Transcriptome Profiles Reveals ScDREB10 from Syntrichia caninervis Regulated Phenylpropanoid Biosynthesis and Starch/Sucrose Metabolism to Enhance Plant Stress Tolerance
Abstract
1. Introduction
2. Results
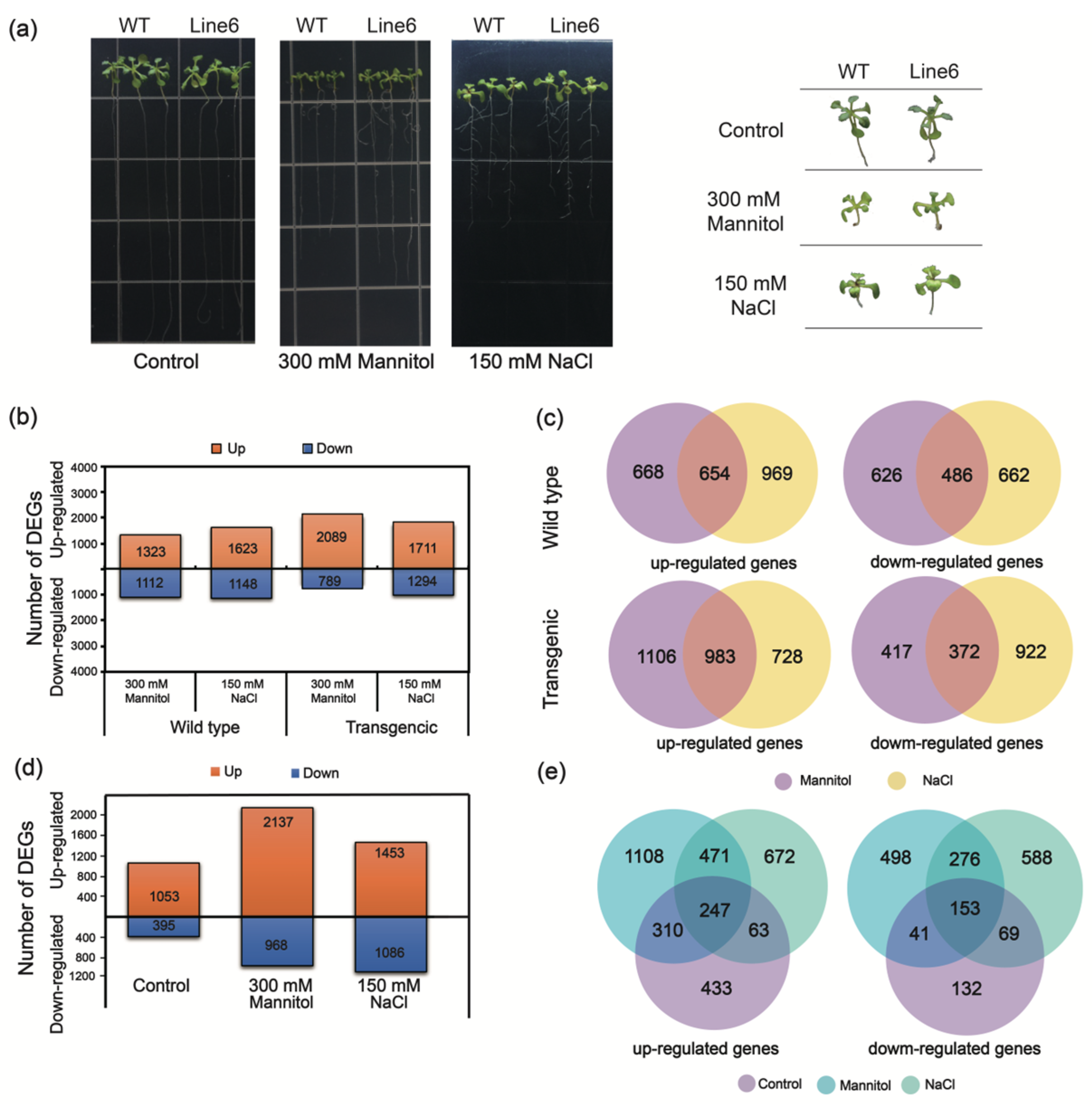
2.1. The DEGs Analysis of Transcriptomes between Overexpression-ScDREB10 and WT Plants in Response to Osmotic and Salt Stresses
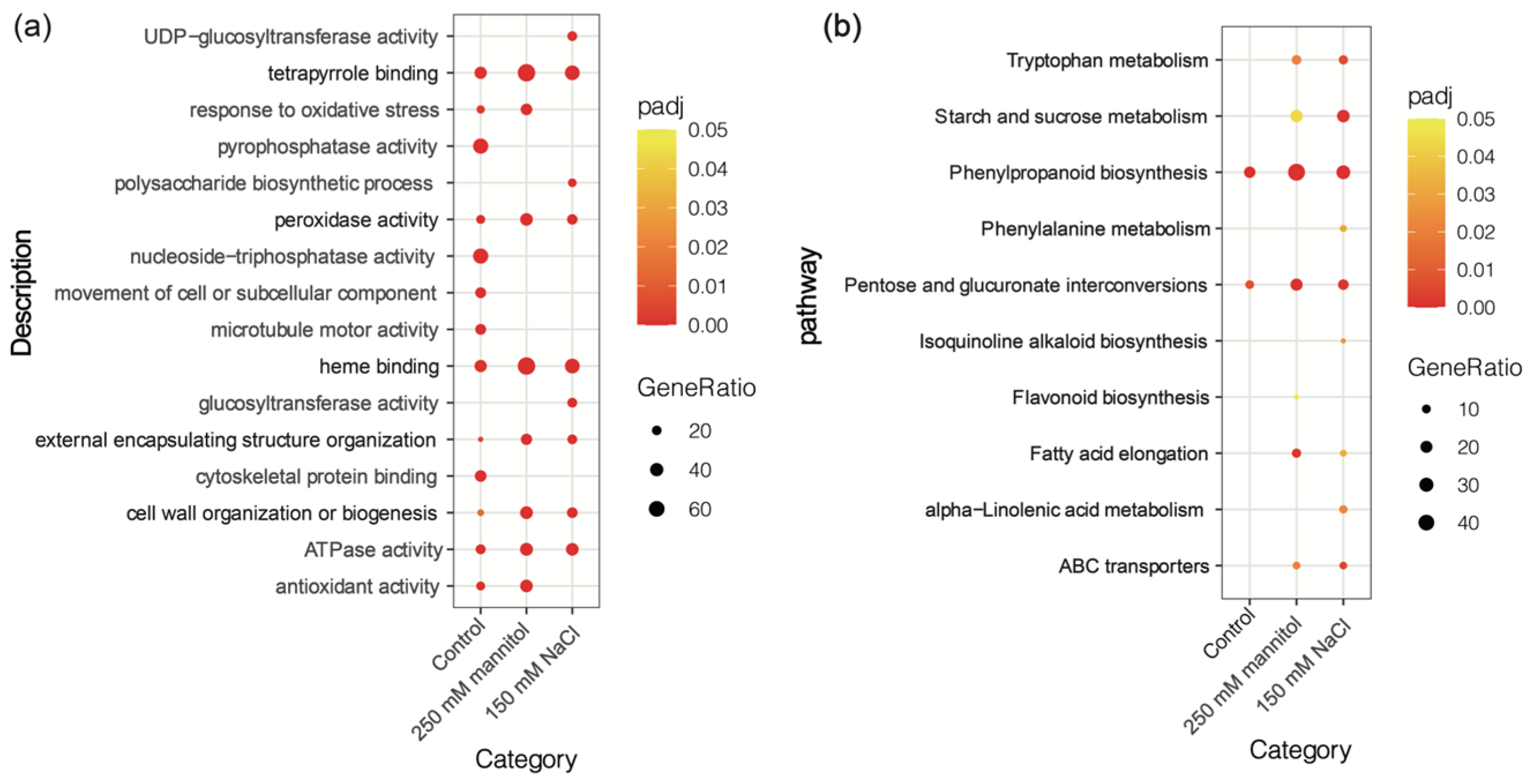
2.2. GO and KEGG Analyses between Overexpression-ScDREB10 and WT Transcriptomes in Response to Osmotic and Salt Stresses
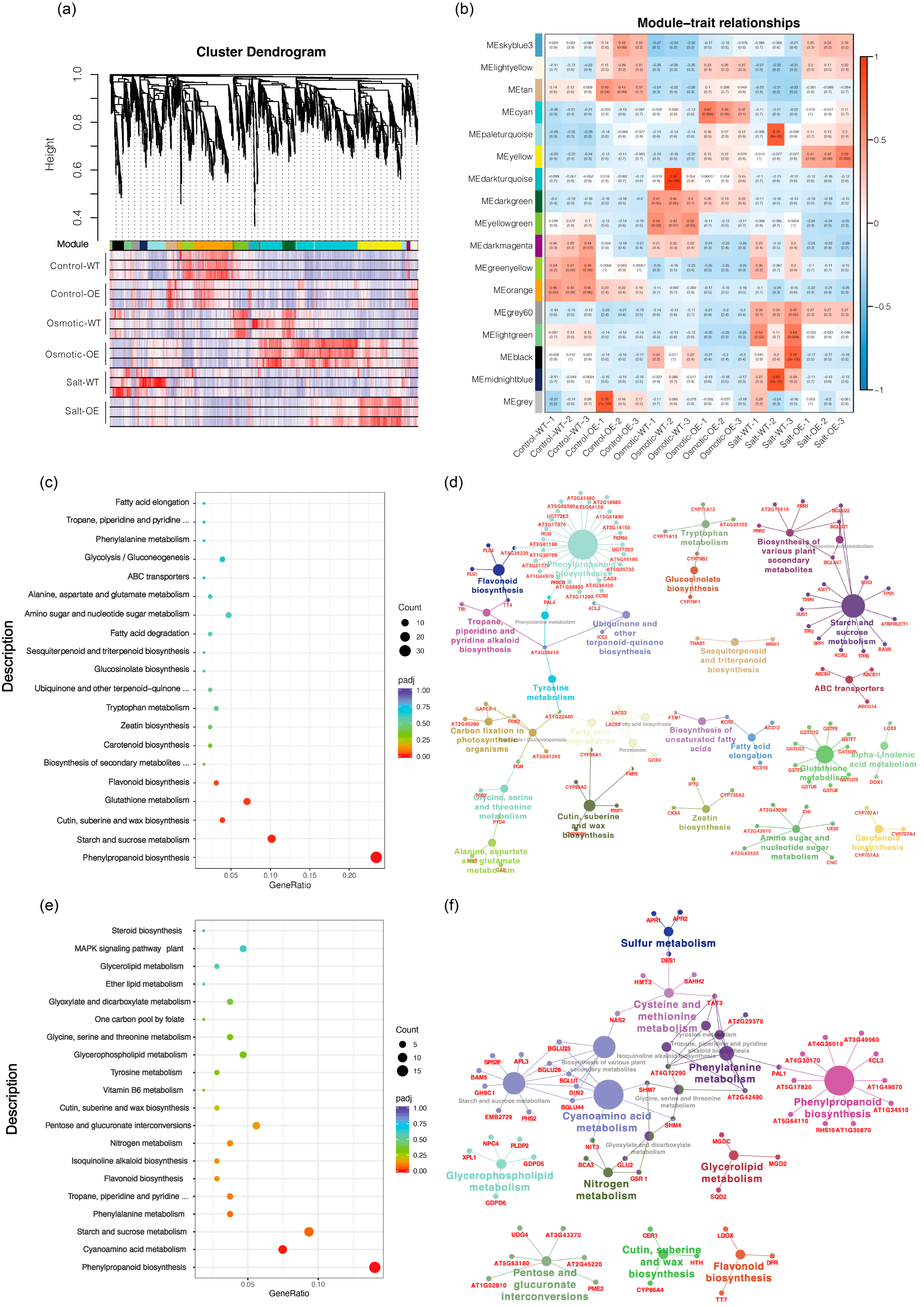
2.3. WGCNA of All DEGs between Overexpression-ScDREB10 and WT Transcriptomes in Response to Osmotic and Salt Stresses
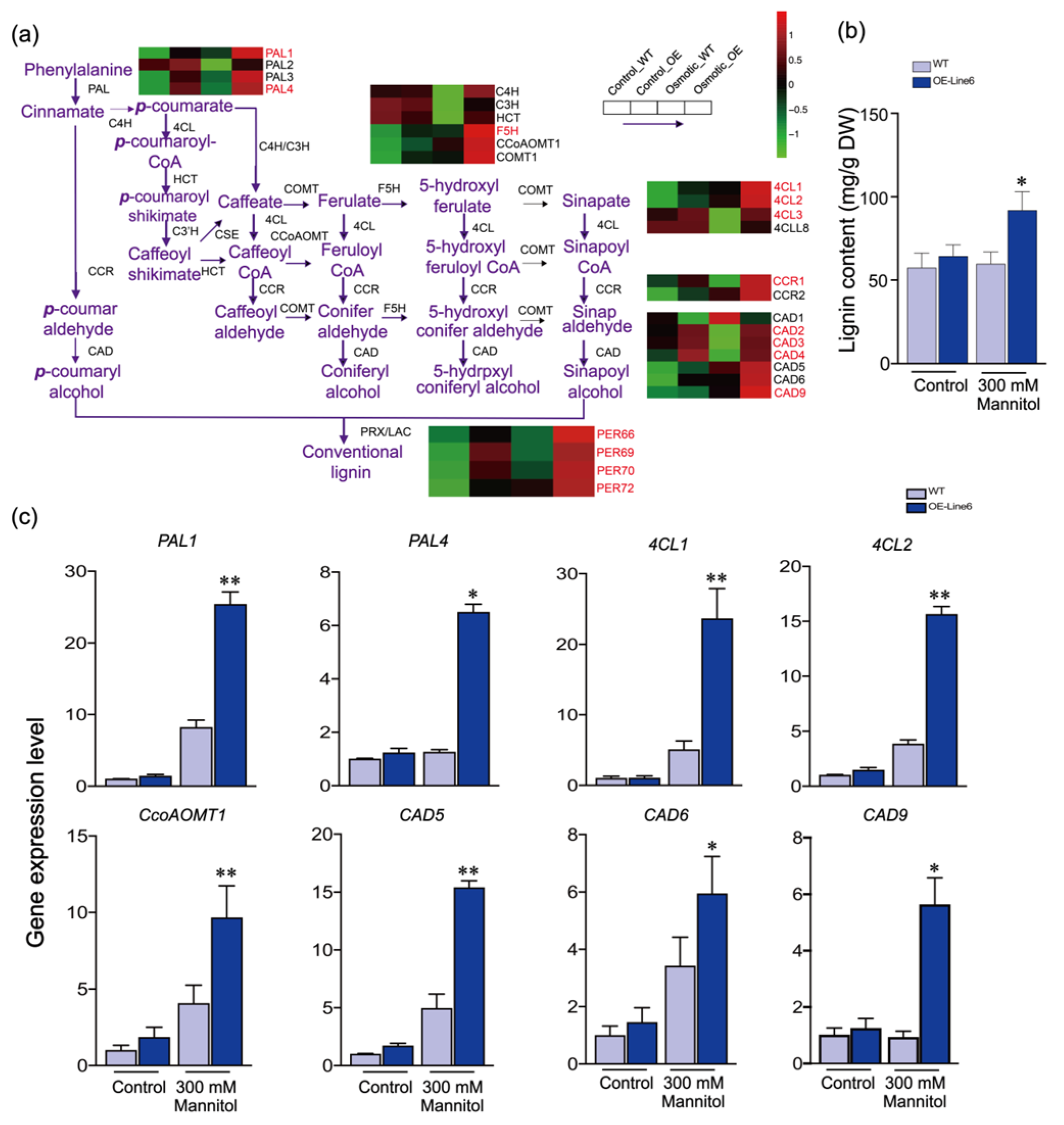
2.4. ScDREB10 May Participate in Regulating Phenylpropanoid Biosynthesis in Response to Plant Osmotic Stress
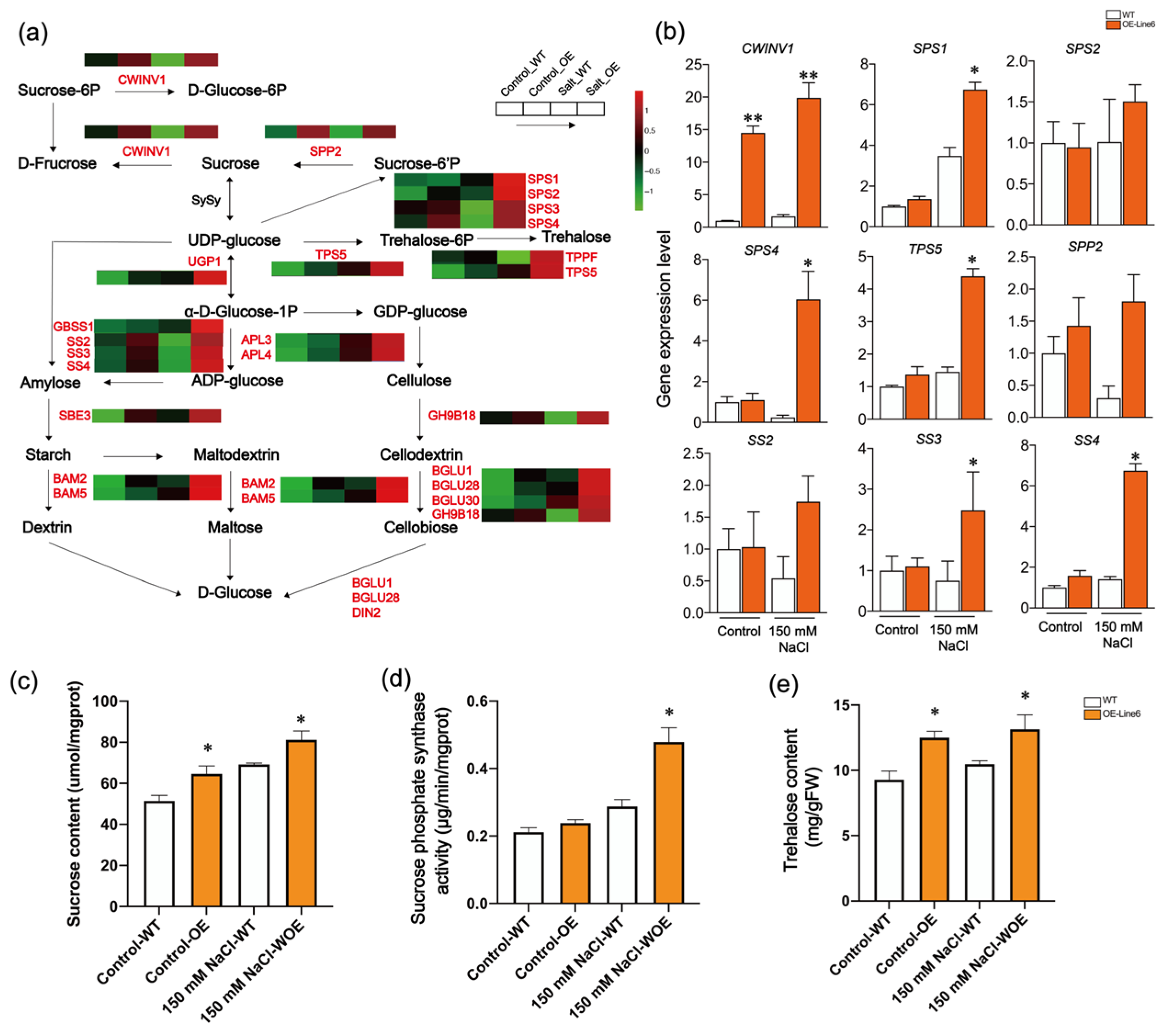
2.5. ScDREB10 May Participate in Regulating Starch and Sucrose Metabolism in Response to Salt Stress
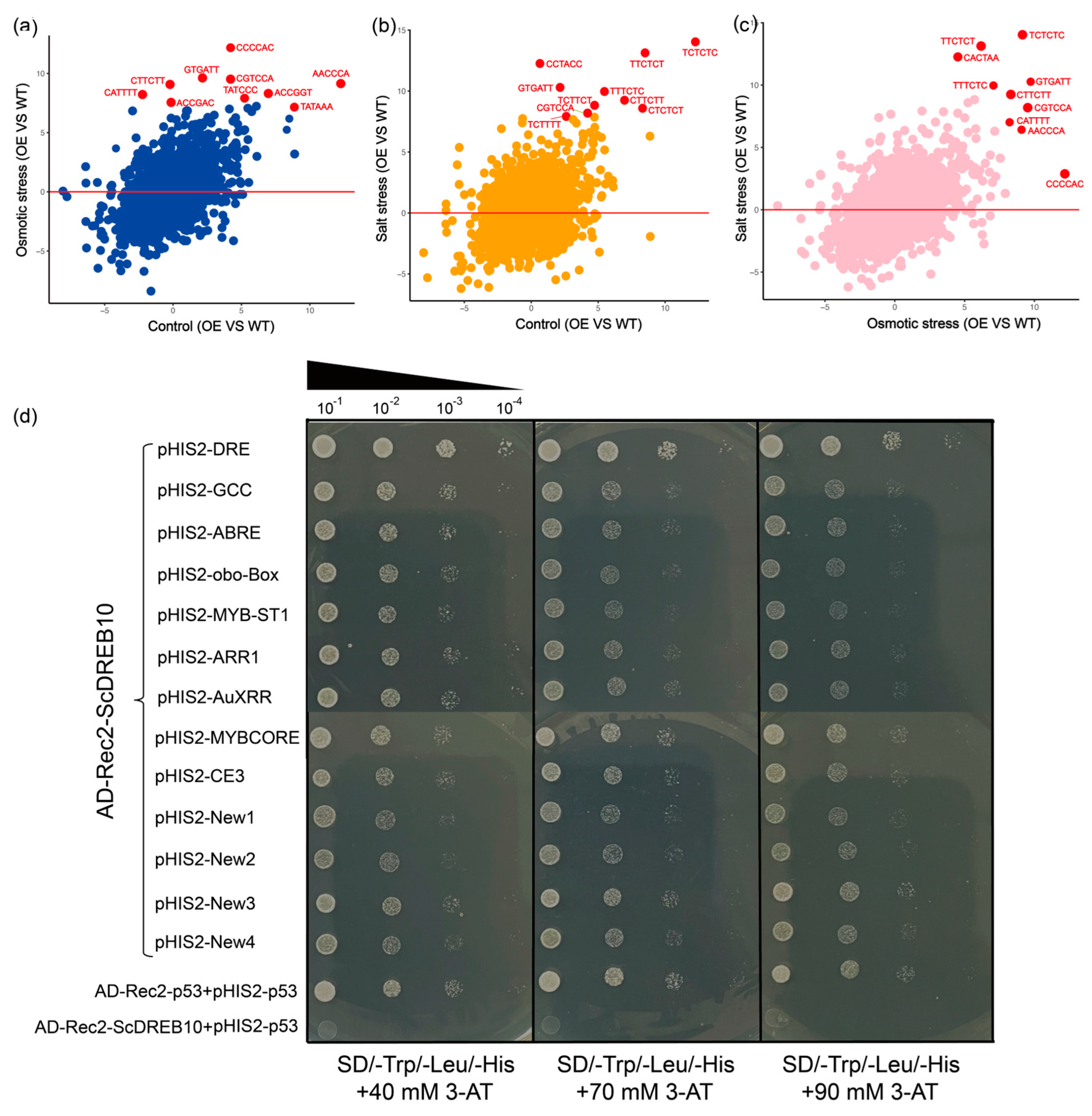
2.6. Cis-Acting Promoter Element Analysis of ScDREB10 in Response to Osmotic and Salt Stresses
3. Discussion
3.1. ScDREB10 Contributes to Drought Tolerance by Activating the Lignin Biosynthesis Pathway
3.2. ScDREB10 Contributes to Salt Tolerance by Activating the Starch and Sucrose Metabolism
3.3. ScDREB10 Was Not Only Able to Bind with Classical Cis-Elements, but Also Bind with Unknown Elements
4. Materials and Methods
4.1. Plant Materials and Methods
4.2. RNA Sequencing (RNA-Seq) Analysis
4.3. RNA Sequencing (RNA-Seq) Analysis
4.4. Cis-Acting Promoter Element Analysis
4.5. Yeast One-Hybrid Assay
4.6. Quantification and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Boyer, J.S. Plant Productivity and Environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.Z.; Xiong, L.M.; Shi, H.Z.; Yang, S.H.; Herrera-Estrella, L.R.; Xu, G.H.; Chao, D.Y.; Li, J.R.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Rejeb, I.B.; Pastor, V.; Mauch-Mani, B. Plant Responses to Simultaneous Biotic and Abiotic Stress: Molecular Mechanisms. Plants 2014, 3, 458–475. [Google Scholar] [CrossRef]
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef]
- Hussain, S.S.; Kayani, M.A.; Amjad, M. Transcription Factors as Tools to Engineer Enhanced Drought Stress Tolerance in Plants. Biotechnol. Progr. 2011, 27, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.P.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006, 18, 1292–1309. [Google Scholar] [CrossRef] [PubMed]
- Augustine, S.M.; Narayan, J.A.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Tuteja, N.; Subramonian, N. Overexpression of EaDREB2 and pyramiding of EaDREB2 with the pea DNA helicase gene (PDH45) enhance drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Cell. Rep. 2015, 34, 247–263. [Google Scholar] [CrossRef]
- Gupta, K.; Agarwal, P.K.; Reddy, M.K.; Jha, B. SbDREB2A, an A-2 type DREB transcription factor from extreme halophyte Salicornia brachiata confers abiotic stress tolerance in Escherichia coli. Plant Cell. Rep. 2010, 29, 1131–1137. [Google Scholar] [CrossRef]
- Li, X.S.; Zhang, D.Y.; Li, H.Y.; Wang, Y.C.; Zhang, Y.M.; Wood, A.J. EsDREB2B, a novel truncated DREB2-type transcription factor in the desert legume Eremosparton songoricum, enhances tolerance to multiple abiotic stresses in yeast and transgenic tobacco. BMC Plant Biol. 2014, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.G.; Zhang, W.K.; He, S.J.; Zhang, J.S.; Liu, Q.; Chen, S.Y. An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor. Appl. Genet. 2003, 106, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Gupta, K.; Lopato, S.; Agarwal, P. Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J. Exp. Bot. 2017, 68, 2135–2148. [Google Scholar] [CrossRef] [PubMed]
- Bouaziz, D.; Pirrello, J.; Ben Amor, H.; Hammami, A.; Charfeddine, M.; Dhieb, A.; Bouzayen, M.; Gargouri-Bouzid, R. Ectopic expression of dehydration responsive element binding proteins (StDREB2) confers higher tolerance to salt stress in potato. Plant Physiol. Biochem. 2012, 60, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Q.Y.; Cheng, X.G.; Xu, Z.S.; Li, L.C.; Ye, X.G.; Xia, L.Q.; Ma, Y.Z. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 2007, 353, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, Z.; Xia, L.; Li, L.; Cheng, X.; Dong, J.; Wang, Q.; Ma, Y. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J. Exp. Bot. 2009, 60, 121–135. [Google Scholar] [CrossRef]
- Liu, N.; Zhong, N.Q.; Wang, G.L.; Li, L.J.; Liu, X.L.; He, Y.K.; Xia, G.X. Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens. Planta 2007, 226, 827–838. [Google Scholar] [CrossRef]
- do Rego, T.F.C.; Santos, M.P.; Cabral, G.B.; Cipriano, T.D.; de Sousa, N.L.; Neto, O.A.D.; Aragao, F.J.L. Expression of a DREB 5-A subgroup transcription factor gene from Ricinus communis (RcDREB1) enhanced growth, drought tolerance and pollen viability in tobacco. Plant Cell Tiss. Org. 2021, 146, 493–504. [Google Scholar] [CrossRef]
- Chen, H.Y.; Hsieh, E.J.; Cheng, M.C.; Chen, C.Y.; Hwang, S.Y.; Lin, T.P. ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 2016, 211, 599–613. [Google Scholar] [CrossRef]
- Olsen, K.M.; Lea, U.S.; Slimestad, R.; Verheul, M.; Lillo, C. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 2008, 165, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Zhang, D.; Li, X.; Yang, H.; Wood, A.J. De novo assembly and characterization of the transcriptome in the desiccation-tolerant moss Syntrichia caninervis. BMC Res. Notes 2014, 7, 490. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Zhang, D.Y.; Li, X.S.; Guan, K.Y.; Yang, H.L. Novel DREB A-5 subgroup transcription factors from desert moss (Syntrichia caninervis) confers multiple abiotic stress tolerance to yeast. J. Plant Physiol. 2016, 194, 45–53. [Google Scholar] [CrossRef]
- Li, X.S.; Liang, Y.Q.; Gao, B.; Mijiti, M.; Bozorov, T.A.; Yang, H.L.; Zhang, D.Y.; Wood, A.J. ScDREB10, an A-5c type of DREB Gene of the Desert Moss Syntrichia caninervis, Confers Osmotic and Salt Tolerances to Arabidopsis. Genes 2019, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.Q.; Li, X.S.; Zhang, D.Y.; Gao, B.; Yang, H.L.; Wang, Y.C.; Guan, K.Y.; Wood, A.J. ScDREB8, a novel A-5 type of DREB gene in the desert moss Syntrichia caninervis, confers salt tolerance to Arabidopsis. Plant Physiol. Biochem. 2017, 120, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, D.; Gao, B.; Liang, Y.; Yang, H.; Wang, Y.; Wood, A.J. Transcriptome-Wide Identification, Classification, and Characterization of AP2/ERF Family Genes in the Desert Moss Syntrichia caninervis. Front. Plant Sci. 2017, 8, 262. [Google Scholar] [CrossRef]
- Liu, J.Y.; Yang, R.R.; Liang, Y.Q.; Wang, Y.; Li, X.S. The DREB A-5 Transcription Factor ScDREB5 From Syntrichia caninervis Enhanced Salt Tolerance by Regulating Jasmonic Acid Biosynthesis in Transgenic Arabidopsis. Front. Plant Sci. 2022, 13, 857396. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Maruyama, K.; Todaka, D.; Mizoi, J.; Yoshida, T.; Kidokoro, S.; Matsukura, S.; Takasaki, H.; Sakurai, T.; Yamamoto, Y.Y.; Yoshiwara, K.; et al. Identification of Cis-Acting Promoter Elements in Cold- and Dehydration-Induced Transcriptional Pathways in Arabidopsis, Rice, and Soybean. DNA Res. 2012, 19, 37–49. [Google Scholar] [CrossRef]
- Lata, C.; Mishra, A.K.; Muthamilarasan, M.; Bonthala, V.S.; Khan, Y.; Prasad, M. Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS ONE 2014, 9, e113092. [Google Scholar] [CrossRef]
- Mizoi, J.; Ohori, T.; Moriwaki, T.; Kidokoro, S.; Todaka, D.; Maruyama, K.; Kusakabe, K.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. GmDREB2A;2, a Canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-Type Transcription Factor in Soybean, Is Posttranslationally Regulated and Mediates Dehydration-Responsive Element-Dependent Gene Expression. Plant Physiol. 2013, 161, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Luo, T.; Zhao, H.; Su, Y.; Ji, W.; Li, H. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene 2020, 740, 144514. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, P. The DREB transcription factor, a biomacromolecule, responds to abiotic stress by regulating the expression of stress-related genes. Int. J. Biol. Macromol. 2023, 243, 125231. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Z.; Dixon, R.A. On-Off Switches for Secondary Cell Wall Biosynthesis. Mol. Plant 2012, 5, 297–303. [Google Scholar] [CrossRef]
- Li, T.; Wang, W.J.; Chen, Q.; Chen, J.L.; Zhang, X.; Wei, L.L.; Deng, L.L.; Yao, S.X.; Zeng, K.F. Transcription factor CsERF1B regulates postharvest citrus fruit resistance to Penicillium digitatum. Postharvest Biol. Tec. 2023, 198, 112260. [Google Scholar] [CrossRef]
- Ma, R.F.; Xiao, Y.; Lv, Z.Y.; Tan, H.X.; Chen, R.B.; Li, Q.; Chen, J.F.; Wang, Y.; Yin, J.; Zhang, L.; et al. AP2/ERF Transcription Factor, Ii049, Positively Regulates Lignan Biosynthesis in Isatis indigotica through Activating Salicylic Acid Signaling and Lignan/Lignin Pathway Genes. Front. Plant Sci. 2017, 8, 1361. [Google Scholar] [CrossRef]
- Moffat, C.S.; Ingle, R.A.; Wathugala, D.L.; Saunders, N.J.; Knight, H.; Knight, M.R. ERF5 and ERF6 Play Redundant Roles as Positive Regulators of JA/Et-Mediated Defense against Botrytis cinerea in Arabidopsis. PLoS ONE 2012, 7, e35995. [Google Scholar] [CrossRef]
- Zeng, J.K.; Li, X.; Xu, Q.; Chen, J.Y.; Yin, X.R.; Ferguson, I.B.; Chen, K.S. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 2015, 13, 1325–1334. [Google Scholar] [CrossRef]
- Zhang, J.; Yin, X.R.; Li, H.; Xu, M.; Zhang, M.X.; Li, S.J.; Liu, X.F.; Shi, Y.N.; Grierson, D.; Chen, K.S. ETHYLENE RESPONSE FACTOR39-MYB8 complex regulates low-temperature-induced lignification of loquat fruit. J. Exp. Bot. 2020, 71, 3172–3184. [Google Scholar] [CrossRef]
- Zhao, X.W.; Wang, Q.; Wang, D.; Guo, W.; Hu, M.X.; Liu, Y.L.; Zhou, G.K.; Chai, G.H.; Zhao, S.T.; Lu, M.Z. PagERF81 regulates lignin biosynthesis and xylem cell differentiation in poplar. J. Integr. Plant Biol. 2023, 65, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Li, X.S.; Yang, R.R.; Liang, Y.Q.; Gao, B.; Li, S.M.; Bai, W.W.; Oliver, M.J.; Zhang, D.Y. The ScAPD1-like gene from the desert moss Syntrichia caninervis enhances resistance to Verticillium dahliae via phenylpropanoid gene regulation. Plant J. 2023, 113, 75–91. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.Q.; Li, X.S.; Yang, R.R.; Gao, B.; Yao, J.X.; Oliver, M.J.; Zhang, D.Y. BaDBL1, a unique DREB gene from desiccation tolerant moss Bryum argenteum, confers osmotic and salt stress tolerances in transgenic Arabidopsis. Plant Sci. 2021, 313, 111047. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Jung, H.; Jang, G.; Jeong, J.S.; Kim, Y.S.; Ha, S.H.; Do Choi, Y.; Kim, J.K. Overexpression of the OsERF71 Transcription Factor Alters Rice Root Structure and Drought Resistance. Plant Physiol. 2016, 172, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.E.; Bang, S.W.; Kim, S.H.; Seo, J.S.; Yoon, H.B.; Kim, Y.S.; Kim, J.K. Overexpression of OsERF83, a Vascular Tissue-Specific Transcription Factor Gene, Confers Drought Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 7656. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Huang, Y.; Khadr, A.; Wang, Y.H.; Xu, Z.S.; Xiong, A.S. DcDREB1A, a DREB-binding transcription factor from Daucus carota, enhances drought tolerance in transgenic Arabidopsis thaliana and modulates lignin levels by regulating lignin-biosynthesis-related genes. Environ. Exp. Bot. 2020, 169, 103896. [Google Scholar] [CrossRef]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef]
- Rohde, A.; Morreel, K.; Ralph, J.; Goeminne, G.; Hostyn, V.; De Rycke, R.; Kushnir, S.; Van Doorsselaere, J.; Joseleau, J.P.; Vuylsteke, M.; et al. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino Acid, and carbohydrate metabolism. Plant Cell. 2004, 16, 2749–2771. [Google Scholar] [CrossRef]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar] [CrossRef]
- Huang, J.L.; Gu, M.; Lai, Z.B.; Fan, B.F.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z.X. Functional Analysis of the Arabidopsis PAL Gene Family in Plant Growth, Development, and Response to Environmental Stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Hare, P.D.; Cress, W.A.; Van Staden, J. Dissecting the roles of osmolyte accumulation during stress. Plant Cell. Environ. 1998, 21, 535–553. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, L.; Chen, M.; Zhou, W.; Dong, Q.; Jiang, H.; Cheng, B. The DOF-Domain Transcription Factor ZmDOF36 Positively Regulates Starch Synthesis in Transgenic Maize. Front. Plant Sci. 2019, 10, 465. [Google Scholar] [CrossRef] [PubMed]
- Zahoor, R.; Dong, H.R.; Abid, M.; Zhao, W.Q.; Wang, Y.H.; Zhou, Z.G. Potassium fertilizer improves drought stress alleviation potential in cotton by enhancing photosynthesis and carbohydrate metabolism. Environ. Exp. Bot. 2017, 137, 73–83. [Google Scholar] [CrossRef]
- Saad, A.S.; Li, X.; Li, H.P.; Huang, T.; Gao, C.S.; Guo, M.W.; Cheng, W.; Zhao, G.Y.; Liao, Y.C. A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci. 2013, 203–204, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Zhang, F.; Pakrasi, H.B. Enhanced production of sucrose in the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Sci. Rep. 2020, 10, 390. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Q.; Yu, M.; Zhang, Y.; Wu, Y.; Zhang, H. Transgenic salt-tolerant sugar beet (Beta vulgaris L.) constitutively expressing an Arabidopsis thaliana vacuolar Na/H antiporter gene, AtNHX3, accumulates more soluble sugar but less salt in storage roots. Plant Cell Environ. 2008, 31, 1325–1334. [Google Scholar] [CrossRef]
- Huang, H.; Xie, S.; Xiao, Q.; Wei, B.; Zheng, L.; Wang, Y.; Cao, Y.; Zhang, X.; Long, T.; Li, Y.; et al. Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156. Sci. Rep. 2016, 6, 27590. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef]
- Qin, L.P.; Wang, L.Q.; Guo, Y.; Li, Y.; Umut, H.; Wang, Y.C. An ERF transcription factor from Tamarix hispida, ThCRF1, can adjust osmotic potential and reactive oxygen species scavenging capability to improve salt tolerance. Plant. Sci. 2017, 265, 154–166. [Google Scholar] [CrossRef]
- Mijiti, M.; Wang, Y.; Wang, L.; Habuding, X. Tamarix hispida NAC Transcription Factor ThNAC4 Confers Salt and Drought Stress Tolerance to Transgenic Tamarix and Arabidopsis. Plants 2022, 11, 2647. [Google Scholar] [CrossRef] [PubMed]
- Yanez, M.; Caceres, S.; Orellana, S.; Bastias, A.; Verdugo, I.; Ruiz-Lara, S.; Casaretto, J.A. An abiotic stress-responsive bZIP transcription factor from wild and cultivated tomatoes regulates stress-related genes. Plant Cell. Rep. 2009, 28, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Qiu, X.J.; Wang, Y.; Abubakar, A.S.; Chen, P.; Chen, J.K.; Chen, K.M.; Yu, C.M.; Wang, X.F.; Gao, G.; et al. Genome-Wide Investigation of the NAC Transcription Factor Family in Apocynum venetum Revealed Their Synergistic Roles in Abiotic Stress Response and Trehalose Metabolism. Int. J. Mol. Sci. 2023, 24, 4578. [Google Scholar] [CrossRef] [PubMed]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wei, X.; Tong, X.; Zhao, J.; Liu, X.; Wang, H.; Tang, L.; Shu, Y.; Li, G.; Wang, Y.; et al. The OsNAC23-Tre6P-SnRK1a feed-forward loop regulates sugar homeostasis and grain yield in rice. Mol. Plant 2022, 15, 706–722. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophs. Res. Co. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Yamaguchiz, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef]
- Zhu, L.; Shan, H.; Chen, S.M.; Jiang, J.F.; Gu, C.S.; Zhou, G.Q.; Chen, Y.; Song, A.P.; Chen, F.D. The Heterologous Expression of the Chrysanthemum R2R3-MYB Transcription Factor Alters Lignin Composition and Represses Flavonoid Synthesis in. PLoS ONE 2013, 8, e65680. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y.; Li, X.; Lei, F.; Yang, R.; Bai, W.; Yang, Q.; Zhang, D. Transcriptome Profiles Reveals ScDREB10 from Syntrichia caninervis Regulated Phenylpropanoid Biosynthesis and Starch/Sucrose Metabolism to Enhance Plant Stress Tolerance. Plants 2024, 13, 205. https://doi.org/10.3390/plants13020205
Liang Y, Li X, Lei F, Yang R, Bai W, Yang Q, Zhang D. Transcriptome Profiles Reveals ScDREB10 from Syntrichia caninervis Regulated Phenylpropanoid Biosynthesis and Starch/Sucrose Metabolism to Enhance Plant Stress Tolerance. Plants. 2024; 13(2):205. https://doi.org/10.3390/plants13020205
Chicago/Turabian StyleLiang, Yuqing, Xiaoshuang Li, Feiya Lei, Ruirui Yang, Wenwan Bai, Qilin Yang, and Daoyuan Zhang. 2024. "Transcriptome Profiles Reveals ScDREB10 from Syntrichia caninervis Regulated Phenylpropanoid Biosynthesis and Starch/Sucrose Metabolism to Enhance Plant Stress Tolerance" Plants 13, no. 2: 205. https://doi.org/10.3390/plants13020205
APA StyleLiang, Y., Li, X., Lei, F., Yang, R., Bai, W., Yang, Q., & Zhang, D. (2024). Transcriptome Profiles Reveals ScDREB10 from Syntrichia caninervis Regulated Phenylpropanoid Biosynthesis and Starch/Sucrose Metabolism to Enhance Plant Stress Tolerance. Plants, 13(2), 205. https://doi.org/10.3390/plants13020205





