Identification of Transcription Factors of Santalene Synthase Gene Promoters and SaSSY Cis-Elements through Yeast One-Hybrid Screening in Santalum album L.
Abstract
1. Introduction
2. Results
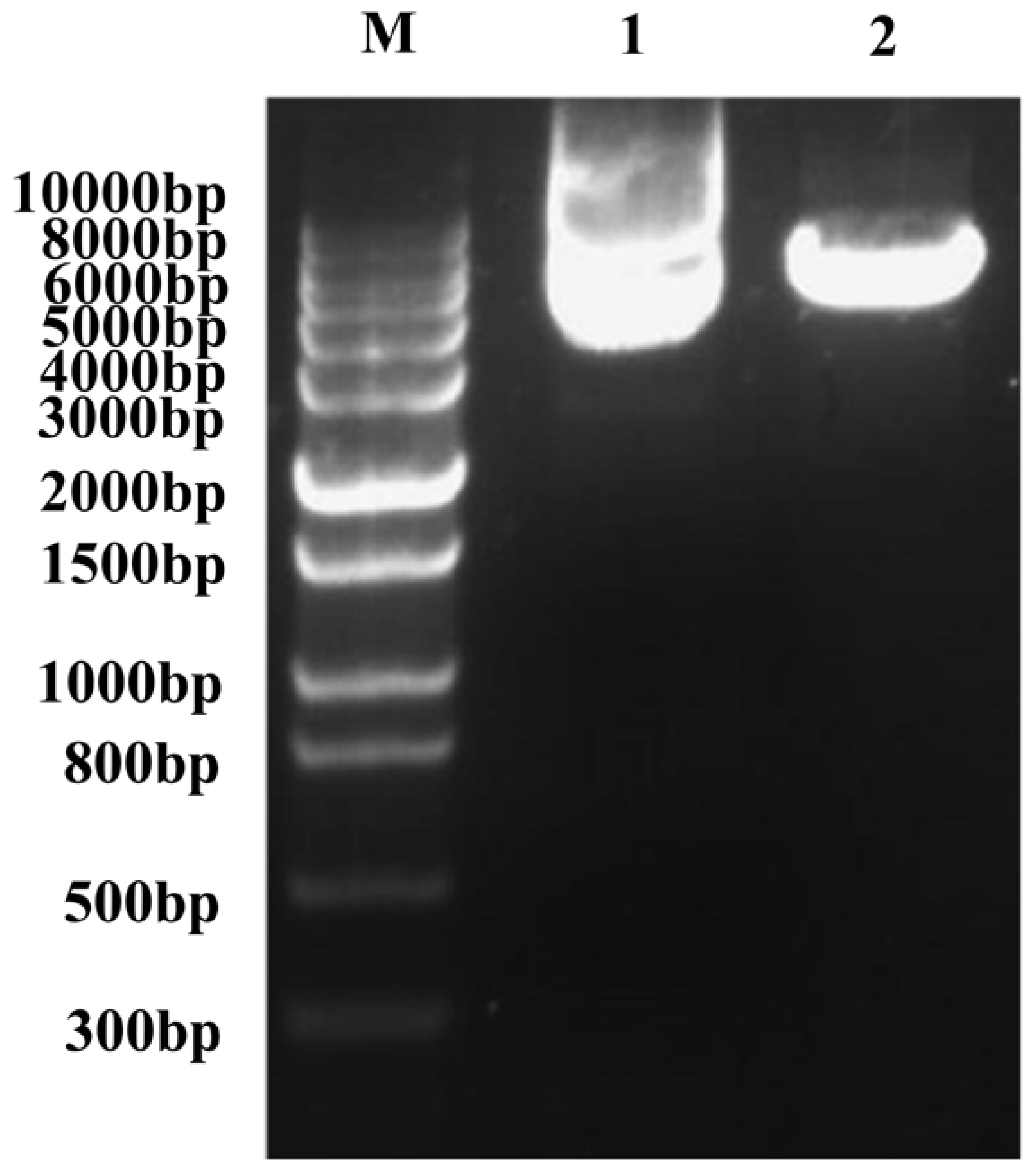
2.1. RNA Extraction and Homogenization of Double-Stranded cDNA
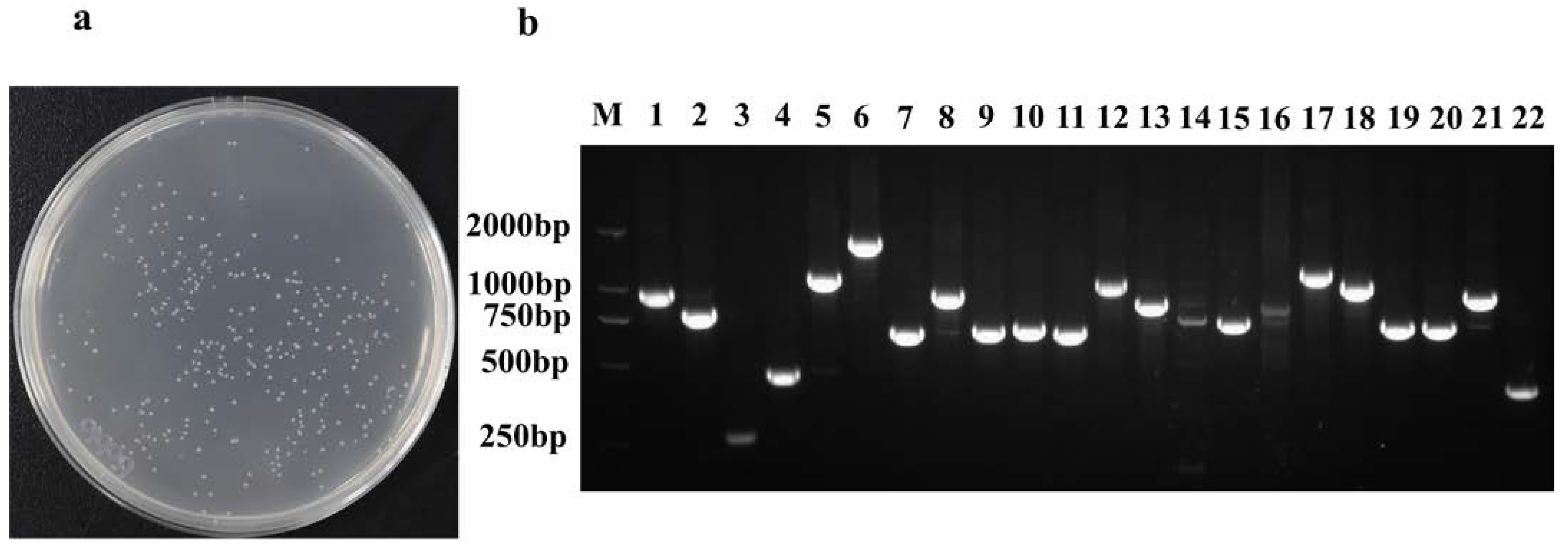
2.2. Library Identification and Titer Determination
2.3. Promoter Prediction Results
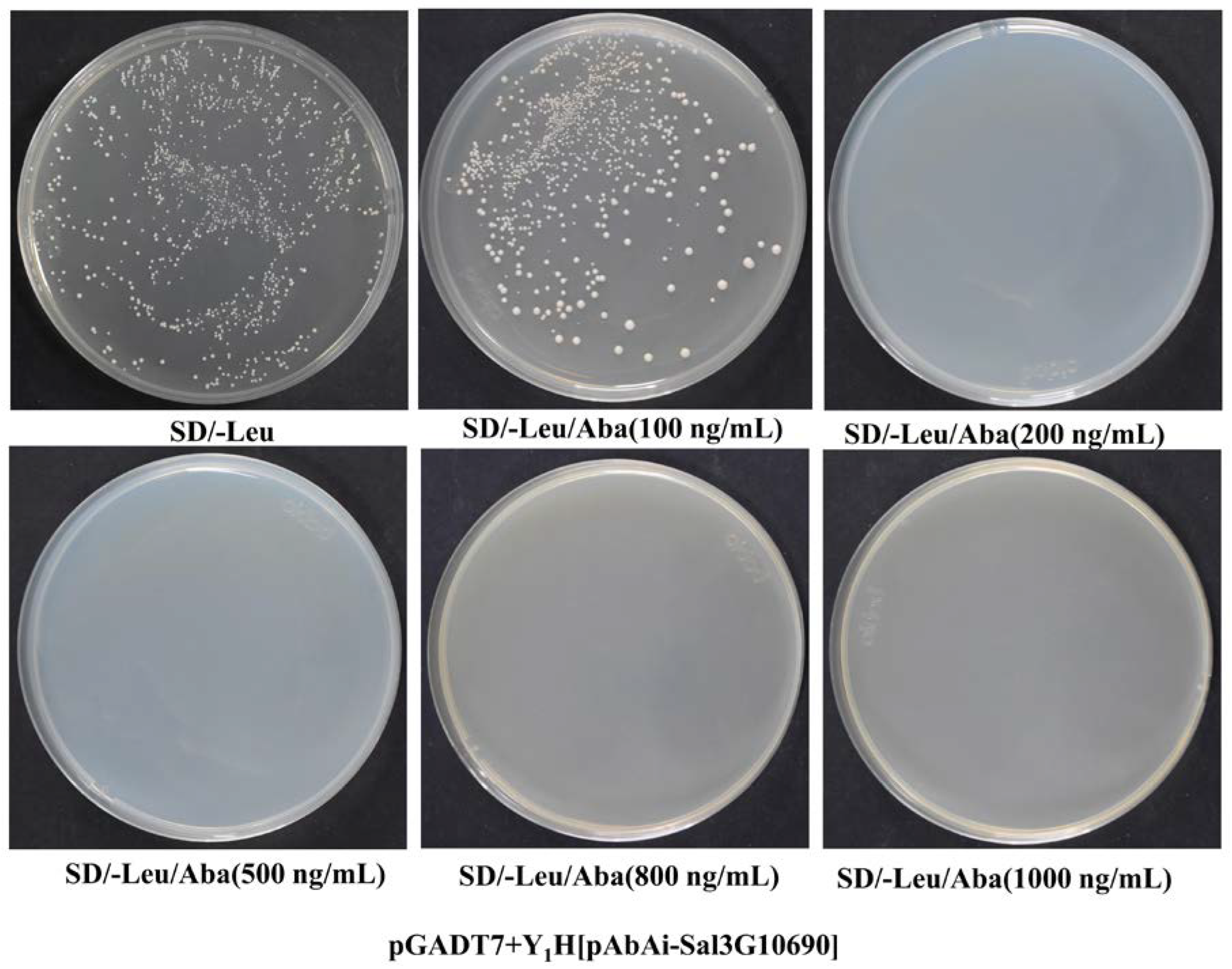
2.4. Construction of Bait-Reporter Strains and Determination of the AbA Concentration
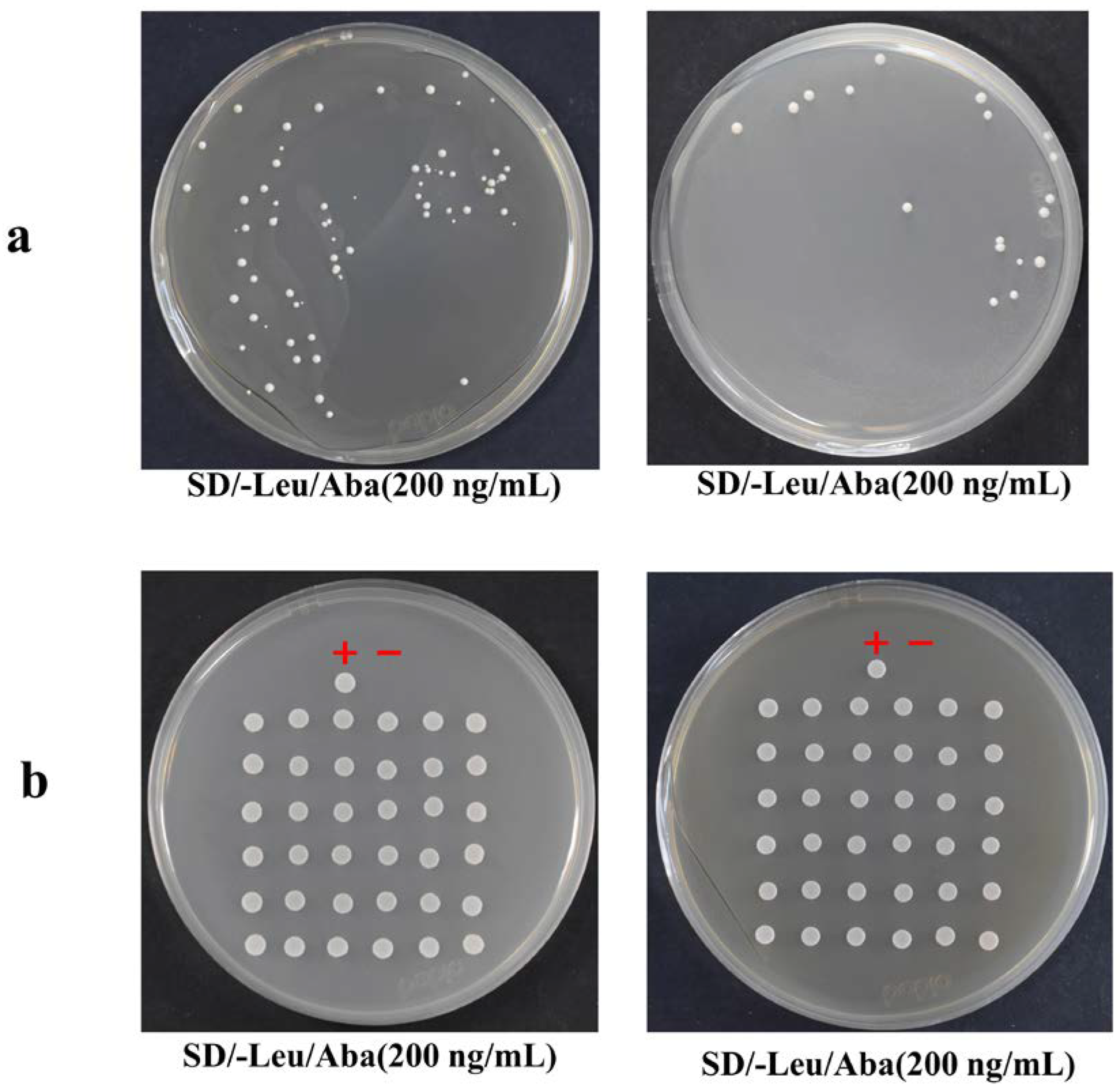
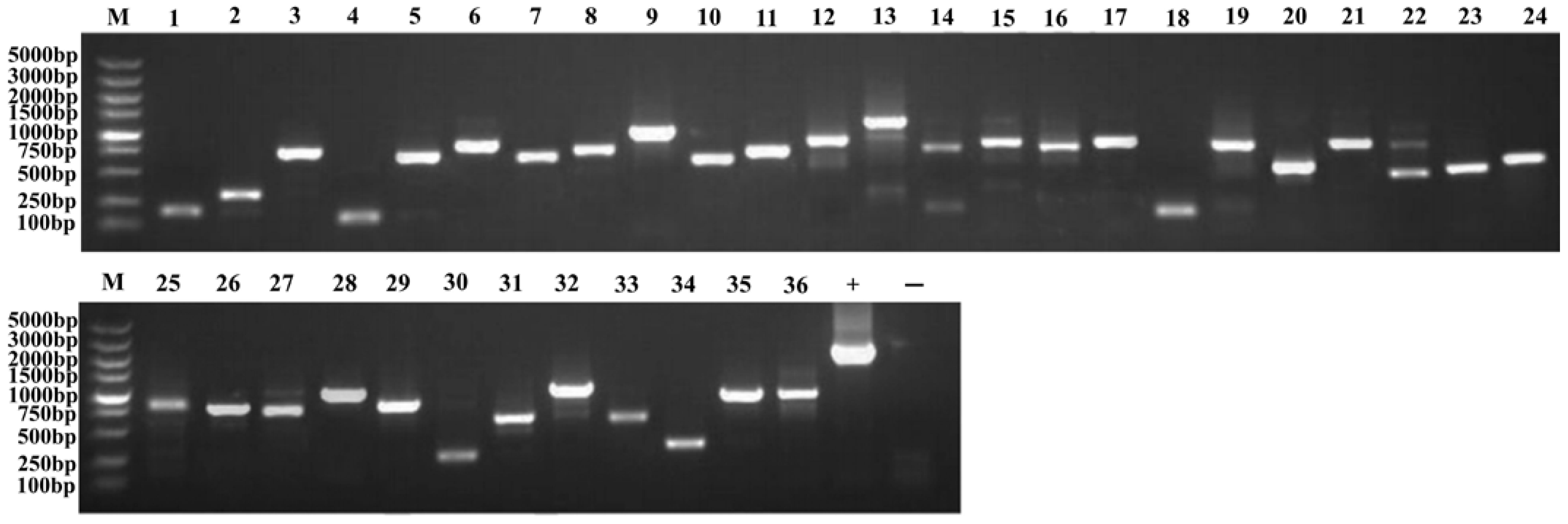
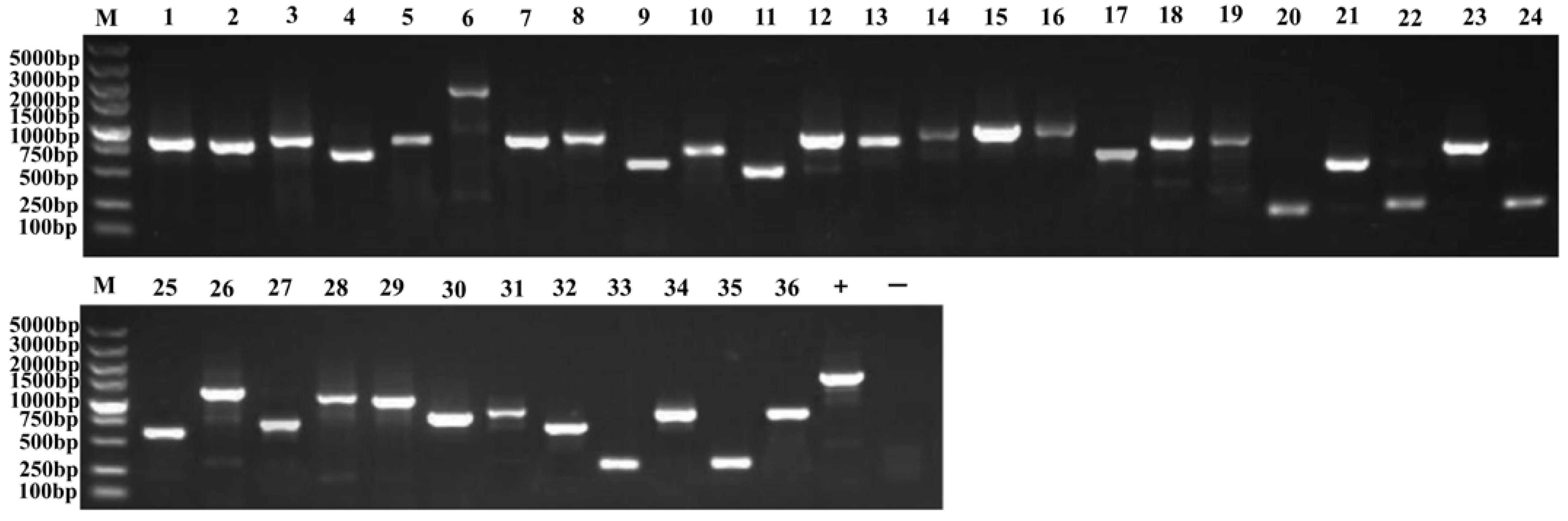
2.5. Transformation of the Plasmid Library and Screening Results of Positive Clones
3. Discussion
4. Materials and Methods
4.1. Experimental Material
4.2. RNA Extraction and mRNA Purification
4.3. Purification and Homogenization of Double-Stranded cDNA
4.4. Construction of a cDNA Library
4.5. Promoter Prediction
4.6. Preparation of Receptive Cells
4.7. Transformation of Linearized Bait Plasmids into Yeast Cells
4.8. Screening of the Y1H Library and Extraction of the Prey Plasmids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Da Silva, J.A.T.; Kher, M.M.; Soner, D.; Page, T.; Zhang, X.; Nataraj, M.; Ma, G. Sandalwood: Basic biology, tissue culture, and genetic transformation. Planta 2016, 243, 847–887. [Google Scholar] [CrossRef]
- Misra, B.; Dey, S. Comparative phytochemical analysis and antibacterial efficacy of in vitro and in vivo extracts from East Indian sandalwood tree (Santalum album L.). Lett. Appl. Microbiol. 2012, 55, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Ravikumar, P.; Reddy, M.D.; Kush, A. Molecular regulation of santalol biosynthesis in Santalum album L. Gene 2013, 527, 642–648. [Google Scholar] [CrossRef]
- Jones, C.G.; Moniodis, J.; Zulak, K.G.; Scaffidi, A.; Plummer, J.A.; Ghisalberti, E.L.; Barbour, E.L.; Bohlmann, J. Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases. J. Biol. Chem. 2011, 286, 17445–17454. [Google Scholar] [CrossRef]
- Zhang, X.; Niu, M.; da Silva, J.A.T.; Zhang, Y.; Yuan, Y.; Jia, Y.; Xiao, Y.; Li, Y.; Fang, L.; Zeng, S.; et al. Identification and functional characterization of three new terpene synthase genes involved in chemical defense and abiotic stresses in Santalum album. BMC Plant Biol. 2019, 19, 115. [Google Scholar] [CrossRef] [PubMed]
- Abbas, F.; Ke, Y.; Yu, R.; Yue, Y.; Amanullah, S.; Jahangir, M.M.; Fan, Y. Volatile terpenoids: Multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta 2017, 246, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Unsicker, S.B.; Kunert, G.; Gershenzon, J. Protective perfumes: The role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 2009, 12, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, I.T.; Halitschke, R.; Paschold, A.; von Dahl, C.C.; Preston, C.A. Volatile signaling in plant-plant interactions: “Talking Trees” in the genomics era. Science 2006, 311, 812–815. [Google Scholar] [CrossRef]
- Yu, F.; Utsumi, R. Diversity, regulation, and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cell. Mol. Life Sci. 2009, 66, 3043–3052. [Google Scholar] [CrossRef]
- Wei, J.; Yang, Y.; Peng, Y.; Wang, S.; Zhang, J.; Liu, X.; Liu, J.; Wen, B.; Li, M. Biosynthesis and the Transcriptional Regulation of Terpenoids in Tea Plants (Camellia sinensis). Int. J. Mol. Sci. 2023, 24, 6937. [Google Scholar] [CrossRef]
- Gong, Z.; Luo, Y.; Zhang, W.; Jian, W.; Zhang, L.; Gao, X.; Hu, X.; Yuan, Y.; Wu, M.; Xu, X.; et al. A SlMYB75-centred transcriptional cascade regulates trichome formation and sesquiterpene accumulation in tomato. J. Exp. Bot. 2021, 72, 3806–3820. [Google Scholar] [CrossRef]
- Hong, G.-J.; Xue, X.-Y.; Mao, Y.-B.; Wang, L.-J.; Chen, X.-Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Li, M.; Cao, X.; Jin, Z.; Zhang, C.; Xu, M.; Chen, K.; Zhang, B. Linalool synthesis related PpTPS1 and PpTPS3 are activated by transcription factor PpERF61 whose expression is associated with DNA methylation during peach fruit ripening. Plant Sci. 2022, 317, 111200. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Liu, H.; Cao, X.; Zhang, M.; Li, X.; Chen, K.; Zhang, B. Synthesis of flavour-related linalool is regulated by PpbHLH1 and associated with changes in DNA methylation during peach fruit ripening. Plant Biotechnol. J. 2021, 19, 2082–2096. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhou, Y.; Zhang, Y.; Wang, J.; Shi, H. Identification of transcription factors of nitrate reductase gene promoters and NRE2 cis-element through yeast one-hybrid screening in Nicotiana tabacum. BMC Plant Biol. 2019, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-P.; Gubler, F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Kamiya, Y. Gibberellin biosynthesis: Its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000, 41, 251–257. [Google Scholar] [CrossRef]
- Rabiei, B.; Bahador, S.; Kordrostami, M. The expression of monoterpene synthase genes and their respective end products are affected by gibberellic acid in Thymus vulgaris. J. Plant Physiol. 2018, 230, 101–108. [Google Scholar] [CrossRef]
- Yang, M.; Liu, G.; Yamamura, Y.; Chen, F.; Fu, J. Divergent Evolution of the Diterpene Biosynthesis Pathway in Tea Plants (Camellia sinensis) Caused by Single Amino Acid Variation of ent-Kaurene Synthase. J. Agric. Food Chem. 2020, 68, 9930–9939. [Google Scholar] [CrossRef]
- Zhang, K.; Logacheva, M.D.; Meng, Y.; Hu, J.; Wan, D.; Li, L.; Janovská, D.; Wang, Z.; Georgiev, M.I.; Yu, Z.; et al. Jasmonate-responsive MYB factors spatially repress rutin biosynthesis in Fagopyrum tataricum. J. Exp. Bot. 2018, 69, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-H.; Liao, Y.-C.; Lv, F.-F.; Zhang, Z.; Sun, P.-W.; Gao, Z.-H.; Hu, K.-P.; Sui, C.; Jin, Y.; Wei, J.-H. Transcription Factor AsMYC2 Controls the Jasmonate-Responsive Expression of ASS1 Regulating Sesquiterpene Biosynthesis in Aquilaria sinensis (Lour.) Gilg. Plant Cell Physiol. 2017, 58, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Li, X.; Li, Y.; Yang, G.; Liu, W.; Shao, B.; Zhong, J.; Huang, P.; Han, D. Overexpression of a Malus baccata MYB transcription factor gene MbMYB4 increases cold and drought tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 1794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, H.; Chen, Y.; Huang, M.; Zhu, S. Comprehensive genome-wide analyses of poplar R2R3-MYB transcription factors and tissue-specific expression patterns under drought stress. Int. J. Mol. Sci. 2023, 24, 5389. [Google Scholar] [CrossRef]
- Tang, M.; Liu, L.; Hu, X.; Zheng, H.; Wang, Z.; Liu, Y.; Zhu, Q.; Cui, L.; Xie, S. Genome-wide characterization of R2R3-MYB gene family in Santalum album and their expression analysis under cold stress. Front. Plant Sci. 2023, 14, 1142562. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, Q.; Gao, X.; Wu, G.; Wu, M.; Yuan, Y.; Zheng, X.; Gong, Z.; Hu, X.; Gong, M.; et al. Auxin and abscisic acid antagonistically regulate ascorbic acid production via the SlMAPK8–SlARF4–SlMYB11 module in tomato. Plant Cell 2022, 34, 4409–4427. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Huang, J.; Hai, Z.; Wang, R.; Yu, Y.; Chen, X.; Liang, W.; Wang, H. Genome-wide analysis of HSP20 gene family and expression patterns under heat stress in cucumber (Cucumis sativus L.). Front. Plant Sci. 2022, 13, 968418. [Google Scholar] [CrossRef]
- Haq, S.U.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Gong, Z.-H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Shi, Y.; Xu, S.; Xie, S.; Huang, X.; Li, Y.; Qu, C.; Liu, J.; Liao, J.; Huang, Y.; et al. Heterologous overexpression of heat shock protein 20 genes of different species of yellow Camellia in Arabidopsis thaliana reveals their roles in high calcium resistance. BMC Plant Biol. 2024, 24, 5. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Wang, Q.; Li, T.; Gao, H.; Li, H.; Zheng, X.; Wang, X.; Zhang, H.; Cheng, J.; Wang, W.; et al. Phylogenetic and Transcriptional Analyses of the HSP20 Gene Family in Peach Revealed That PpHSP20-32 Is Involved in Plant Height and Heat Tolerance. Int. J. Mol. Sci. 2022, 23, 10849. [Google Scholar] [CrossRef]
- Xue, D.; Chen, Y.; Li, J.; Han, J.; Liu, Y.; Jiang, S.; Zhou, Z.; Zhang, W.; Chen, M.; Lin, M.; et al. Targeting Hsp20 using the novel small non-coding RNA DnrH regulates heat tolerance in Deinococcus radiodurans. Front. Microbiol. 2019, 10, 2354. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.H.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Liang, B.; Yang, L.; Hu, W.; Kuang, L.; Song, J.; Xie, J.; Huang, Y.; Liu, D.; Liu, Y. The MADS-box family gene PtrANR1 encodes a transcription activator promoting root growth and enhancing plant tolerance to drought stress. Plant Cell Rep. 2023, 43, 16. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Wu, Z.; Song, Z.; Abbas, F.; Chen, W.; Li, X.; Zhu, X. ATAC-seq and RNA-seq reveal the role of AGL18 in regulating fruit ripening via ethylene-auxin crosstalk in papaya. Postharvest Biol. Technol. 2022, 191, 111984. [Google Scholar] [CrossRef]
- Sharif, R.; Raza, A.; Chen, P.; Li, Y.; El-Ballat, E.M.; Rauf, A.; Hano, C.; El-Esawi, M.A. HD-ZIP Gene Family: Potential Roles in Improving Plant Growth and Regulating Stress-Responsive Mechanisms in Plants. Genes 2021, 12, 1256. [Google Scholar] [CrossRef] [PubMed]
- Turchi, L.; Carabelli, M.; Ruzza, V.; Possenti, M.; Sassi, M.; Peñalosa, A.; Sessa, G.; Salvi, S.; Forte, V.; Morelli, G.; et al. Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development 2013, 140, 2118–2129. [Google Scholar] [CrossRef]
- Guo, Q.; Jiang, J.; Yao, W.; Li, L.; Zhao, K.; Cheng, Z.; Han, L.; Wei, R.; Zhou, B.; Jiang, T. Genome-wide analysis of poplar HD-Zip family and over-expression of PsnHDZ63 confers salt tolerance in transgenic Populus simonii × P. nigra. Plant Sci. 2021, 311, 111021. [Google Scholar] [CrossRef]
- Wu, Z.; Li, T.; Zhang, D.; Teng, N. Lily HD-Zip I transcription factor LlHB16 promotes thermotolerance by activating LlHSFA2 and LlMBF1c. Plant Cell Physiol. 2022, 63, 1729–1744. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.; Meng, S.; Ye, Y.; Han, L.; He, Y.; Cui, Y.; Tan, C.; Ma, J.; Qi, M.; Li, T. A molecular framework for lc controlled locule development of the floral meristem in tomato. Front. Plant Sci. 2023, 14, 1249760. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, J.; Liu, Q.; Li, K.; Zhou, Y. Construction and characterization of a high-quality cDNA library of Cymbidium faberi suitable for yeast one- and two-hybrid assays. BMC Biotechnol. 2020, 20, 4. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Zhang, H.; Zhao, Q.; Cui, S.; Yu, K.; Sun, R.; Yu, Y. Construction of Yeast One-Hybrid Library of Alternaria oxytropis and Screening of Transcription Factors Regulating swnK Gene Expression. J. Fungi 2023, 9, 822. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]

| Site Name | Sequence | Position | Function | Number of Repeats |
|---|---|---|---|---|
| AAGAA-motif | gGTAAAGAAA | −302 | Function unknown | 1 |
| ABRE | ACGTG | +1840 | cis-acting element involved in the abscisic acid responsiveness | 1 |
| as-1 | TGACG | +1336 | Function unknown | 1 |
| AT~TATA-box | TATATA | +442, +444, +446, +448, +450, +452, +454, +456, +458, +888, −1259, −1537, −1906, −1908 | Function unknown | 14 |
| ATCT-motif | AATCTAATCC | −26 | cis-acting element involved in light responsiveness | 1 |
| Box 4 | ATTAAT | +336, +354, +1042 | cis-acting element involved in light responsiveness | 3 |
| CAAT-box | CAAAT | −22, +53, +221, −227, −317, +592, +680, +899, +907, +1353, −1356, +1700, +1780 | common cis-acting element in promoter and enhancer regions | 13 |
| CAAT-box | CAAT | +48, −157, +164, +273, +323, −327, −463, −635, −657, −683, −722, +749, −952, +985, −1204, +1224, −1297, −1317, +1625, +1781, +1878 | Function unknown | 21 |
| CGTCA-motif | CGTCA | −1336 | cis-acting regulatory element involved in the MeJA responsiveness | 1 |
| circadian | CAAAGATATC | −404 | cis-acting regulatory element involved in circadian control | 1 |
| ERE | ATTTTAAA | +1640 | Function unknown | 1 |
| G-Box | CACGTT | −1839 | cis-acting regulatory element involved in light responsiveness | 1 |
| GCN4_motif | TGAGTCA | +1421 | cis-regulatory element involved in endosperm expression | 1 |
| GT1-motif | GGTTAA(T) | −548, −1113, −1114, +1792 | Light-responsive element | 2 |
| I-box | AAGATAAGGCT | −555 | part of a light-responsive element | 1 |
| LTR | CCGAAA | +1818 | cis-acting element involved in low-temperature responsiveness | 1 |
| MYB | CAACCA | −1433, −1437 | Function unknown | 2 |
| MYB-like | TAACCA | −1791 | Function unknown | 1 |
| Myc | TCTCTTA | −1087 | Function unknown | 1 |
| MYC | CAT(T/G) TG | +316, −810, −1700, +1477 | Function unknown | 2 |
| P-box | CCTTTTG | +1931 | gibberellin-responsive element | 1 |
| STRE | AGGGG | −1140, −1929 | Function unknown | 2 |
| TATA | TATAAAAT | −575, +1763 | Function unknown | 2 |
| TATA-box | AT(T)ATA(A/T)/TATA(AAAA)/TATATA(A)/TATTTAAA | −341, −578, −1675, −1980, +340, +440, +886, +342, +379, +428, +460, +579, +890, +911, −1082, −1095, −1261, −1491, −1539, −1676, −1763, −1910, −1981, +378, +443, +445, +447, +449, +451, +453, +455, +457, +459, −1094, −1258, −1536, −1907, −441, −887, +442, +444, +446, +448, +450, +452, +454, +456, +458, +888, −1259, −1537, −1906, −1908, −576, −1978, −577, −1979, +1081, +1399 | core promoter element around −30 of transcription start | 59 |
| TATC-box | TATCCCA | +1727 | cis-acting element involved in gibberellin responsiveness | 1 |
| TCT-motif | TCTTAC | +123, −706 | part of a light-responsive element | 2 |
| TGACG-motif | TGACG | +1336 | cis-acting regulatory element involved in the MeJA responsiveness | 1 |
| Unnamed-1 | CGTGG | +1841 | Function unknown | 1 |
| Unnamed-16 | GCTGCCCGTC | −1864 | Function unknown | 1 |
| Unnamed-4 | CTCC | +83, +201, −743, −781, +948, −962, +1153, −1360, +1735 | Function unknown | 9 |
| WRE3 | CCACCT | +1660, −1871 | Function unknown | 2 |
| Name | Fragment Length (bp) | Blastx/Blastn | NCBI Number | Description | Scientific Name |
|---|---|---|---|---|---|
| Sal6G03620.1 | 1153/744 | Blastn | XM_058111947.1 | PREDICTED: Malania oleifera transcription factor MYB36-like (LOC131157642), mRNA | Malania oleifera |
| Sal8G07920.2 | 1225/666 | Blastn | XM_058095332.1 | PREDICTED: Malania oleifera small heat shock protein, chloroplastic (LOC131146053), mRNA | Malania oleifera |
| Sal1G00910.1 | 597/206 | Blastx | KAI6692205.1 | hypothetical protein NL676_019915 | Syzygium grande |
| Sal4G10880.1 | 2514/836 | Blastx | XP_057976689.1 | homeobox-leucine zipper protein ATHB-15 | Malania oleifera |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Li, X.; Wang, D.; Yu, Z.; Liu, Y.; Hu, L.; Bian, Z. Identification of Transcription Factors of Santalene Synthase Gene Promoters and SaSSY Cis-Elements through Yeast One-Hybrid Screening in Santalum album L. Plants 2024, 13, 1882. https://doi.org/10.3390/plants13131882
Zhou Y, Li X, Wang D, Yu Z, Liu Y, Hu L, Bian Z. Identification of Transcription Factors of Santalene Synthase Gene Promoters and SaSSY Cis-Elements through Yeast One-Hybrid Screening in Santalum album L. Plants. 2024; 13(13):1882. https://doi.org/10.3390/plants13131882
Chicago/Turabian StyleZhou, Yunqing, Xiang Li, Dongli Wang, Zequn Yu, Yunshan Liu, Lipan Hu, and Zhan Bian. 2024. "Identification of Transcription Factors of Santalene Synthase Gene Promoters and SaSSY Cis-Elements through Yeast One-Hybrid Screening in Santalum album L." Plants 13, no. 13: 1882. https://doi.org/10.3390/plants13131882
APA StyleZhou, Y., Li, X., Wang, D., Yu, Z., Liu, Y., Hu, L., & Bian, Z. (2024). Identification of Transcription Factors of Santalene Synthase Gene Promoters and SaSSY Cis-Elements through Yeast One-Hybrid Screening in Santalum album L. Plants, 13(13), 1882. https://doi.org/10.3390/plants13131882






