, R.N. The distribution of B chromosomes across species. Cytogenet. Genome Res. 2004, 106, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Trivers, R.; Burt, A.; Palestis, B.G. B chromosomes and genome size in flowering plants. Genome 2004, 47, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Puertas, M.J.; Romera, F.; de la Peña, A. Comparison of B chromosome effects on Secale cereale and Secale vavilovii. Heredity 1985, 55, 229–234. [Google Scholar] [CrossRef]
- Bougourd, S.M.; Jones, N. B chromosomes: A physiological enigma. New Phytol. 1997, 137, 43–54. [Google Scholar] [CrossRef]
- Camacho, J.P.M.; Sharbel, T.F.; Beukeboom, L.W. B–chromosome evolution. Philos. Trans. R. Soc. Lond. B Bios. Sci. 2000, 355, 163–178. [Google Scholar] [CrossRef]
- Puertas, M.J. Nature and evolution of B chromosomes in plants: A non-coding but information-rich part of plant genomes. Cytogenet. Genome. Res. 2002, 96, 198–205. [Google Scholar] [CrossRef]
- Jones, R.N.; Viegas, W.; Houben, A. A century of B chromosomes in plants: So what? Ann. Bot. 2008, 101, 767–775. [Google Scholar] [CrossRef]
- Dhar, M.K.; Kour, J.; Kaul, S. Origin, behaviour, and transmission of B chromosome with special reference to Plantago lagopus. Genes 2019, 10, 152. [Google Scholar] [CrossRef]
- Bednářová, M.; Karafiátová, M.; Hřibová, E.; Bartoš, J. B chromosomes in genus Sorghum (Poaceae). Plants 2021, 10, 505. [Google Scholar] [CrossRef] [PubMed]
- Rajpal, V.R.; Sharma, S.; Sehgal, D.; Sharma, P.; Wadhwa, N.; Dhakate, P.; Chandra, A.; Thakur, R.K.; Deb, S.; Rama Rao, S.; et al. Comprehending the dynamism of B chromosomes in their journey towards becoming unselfish. Front. Cell Dev. Biol. 2023, 10, 1072716. [Google Scholar] [CrossRef] [PubMed]
- Plowman, A.; Bougourd, S. Selectively advantageous effects of B chromosomes on germination behaviour in Allium schoenoprasum L. Heredity 1994, 72, 587–593. [Google Scholar] [CrossRef]
- Holmes, D.S.; Bougourd, S.M. B–chromosome selection in Allium schoenoprasum. II. Experimental populations. Heredity 1991, 67, 117–122. [Google Scholar] [CrossRef]
- Karafiátová, M.; Bednářová, M.; Said, M.; Čížková, J.; Holušová, K.; Blavet, N.; Bartoš, J. The B chromosome of Sorghum purpureosericeum reveals the first pieces of its sequence. J. Exp. Bot. 2021, 72, 1606–1616. [Google Scholar] [CrossRef] [PubMed]
- Leitch, I.J.; Bennett, M.D. Genome downsizing in polyploidy plants. Biol. J. Linn. Soc. Lond. 2004, 82, 651–663. [Google Scholar] [CrossRef]
- Knight, C.A.; Molinari, N.A.; Petrov, D.A. The large genome constraint hypothesis: Evolution, ecology and phenotype. Ann. Bot. 2005, 95, 177–190. [Google Scholar] [CrossRef]
- Vidic, T.; Greilhuber, J.; Vilhar, B.; Dermastia, M. Selective significance of genome size in a plant community with heavy metal pollution. Ecol. Appl. 2009, 19, 1515–1521. [Google Scholar] [CrossRef]
- Pustahija, F.; Brown, S.C.; Bogunić, F.; Bašić, N.; Muratović, E.; Ollier, S.; Hidalgo, O.; Bourge, M.; Stevanović, V.; Siljak-Yakovlev, S. Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa. Plant Soil 2013, 373, 427–453. [Google Scholar] [CrossRef]
- Gregory, T.R. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. 2001, 76, 65–101. [Google Scholar] [CrossRef]
- Siljak–Yakovlev, S.; Pustahija, F.; Šolić, E.M.; Bogunić, F.; Muratović, E.; Bašić, N.; Catrice, O.; Brown, S.C. Towards a genome size and chromosome number database of Balkan flora: C–values in 343 taxa with novel values for 242. Adv. Sci. Lett. 2010, 3, 190–224. [Google Scholar] [CrossRef]
- Olszewska, M.J.; Osiecka, R. The relationship between 2C DNA content, life–cycle type, systematic position, and the level of DNA endoreplication in nuclei of parenchyma cells during growth and differentiation of roots in some monocotyledonous species. Biochem. Physiol. Pflanz. 1982, 177, 319–336. [Google Scholar] [CrossRef]
- D’Amato, G. Karyotype and heterochromatin characterization in some species of Narcissus (Amaryllidaceae). Caryologia 2004, 57, 99–105. [Google Scholar] [CrossRef]
- Maugini, E. Ricerche citosistematiche su Narcissus puccinellii Parl. (N. jonquilla L. × N. poeticus L.). Caryologia 1962, 15, 485–506. [Google Scholar] [CrossRef]
- De Dominicis, R.I.; D’Amato, G.; Tucci, G.F. On the hybrid origin of Narcissus biflorus (Amaryllidaceae): Analysis of C–banding and rDNA structure. Caryologia 2002, 55, 129–134. [Google Scholar] [CrossRef]
- Robert, T.; Khalfallah, N.; Martel, E.; Lamy, F.; Poncet, V.; Alline, C.; Remigereau, M.S.; Rekima, S.; Leveugle, M.; Lakis, G.; et al. Pennisetum. In Wild Crop Relatives: Genomic and Breeding Resources, 2011th ed.; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 217–255. [Google Scholar] [CrossRef]
- Porter, H.L.; Rayburn, A.L. B chromosome and C-band heterochromatin variation in Arizona maize populations adapted to different altitudes. Genome 1990, 33, 659–662. [Google Scholar] [CrossRef]
- Parker, J.S.; Lozano, R.; Taylor, S.; Rejon, M.R. Chromosomal structure of populations of Scilla autumnalis in the Iberian Peninsula. Heredity 1991, 67, 287–297. [Google Scholar] [CrossRef]
- Jang, T.-S.; Parker, J.S.; Weiss-Schneeweiss, H. Structural polymorphisms and distinct genomic composition suggest recurrent origin and ongoing evolution of B chromosomes in the Prospero autumnale complex (Hyacinthaceae). New Phytol. 2016, 210, 669–679. [Google Scholar] [CrossRef]
- Johnson Pokorná, M.; Reifová, R. Evolution of B chromosomes: From dispensable parasitic chromosomes to essential genomic players. Front. Genet. 2021, 12, 727570. [Google Scholar] [CrossRef]
- Bougourd, S.M.; Parker, J.S. The B–chromosome system of Allium schoenoprasum. III. An abrupt change in B–frequency. Chromosoma 1979, 75, 385–392. [Google Scholar] [CrossRef]
- Maluszynska, J.; Schweizer, D. Ribosomal RNA genes in B chromosomes of Crepis capillaris detected by non-radioactive in situ hybridization. Heredity 1989, 62, 59–65. [Google Scholar] [CrossRef]
- Muratović, E.; Robin, O.; Bogunić, F.; Šoljan, D.; Siljak-Yakovlev, S. Karyotype evolution and speciation of European lilies from Lilium sect. Liriotypus. Taxon 2010, 59, 165–175. [Google Scholar] [CrossRef]
- Huang, W.; Du, Y.; Zhao, X.; Jin, W. B chromosome contains active genes and impacts the transcription of A chromosomes in maize (Zea mays L.). BMC Plant Biol. 2016, 16, 88. [Google Scholar] [CrossRef]
- Marques, A.; Klemme, S.; Houben, A. Evolution of plant B chromosome enriched sequences. Genes 2018, 9, 515. [Google Scholar] [CrossRef]
- Ramsey, J.; Schemske, D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 1998, 29, 467–501. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S.; Tate, J.A. Advances in the study of polyploidy since plant speciation. New Phytol. 2003, 161, 173–191. [Google Scholar] [CrossRef]
- Leitch, A.R.; Leitch, I.J. Genomic plasticity and the diversity of polyploid plants. Science 2008, 320, 481–483. [Google Scholar] [CrossRef]
- Van Tuyl, J.M. Interspecific hybridization of flower bulbs: A review. Acta Hortic. 1997, 430, 465–476. [Google Scholar] [CrossRef]
- Hadač, E. Ecological significance of polyploidy in high mountain plants and plant communities. Folia Geobot. Phytotax. 1989, 24, 51–56. [Google Scholar] [CrossRef]
- Karihaloo, J.L. Variation in the karyotype of three cultivars of Narcissus tazetta L. (Amaryllidaceae). Genetica 1987, 73, 217–221. [Google Scholar] [CrossRef]
- Jones, R.N.; Houben, A. B chromosomes in plants: Escapees from the A chromosome genome? Trends Plant Sci. 2003, 8, 417–423. [Google Scholar] [CrossRef]
- Ribeiro, T.; Pires, B.; Delgado, M.; Viegas, W.; Jones, N.; Morais–Cecílio, L. Evidence for ‘cross–talk’ between A and B chromosomes of rye. Proc. R. Soc. Lond. B 2004, 271 (Suppl. S6), S482–S484. [Google Scholar] [CrossRef]
- Marie, D.; Brown, S.C. A cytometric exercise in plant DNA histograms with 2C values for 70 species. Biol. Cell 1993, 78, 41–51. [Google Scholar] [CrossRef]
- Galbraith, D.; Harkins, K.; Maddoks, J.; Ayres, N.; Sharma, D.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef]
- Siljak-Yakovlev, S.; Pustahija, F.; Vičić-Bočkor, V.; Robin, O. Molecular cytogenetics (Fluorescence In Situ Hybridization-FISH and fluorochrome banding): Resolving species relationships and genome organization. In Molecular Plant Taxonomy: Methods and Protocols, Methods in Molecular Biology; Besse, P., Ed.; Humana Press, Springer Science + Business Media: New York, NY, USA, 2021; Volume 2222, pp. 363–379. [Google Scholar] [CrossRef]
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Arano, H.; Saito, H. Cytological studies in family Umbelliferae 5. Karyotypes of seven species in subtribe Seselinae. La Kromosomo 1980, 2, 471–480. [Google Scholar]
- Hall, K.J.; Parker, J.S. Stable chromosome fission associated with rDNA mobility. Chromosome Res. 1995, 3, 417–422. [Google Scholar] [CrossRef]
- Schweizer, D. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma 1976, 58, 307–324. [Google Scholar] [CrossRef]
- Siljak–Yakovlev, S.; Cerbah, M.; Couland, J.; Stoian, V.; Brown, S.C.; Zoldos, V.; Jelenic, S.; Papes, D. Nuclear DNA content, base composition, heterochromatin and rDNA in Picea omorika and Picea abies. Theor. Appl. Genet. 2002, 104, 505–512. [Google Scholar] [CrossRef]
- Martin, J.; Hesemann, C.U. Evaluation of improved Giemsa C—and fluorochrome banding–techniques in rye chromosome. Heredity 1988, 6, 459–467. [Google Scholar] [CrossRef]
- Heslop–Harrison, L.S.; Schwarzacher, T.; Anamthawat–Jonsson, K.; Leitch, I.J. In situ hybridization with automated chromosome denaturation techniques. Method. Mol. Cell. Biol. 1991, 3, 109–116. [Google Scholar]
- Gerlach, W.T.; Dyer, T.A. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980, 8, 4851–4865. [Google Scholar] [CrossRef] [PubMed]
| Acc. No | 2n | Pair 7 | Bs Type | Number of Nucleoli | Ecological Characteristics | |
|---|---|---|---|---|---|---|
| CMA | 35S | |||||
| 1. | 14 | +/− | +/− | / | 1–2 | A |
| 2. | 14 T | +/− | +/− | / | 1–2 | |
| 3. | 14 | +/− | +/− | / | 1–2 | |
| 4. | 14 | / | 1–2 | |||
| 5. | 14 + 0 − 1B | + | + | Ib, Ic | 1–3 | B |
| 6. | 14 + 0 − 1B | + | + | Ib, Ic | 1–3 | |
| 7. | 14 + 0 − 1B | + | + | Ia, Ib, Ic | 1–3 | |
| 8. | 14 + 0 − 3B 21 + 0 − 3B | + | + | Ib, Ic, Id, II, III | 1–6 | |
| 9. | 14 + 0 − 1B | + | + | Ic, Id | 1–3 | C |
| 10. | 14 + 0 − 1B | + | + | Ia, Id | 1–3 | |
| 11. | 14 T + 0 − 2Bs | + | + | Ib, Ic, Id | 1–4 | D |
| 12. | 14 T + 0 − 1B 21 | + | + | Ib, Ic, Id | 1–3 | |
| 13. | 14 + 0 − 1B | + | + | Ib, Ic, Id | 1–3 | |
| Diploids | Triploids | ||||||
|---|---|---|---|---|---|---|---|
| Translocation | |||||||
| Without Bs (Pops 1–4) | With 1B (Pops 5–7,9,10,12,13) | With 2Bs (Pop 11) | Serpentine (Pop 12) | Limestone (Pop 2) | Without Bs (Pops 8,12) | With 3 Bs (Pop 8) | |
| 2C DNA (pg ± sd) | 25.25 ± 0.39 | 25.81 ± 0.36 | 26.52 ± 0.15 | 25.77 | 24.91 | 34.55 ± 0.59 | 38.80 |
| Bs contribution to total 2C DNA (%; ±sd) | 2.22 ± 0.27 | 5.03 ± 0.09 | 12.30 | ||||
| Acc. No | Locality | Substrate | Latitude (N) | Longitude (E) | Altitude (m) | N |
|---|---|---|---|---|---|---|
| 1. | Mt. Čvrsnica, Mala Čvrsnica, B&H | L | 43°34′38″ | 17°29′33″ | 1384 | 5 |
| 2. | Mt. Čvrsnica, Barice, B&H | L | 43°35′01″ | 17°30′02″ | 1350 | 6 |
| 3. | Glamočko Polje, Mliništa, B&H | L | 44°14′07″ | 16°49′57″ | 1198 | 11 |
| 4. | Mt. Zelengora, Točila, B&H | L | 43°19′51″ | 18°33′10″ | 1470–1570 | 11 |
| 5. | Livanjsko Polje, Peulje, B&H | L | 44°08′42″ | 16°27′56″ | 806–859 | 13 |
| 6. | Livanjsko Polje, Čelebići, B&H | L | 43°56′01″ | 16°40′52″ | 710 | 5 |
| 7. | Mt. Čvrsnica, Rakitno, B&H | L | 43°32′79″ | 17°24′90″ | 895–910 | 8 |
| 8. | Gatačko Polje, Avtovica, B&H | L | 43°09′15″ | 18°31′32″ | 940 | 54 |
| 9. | Mt. Biokovo, Lađena, Cro | L | 43°17′27″ | 17°05′25″ | 1265 | 7 |
| 10. | Mt. Biokovo, Vošac, Cro | L | 43°18′46″ | 17°04′01″ | 1340 | 9 |
| 11. | Kladanj, Katranica, B&H | S | 44°16′09″ | 18°33′15″ | 800–940 | 26 |
| 12. | Žepče, Matinski Vis, B&H | S | 44°28′03″ | 17°58′37″ | 820–900 | 23 |
| 13. | Mt. Zlatibor, Mokra Gora, Srb | S | 43°49′09″ | 19°30′24″ | 810 | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pustahija, F.; Bašić, N.; Siljak-Yakovlev, S. Karyotype Variability in Wild Narcissus poeticus L. Populations from Different Environmental Conditions in the Dinaric Alps. Plants 2024, 13, 208. https://doi.org/10.3390/plants13020208
Pustahija F, Bašić N, Siljak-Yakovlev S. Karyotype Variability in Wild Narcissus poeticus L. Populations from Different Environmental Conditions in the Dinaric Alps. Plants. 2024; 13(2):208. https://doi.org/10.3390/plants13020208
Chicago/Turabian StylePustahija, Fatima, Neđad Bašić, and Sonja Siljak-Yakovlev. 2024. "Karyotype Variability in Wild Narcissus poeticus L. Populations from Different Environmental Conditions in the Dinaric Alps" Plants 13, no. 2: 208. https://doi.org/10.3390/plants13020208
APA StylePustahija, F., Bašić, N., & Siljak-Yakovlev, S. (2024). Karyotype Variability in Wild Narcissus poeticus L. Populations from Different Environmental Conditions in the Dinaric Alps. Plants, 13(2), 208. https://doi.org/10.3390/plants13020208






