Overexpression of AcWRKY31 Increases Sensitivity to Salt and Drought and Improves Tolerance to Mealybugs in Pineapple
Abstract
1. Introduction
2. Results
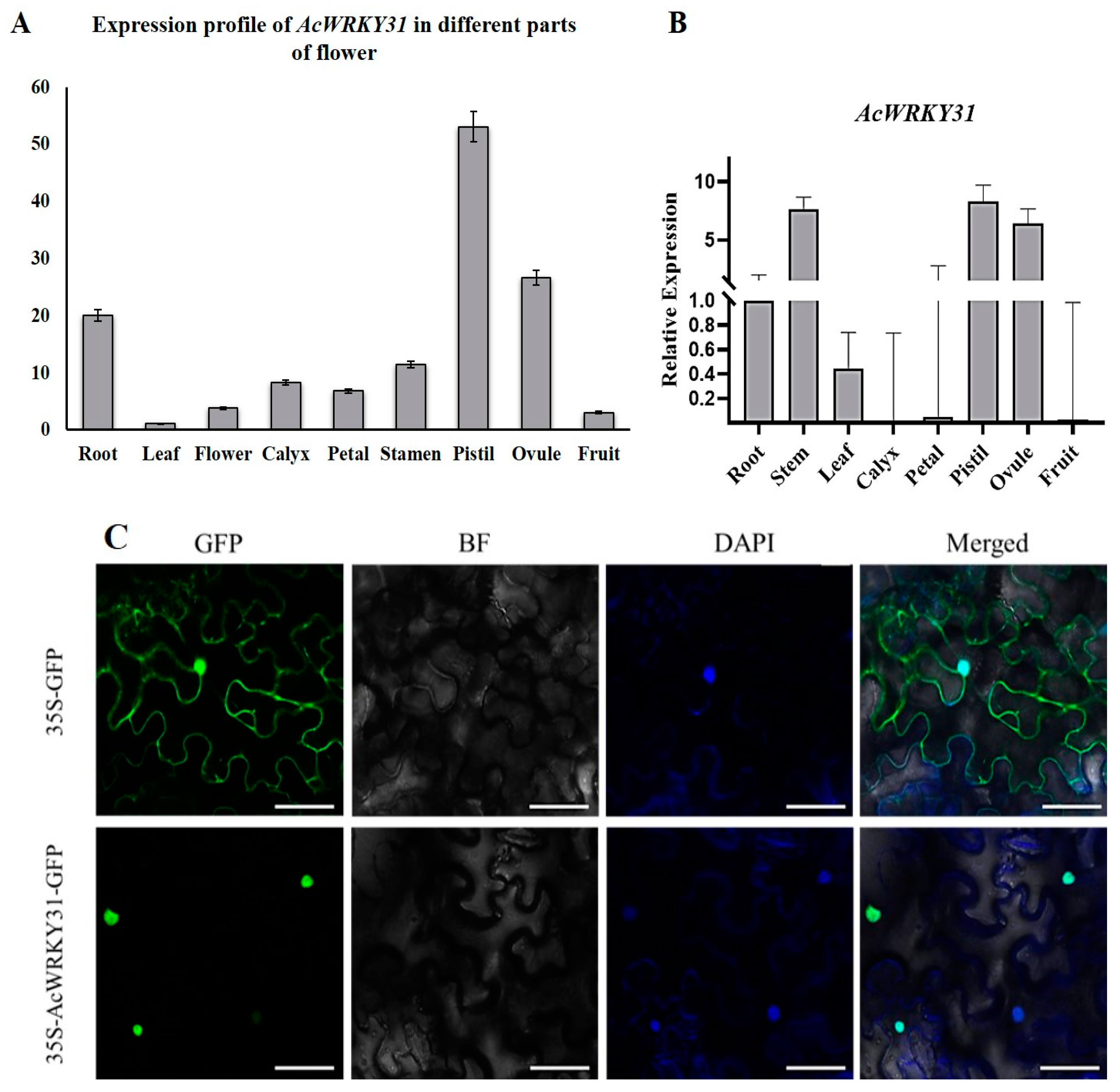
2.1. The Expression Profiles and Subcellular Localization of AcWRKY31
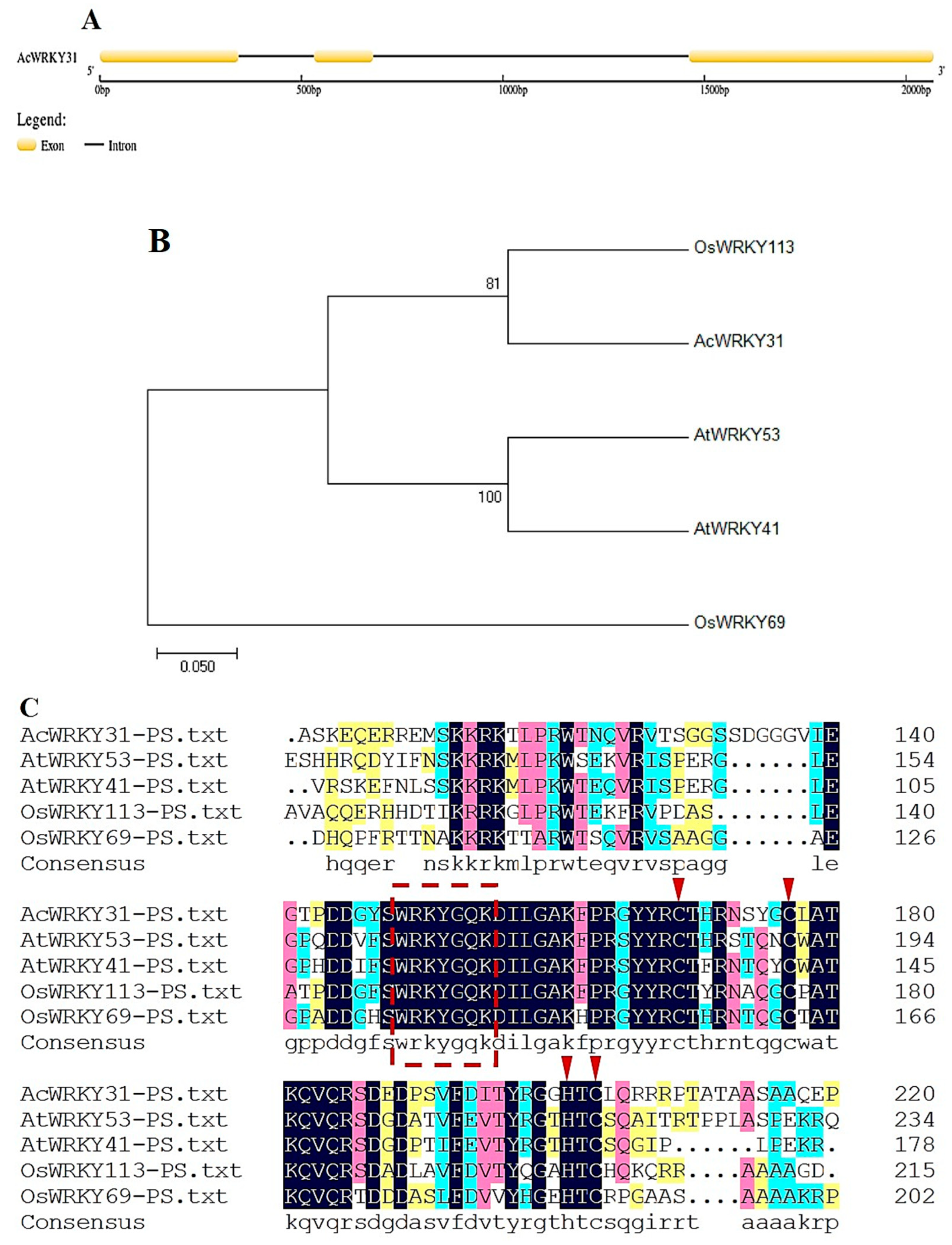
2.2. Phylogenetic Analysis and Sequence Alignment of AcWRKY31
2.3. Transformation and Identification of Overexpressed AcWRKY31 in Pineapple
2.4. Overexpression of AcWRKY31 Reduces Tolerance to Salt and Drought Stresses in Pineapple
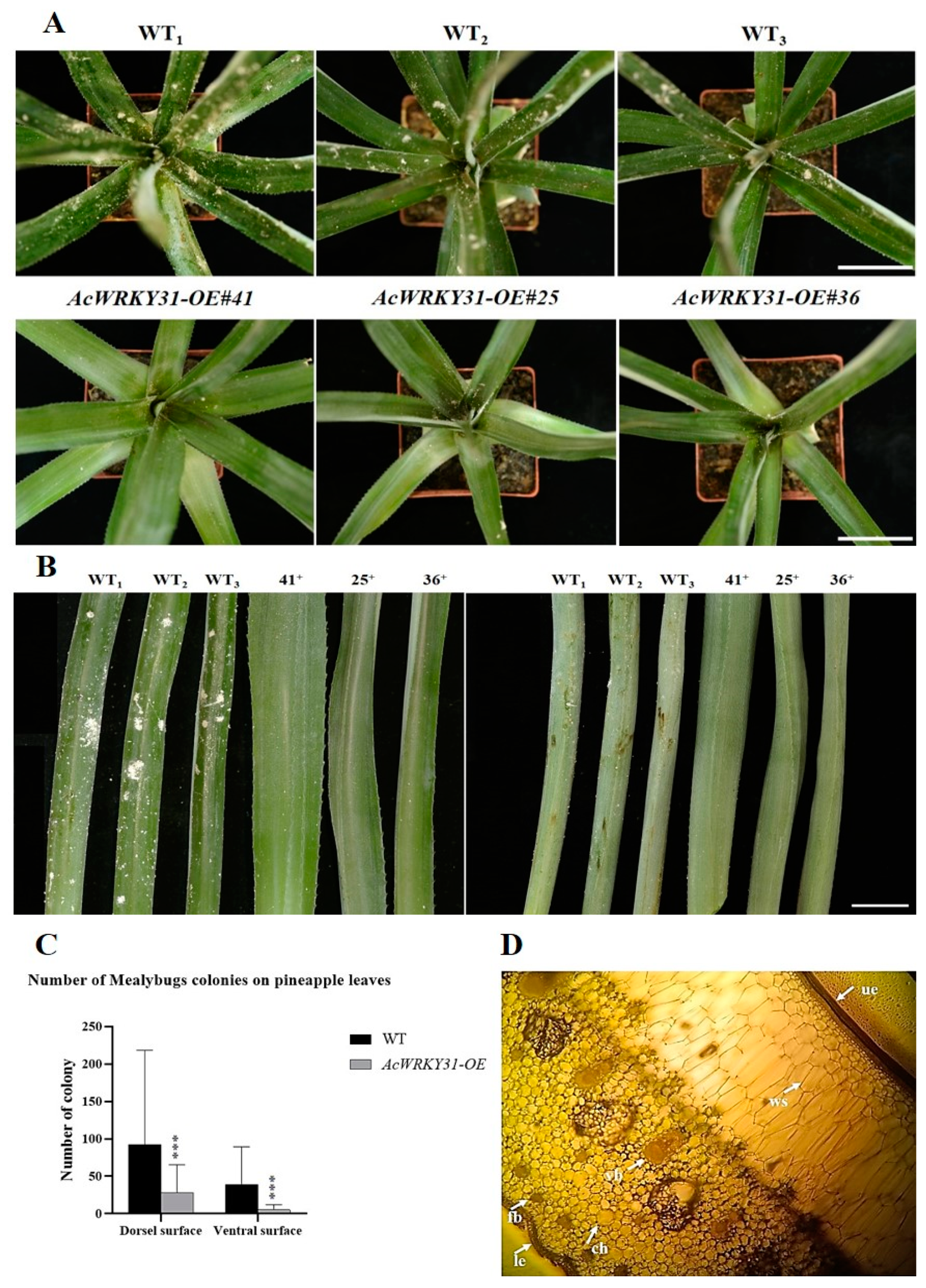
2.5. Overexpression of AcWRKY31 Increases the Resistance to Pineapple Mealybug
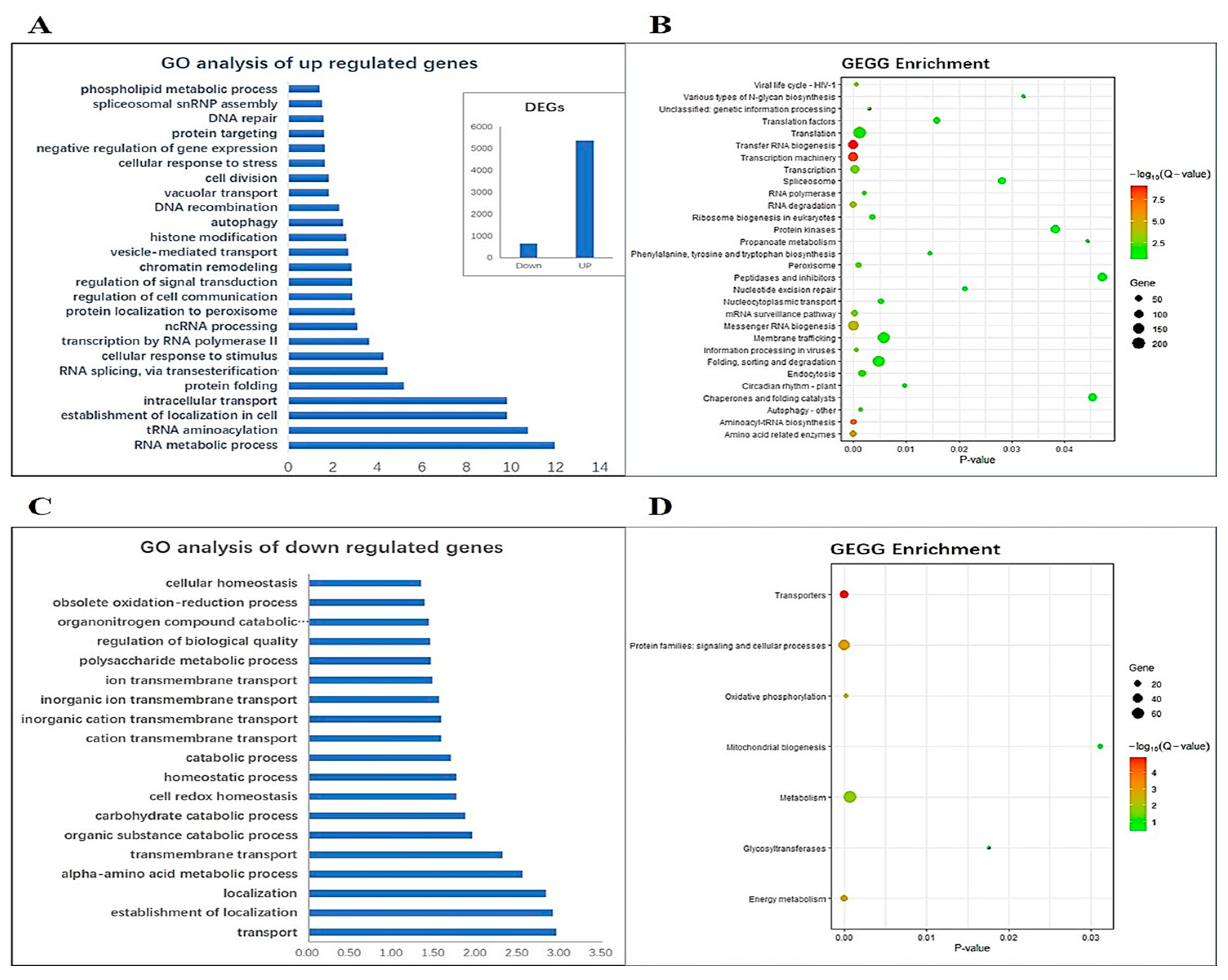
2.6. RNA-Seq Analysis of AcWRKY31-OE Pineapple
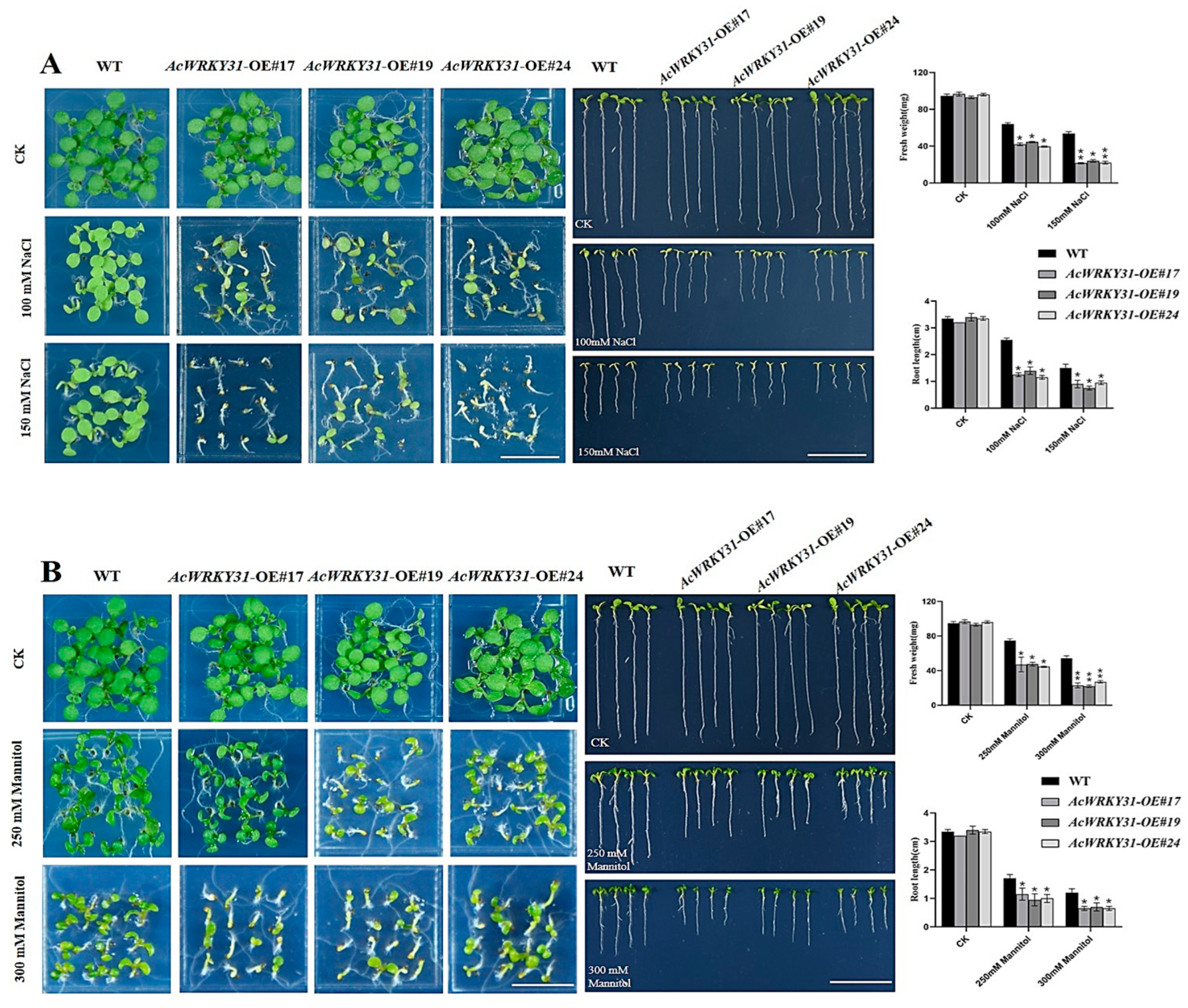
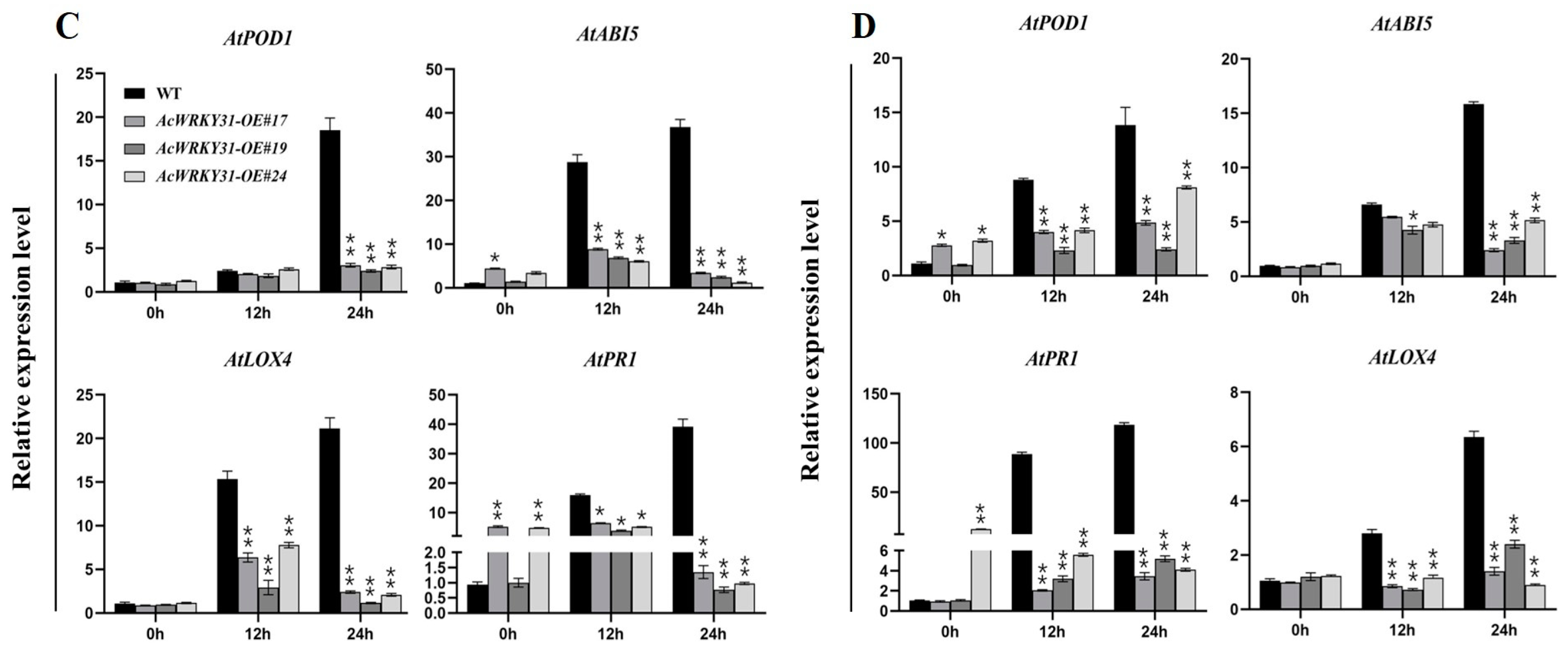
2.7. AcWRKY31-OE Transgenic Arabidopsis Reduces Tolerance to Salt and Drought Stresses
2.8. AcWRKY31-OE Transgenic Arabidopsis Increases Resistance to the Sclerotinia sclerotiorum Pathogen
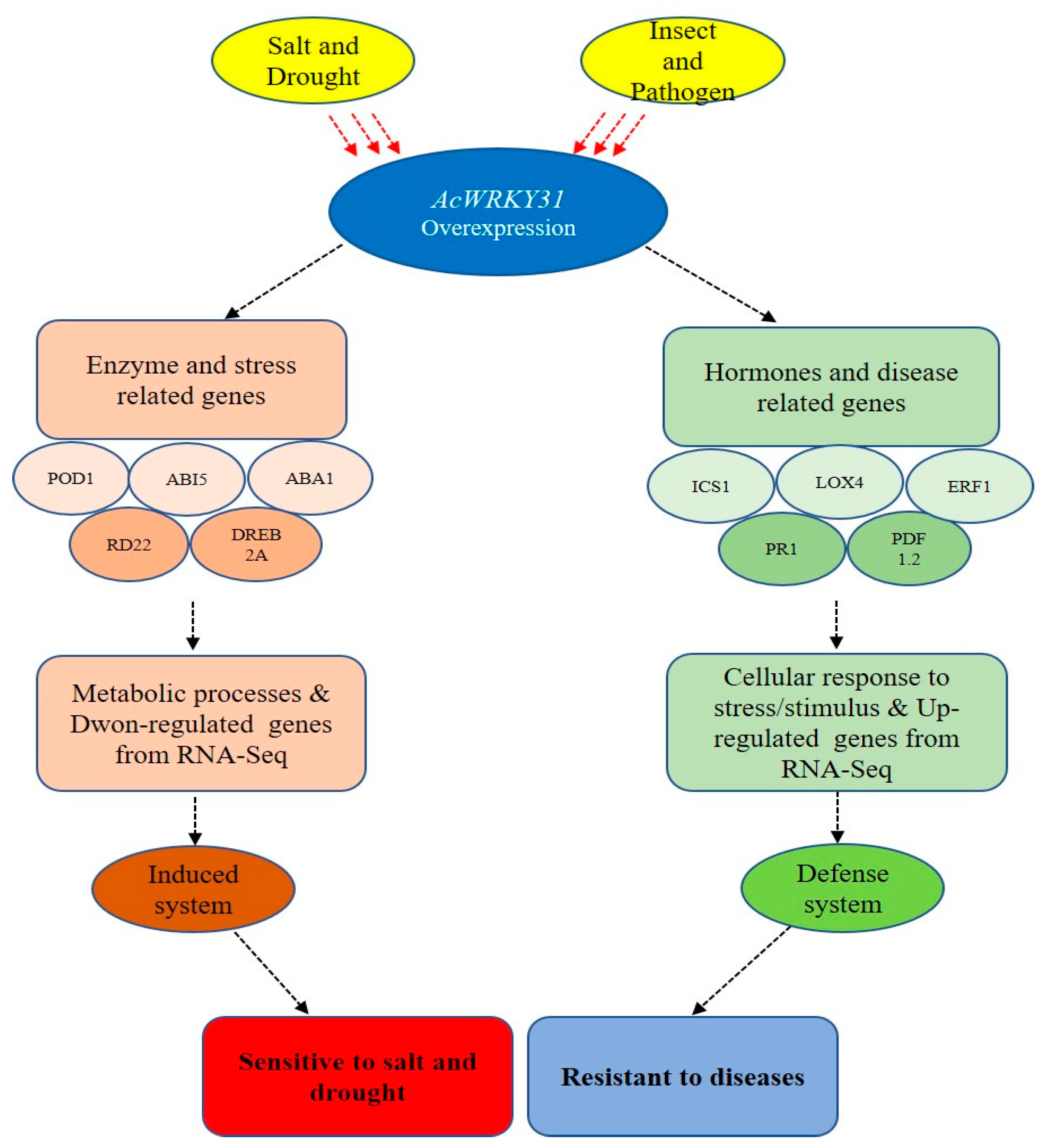
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Pineapple Callus Induction
4.2. Vector Construction, Gene Transformation of AcWRKY31, and Transgenic Plant Selection in Pineapple
4.3. Analysis of Salt and Drought Stress in AcWRKY31-Overexpressed Pineapple
4.4. Analysis of Biotic Stress in AcWRKY31-OE Transgenic Pineapple
4.5. Genetic Transformation of AcWRKY31 and AtWRKY53 in Arabidopsis
4.6. Analysis of Abiotic Treatments in Transgenic Arabidopsis
4.7. Analysis of Sclerotinia sclerotiorum Inoculation on Transgenic Arabidopsis
4.8. Analysis of Transcriptomic Data
4.9. Quantitative Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WT | Wild-type |
| TFs | Transcription factors |
| OE | Overexpression |
| DEGs | Differentially expressed genes |
| PCR | Polymerase chain reaction |
| RT-qPCR | Quantitative real-time polymerase chain reaction |
| PGRs | Plant growth regulators |
| DA-6 | Diethyl aminoethyl hexanoate |
| COS | Chitosan oligosaccharide |
| ABA | Abscisic acid |
| BRs | Brassinosteroids |
| ETH | Ethylene |
| JA | Jasmonate acid |
| SA | Salicylic acid |
| MS | Murishige and Skoog medium |
| BAP | 6-Benzylaminopurine |
| NAA | Naphthalene acetic acid |
| GFP | Green fluorescent protein |
| DNA | Deoxyribonucleic acid |
| cDNA | Complementary DNA |
| NaCl | Sodium chloride |
| ROS | Relative oxygen species |
| Cotton: | |
| GhHBA | High level of beta-amylase activity |
| Tobacco: | |
| NtNCED1 | 9-Cis-Epoxycarotenoid Dioxygenase 1 |
| NtDREB3 | Dehydration Response Element-Binding protein 3 |
| NtLEA5 | Late Embryogenesis Abundant-like 5 |
| Pineapple: | |
| AcPOD | Peroxidase |
| AcABI5 | ABA Insensitive 5 |
| AcABA1 | ABA Deficient 1 |
| AcPR1 | Pathogenesis-Related Gene 1 |
| AcCAT1 | Catalase 1 |
| AcLOX4 | Lipoxygenase 4 |
| AcRD22 | Responsive to Desiccation 22 |
| AcDREB2A | Dehydration Response Element-Binding protein 2 |
| AcCPK | Calcium-dependent Protein Kinase |
| Arabidopsis: | |
| AtSOS1 | Salt Overly Sensitive 1 |
| AtHKT1 | High-Affinity K+ Transporter 1 |
| AtICS1 | Isochorismate Synthase 1 (SA related) |
| AtPDF1.2 | Plant Defensin 1.2 |
| AtPR1 | Pathogenesis-Related Gene 1 |
| AtLOX4 | Lipoxygenase 4 (JA related) |
| AtERF1 | Ethylene Response Factor 1 |
| AtABI5 | ABA Insensitive 5 |
References
- Sinha, A.K.; Jaggi, M.; Raghuram, B.; Tuteja, N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 2011, 6, 196–203. [Google Scholar] [CrossRef]
- Rodziewicz, P.; Swarcewicz, B.; Chmielewska, K.; Wojakowska, A.; Stobiecki, M. Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiol. Plant. 2013, 36, 1–19. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.Y.; Li, J.; Wang, P.Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Boisson-Dernier, A.; Roy, S.; Kritsas, K.; Grobei, M.A.; Jaciubek, M.; Schroeder, J.I.; Grossniklaus, U. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 2009, 136, 3279–3288. [Google Scholar] [CrossRef] [PubMed]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef]
- Shao, H.B.; Chu, L.Y.; Shao, M.A.; Jaleel, C.A.; Mi, H.M. Higher plant antioxidants and redox signaling under environmental stresses. Comptes Rendus Biol. 2008, 331, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.; Song, X.F.; Liu, X.F.; Yan, L.Y.; Zhou, Z.Y.; Zhang, X.L. Genome-wide analysis of CsWOX transcription factor gene family in cucumber (Cucumis sativus L.). Sci. Rep. 2020, 10, 6216. [Google Scholar] [CrossRef]
- Evans, M.; Kermicle, J.L. Interaction between maternal effect and zygotic effect mutations during maize seed development. Genetics 2001, 159, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Westrick, N.M.; Jain, S.; Piotrowski, J.S.; Ranjan, M.; Kessens, R.; Stiegman, L.; Grau, C.R.; Conley, S.P.; Smith, D.L.; et al. Resistance against Sclerotinia sclerotiorum in soybeans involves reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 2019, 17, 1567–1581. [Google Scholar] [CrossRef]
- Caceres, M.; Hidalgo, W.; Stashenko, E.; Torres, R.; Ortiz, C. Essential Oils of Aromatic Plants with Antibacterial, Anti-Biofilm and Anti-Quorum Sensing Activities against Pathogenic Bacteria. Antibiotics 2020, 9, 147. [Google Scholar] [CrossRef]
- Santa-Cecília, L.V.C.; Prado, E.; Souza, B. Probing behavior of Dysmicoccus brevipes mealybug in pineapple plants1. Pesqui. Agropecu. Trop. 2016, 46, 458–463. [Google Scholar] [CrossRef][Green Version]
- Rohrbach, K.G.; Johnson, M.W. Pests, diseases and weeds. In The Pineapple: Botany, Production and Uses; CABI Publishing: Wallingford, UK, 2003; pp. 203–251. [Google Scholar]
- Bolton, M.D.; Thomma, B.P.; Nelson, B.D. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 2006, 7, 1–16. [Google Scholar] [CrossRef]
- Zhou, F.; Zhang, X.L.; Li, J.L.; Zhu, F.X. Dimethachlon Resistance in Sclerotinia sclerotiorum in China. Plant Dis. 2014, 98, 1221–1226. [Google Scholar] [CrossRef] [PubMed]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY Transcription Factors: Molecular Regulation and Stress Responses in Plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef]
- Xu, Y.H.; Sun, P.W.; Tang, X.L.; Gao, Z.H.; Zhang, Z.; Wei, J.H. Genome-wide analysis of WRKY transcription factors in Aquilaria sinensis (Lour.) Gilg. Sci. Rep. 2020, 10, 3018. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef]
- Rushton, P.J.; Bokowiec, M.T.; Han, S.; Zhang, H.; Brannock, J.F.; Chen, X.; Laudeman, T.W.; Timko, M.P. Tobacco transcription factors: Novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 2008, 147, 280–295. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Yang, Z.; Zhang, X.; Zhang, F.; Huang, X.; Han, X. Transcriptome-wide identification of WRKY transcription factors and their expression profiles under different stress in Cynanchum thesioides. PeerJ 2022, 10, e14436. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef]
- Chen, C.; Shang, X.; Sun, M.; Tang, S.; Khan, A.; Zhang, D.; Yan, H.; Jiang, Y.; Yu, F.; Wu, Y.; et al. Comparative Transcriptome Analysis of Two Sweet Sorghum Genotypes with Different Salt Tolerance Abilities to Reveal the Mechanism of Salt Tolerance. Int. J. Mol. Sci. 2022, 23, 2272. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, J.; Sui, Y.; Han, G.; Zhang, Y.; Guo, S.; Sui, N. The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis. Plant Mol. Biol. 2020, 102, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Song, A.; Gao, C.; Wang, L.; Wang, Y.; Sun, J.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 2015, 34, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; He, Y.; Zhang, H.; Wang, Z. Advances in the molecular regulation of seed germination in plants. Seed Biol. 2024, 3, e006. [Google Scholar] [CrossRef]
- Jia, H.; Wang, C.; Wang, F.; Liu, S.; Li, G.; Guo, X. GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PLoS ONE 2015, 10, e0120646, Erratum in PLoS ONE 2019, 14, e0213540. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Hu, W.; Zhou, R.; Wang, L.; Wang, X.; Wang, Q.; Feng, Z.; Li, Y.; Qiu, D.; He, G.; et al. The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 2015, 34, 23–35. [Google Scholar] [CrossRef]
- Cai, H.; Yang, S.; Yan, Y.; Xiao, Z.; Cheng, J.; Wu, J.; Qiu, A.; Lai, Y.; Mou, S.; Guan, D.; et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 2015, 66, 3163–3174. [Google Scholar] [CrossRef]
- Wang, F.; Chen, H.W.; Li, Q.T.; Wei, W.; Li, W.; Zhang, W.K.; Ma, B.; Bi, Y.D.; Lai, Y.C.; Liu, X.L.; et al. GmWRKY27 interacts with GmMYB174 to reduce the expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015, 83, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, F.; Chai, M.; Xi, X.; Zhu, W.; Qi, J.; Liu, K.; Ma, S.; Su, H.; Tian, Y.; et al. Ectopic Overexpression of Pineapple Transcription Factor AcWRKY31 Reduces Drought and Salt Tolerance in Rice and Arabidopsis. Int. J. Mol. Sci. 2022, 23, 6269. [Google Scholar] [CrossRef]
- Zhou, Q.; Priyadarshani, S.V.G.N.; Qin, R.; Cheng, H.; Luo, T.; Wai, M.H.; Mohammadi, M.A.; Liu, Y.; Liu, C.; Cai, H.; et al. AcWRKY28-mediated activation of AcCPK genes confers salt tolerance in pineapple (Ananas comosus). Hortic. Plant J. 2024, 10, 398–412. [Google Scholar] [CrossRef]
- Huang, X.; Rao, G.; Peng, X.; Xue, Y.; Hu, H.; Feng, N.; Zheng, D. Effect of plant growth regulators DA-6 and COS on drought tolerance of pineapple through bromelain and oxidative stress. BMC Plant Biol. 2023, 23, 180. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Wai, C.M.; Guyot, R. Pineapple Genome: A Reference for Monocots and CAM Photosynthesis. Trends Genet. 2016, 32, 690–696. [Google Scholar] [CrossRef]
- Gao, Y.; Yao, Y.; Chen, X.; Wu, J.; Wu, Q.; Liu, S.; Guo, A.; Zhang, X. Metabolomic and transcriptomic analyses reveal the mechanism of sweet-acidic taste formation during pineapple fruit development. Front. Plant Sci. 2022, 13, 971506. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Zhang, J.; Luan, A.; Zhang, W.; Wu, J.; Cai, Z.; He, Y. Comparative transcriptome analysis of a fan-shaped inflorescence in pineapple using RNA-seq. Genomics 2021, 113, 3653–3665. [Google Scholar] [CrossRef]
- Priyadarshani, S.; Cai, H.; Zhou, Q.; Liu, Y.; Cheng, Y.; Xiong, J.; Patson, D.L.; Cao, S.; Zhao, H.; Qin, Y. An Efficient Agrobacterium Mediated Transformation of Pineapple with GFP-Tagged Protein Allows Easy, Non-Destructive Screening of Transgenic Pineapple Plants. Biomolecules 2019, 9, 617. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yu, D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 2015, 34, 1295–1306. [Google Scholar] [CrossRef]
- Cai, J.; Liu, T.; Li, Y.; Ow, D.W. A C-terminal fragment of Arabidopsis OXIDATIVE STRESS 2 can play a positive role in salt tolerance. Biochem. Biophys. Res. Commun. 2021, 556, 23–30. [Google Scholar] [CrossRef]
- Xiang, S.; Wu, S.; Zhang, H.; Mou, M.; Chen, Y.; Li, D.; Wang, H.; Chen, L.; Yu, D. The PIFs Redundantly Control Plant Defense Response against Botrytis cinerea in Arabidopsis. Plants 2020, 9, 1246. [Google Scholar] [CrossRef]
- Grebner, W.; Stingl, N.E.; Oenel, A.; Mueller, M.J.; Berger, S. Lipoxygenase6-dependent oxylipin synthesis in roots is required for abiotic and biotic stress resistance of Arabidopsis. Plant Physiol. 2013, 161, 2159–2170. [Google Scholar] [CrossRef]
- Zentgraf, U.; Laun, T.; Miao, Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 2010, 89, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhao, J.; Huang, W.; Liu, C.; Hao, X.; Gao, C.; Deng, M.; Wen, J. Genome-Wide Identification of WRKY Transcription Factor Family in Chinese Rose and Response to Drought, Heat, and Salt Stress. Genes 2024, 15, 800. [Google Scholar] [CrossRef]
- Baillo, E.H.; Hanif, M.S.; Guo, Y.; Zhang, Z.; Xu, P.; Algam, S.A. Genome-wide Identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L.) Moench). PLoS ONE 2020, 15, e0236651. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhang, Z.; Tong, T.; Fang, Y.; Zhang, X.; Niu, C.; Li, J.; Wu, Y.; Xue, D.; Zhang, X. Genome-Wide Identification of WRKY Gene Family and Expression Analysis under Abiotic Stress in Barley. Agronomy 2021, 11, 521. [Google Scholar] [CrossRef]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef]
- Jeyasri, R.; Muthuramalingam, P.; Satish, L.; Adarshan, S.; Lakshmi, M.A.; Pandian, S.K.; Chen, J.-T.; Ahmar, S.; Wang, X.; Mora-Poblete, F.; et al. The Role of OsWRKY Genes in Rice When Faced with Single and Multiple Abiotic Stresses. Agronomy 2021, 11, 1301. [Google Scholar] [CrossRef]
- Song, Y.; Gao, J. Genome-wide analysis of WRKY gene family in Arabidopsis lyrata and comparison with Arabidopsis thaliana and Populus trichocarpa. Chin. Sci. Bull. 2014, 59, 754–765. [Google Scholar] [CrossRef]
- Fei, X.; Hou, L.; Shi, J.; Yang, T.; Liu, Y.; Wei, A. Patterns of Drought Response of 38 WRKY Transcription Factors of Zanthoxylum bungeanum Maxim. Int. J. Mol. Sci. 2018, 20, 68. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, Y.; Sun, J.; Liao, Z.; Ye, K.; Hu, B.; Dong, C.; Li, Z.; Deng, F.; Wang, L.; et al. Unveiling the genomic blueprint of salt stress: Insights from Ipomoea pes-caprae L. Seed Biol. 2023, 2, 21. [Google Scholar] [CrossRef]
- Viana, V.E.; Marini, N.; Finatto, T.; Ezquer, I.; Busanello, C.; Dos Santos, R.S.; Pegoraro, C.; Colombo, L.; Costa de Oliveira, A. Iron excess in rice: From phenotypic changes to functional genomics of WRKY transcription factors. Genet. Mol. Res. 2017, 16, gmr16039694. [Google Scholar] [CrossRef] [PubMed]
- Windram, O.; Madhou, P.; McHattie, S.; Hill, C.; Hickman, R.; Cooke, E.; Jenkins, D.J.; Penfold, C.A.; Baxter, L.; Breeze, E.; et al. Arabidopsis defense against Botrytis cinerea: Chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 2012, 24, 3530–3557. [Google Scholar] [CrossRef] [PubMed]
- Encinas-Villarejo, S.; Maldonado, A.M.; Amil-Ruiz, F.; de los Santos, B.; Romero, F.; Pliego-Alfaro, F.; Munoz-Blanco, J.; Caballero, J.L. Evidence for a positive regulatory role of strawberry (Fragaria × ananassa) Fa WRKY1 and Arabidopsis At WRKY75 proteins in resistance. J. Exp. Bot. 2009, 60, 3043–3065. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Kariola, T.; Palva, E.T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006, 46, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Kloth, K.J.; Wiegers, G.L.; Busscher-Lange, J.; van Haarst, J.C.; Kruijer, W.; Bouwmeester, H.J.; Dicke, M.; Jongsma, M.A. AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signaling. J. Exp. Bot. 2016, 67, 3383–3396. [Google Scholar] [CrossRef]
- Hwang, S.H.; Yie, S.W.; Hwang, D.J. Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense-related genes and enhances resistance to pathogens. Plant Sci. 2011, 181, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Peng, Y.; Guo, Z. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008, 18, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Xiao, J.; Xie, W.; Liu, H.; Li, X.; Xiong, L.; Wang, S. Rice Gene Network Inferred from Expression Profiling of Plants Overexpressing OsWRKY13, a Positive Regulator of Disease Resistance. Mol. Plant 2008, 1, 538–551. [Google Scholar] [CrossRef]
- Dietz, K.J.; Vogel, M.O.; Viehhauser, A. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal, and environmental signals in stress acclimation and retrograde signaling. Protoplasma 2010, 245, 3–14. [Google Scholar] [CrossRef]
- Liu, Z.Q.; Yan, L.; Wu, Z.; Mei, C.; Lu, K.; Yu, Y.T.; Liang, S.; Zhang, X.F.; Wang, X.F.; Zhang, D.P. Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot. 2012, 63, 6371–6392. [Google Scholar] [CrossRef]
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 13, 1039329. [Google Scholar] [CrossRef]
- Shi, W.Y.; Du, Y.T.; Ma, J.; Min, D.H.; Jin, L.G.; Chen, J.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; et al. The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Ao, C.; Wang, X.; Wu, Y.; Du, X. GhWRKY1-like, a WRKY transcription factor, mediates drought tolerance in Arabidopsis via modulating ABA biosynthesis. BMC Plant Biol. 2021, 21, 458. [Google Scholar] [CrossRef]
- Priyadarshani, S.V.G.N.; Hu, B.; Li, W.; Ali, H.; Jia, H.; Zhao, L.; Ojolo, S.P.; Azam, S.M.; Xiong, J.; Yan, M.; et al. Simple protoplast isolation system for gene expression and protein interaction studies in pineapple (Ananas comosus L.). Plant Methods 2018, 14, 95. [Google Scholar] [CrossRef]
- He, Y.; Luan, A.; Wu, J.; Zhang, W.; Lin, W. Overcoming key technical challenges in the genetic transformation of pineapple. Trop. Plants 2023, 2, 6. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Aydemir, B.C.; Ozmen, C.Y.; Kibar, U.; Mutaf, F.; Buyuk, P.B.; Bakir, M.; Ergul, A. Salt stress induces endoplasmic reticulum stress-responsive genes in a grapevine rootstock. PLoS ONE 2020, 15, e0236424. [Google Scholar] [CrossRef]
- Bowler, C.; Benvenuto, G.; Laflamme, P.; Molino, D.; Probst, A.V.; Tariq, M.; Paszkowski, J. Chromatin techniques for plant cells. Plant J. 2004, 39, 776–789. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.; Madhani, H.D.; Garcia, J.F. Total RNA Isolation and Quantification of Specific RNAs in Fission Yeast. Methods Mol. Biol. 2018, 1721, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Goel, S.; Meeley, R.B.; Dantec, C.; Parrinello, H.; Michaud, C.; Leblanc, O.; Grimanelli, D. Production of Viable Gametes without Meiosis in Maize Deficient for an ARGONAUTE Protein. Plant Cell 2011, 23, 443–458. [Google Scholar] [CrossRef]
- Sang, J.; Han, X.; Liu, M.; Qiao, G.; Jiang, J.; Zhuo, R. Selection and validation of reference genes for real-time quantitative PCR in hyperaccumulating ecotype of Sedum alfredo under different heavy metals stresses. PLoS ONE 2013, 8, e82927. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wai, M.H.; Luo, T.; Priyadarshani, S.V.G.N.; Zhou, Q.; Mohammadi, M.A.; Cheng, H.; Aslam, M.; Liu, C.; Chai, G.; Huang, D.; et al. Overexpression of AcWRKY31 Increases Sensitivity to Salt and Drought and Improves Tolerance to Mealybugs in Pineapple. Plants 2024, 13, 1850. https://doi.org/10.3390/plants13131850
Wai MH, Luo T, Priyadarshani SVGN, Zhou Q, Mohammadi MA, Cheng H, Aslam M, Liu C, Chai G, Huang D, et al. Overexpression of AcWRKY31 Increases Sensitivity to Salt and Drought and Improves Tolerance to Mealybugs in Pineapple. Plants. 2024; 13(13):1850. https://doi.org/10.3390/plants13131850
Chicago/Turabian StyleWai, Myat Hnin, Tiantian Luo, S. V. G. N. Priyadarshani, Qiao Zhou, Mohammad Aqa Mohammadi, Han Cheng, Mohammad Aslam, Chang Liu, Gaifeng Chai, Dongping Huang, and et al. 2024. "Overexpression of AcWRKY31 Increases Sensitivity to Salt and Drought and Improves Tolerance to Mealybugs in Pineapple" Plants 13, no. 13: 1850. https://doi.org/10.3390/plants13131850
APA StyleWai, M. H., Luo, T., Priyadarshani, S. V. G. N., Zhou, Q., Mohammadi, M. A., Cheng, H., Aslam, M., Liu, C., Chai, G., Huang, D., Liu, Y., Cai, H., Wang, X., Qin, Y., & Wang, L. (2024). Overexpression of AcWRKY31 Increases Sensitivity to Salt and Drought and Improves Tolerance to Mealybugs in Pineapple. Plants, 13(13), 1850. https://doi.org/10.3390/plants13131850









