Quantitative Trait Loci Sequencing and Genetic Mapping Reveal Two Main Regulatory Genes for Stem Color in Wax Gourds
Abstract
1. Introduction
2. Results
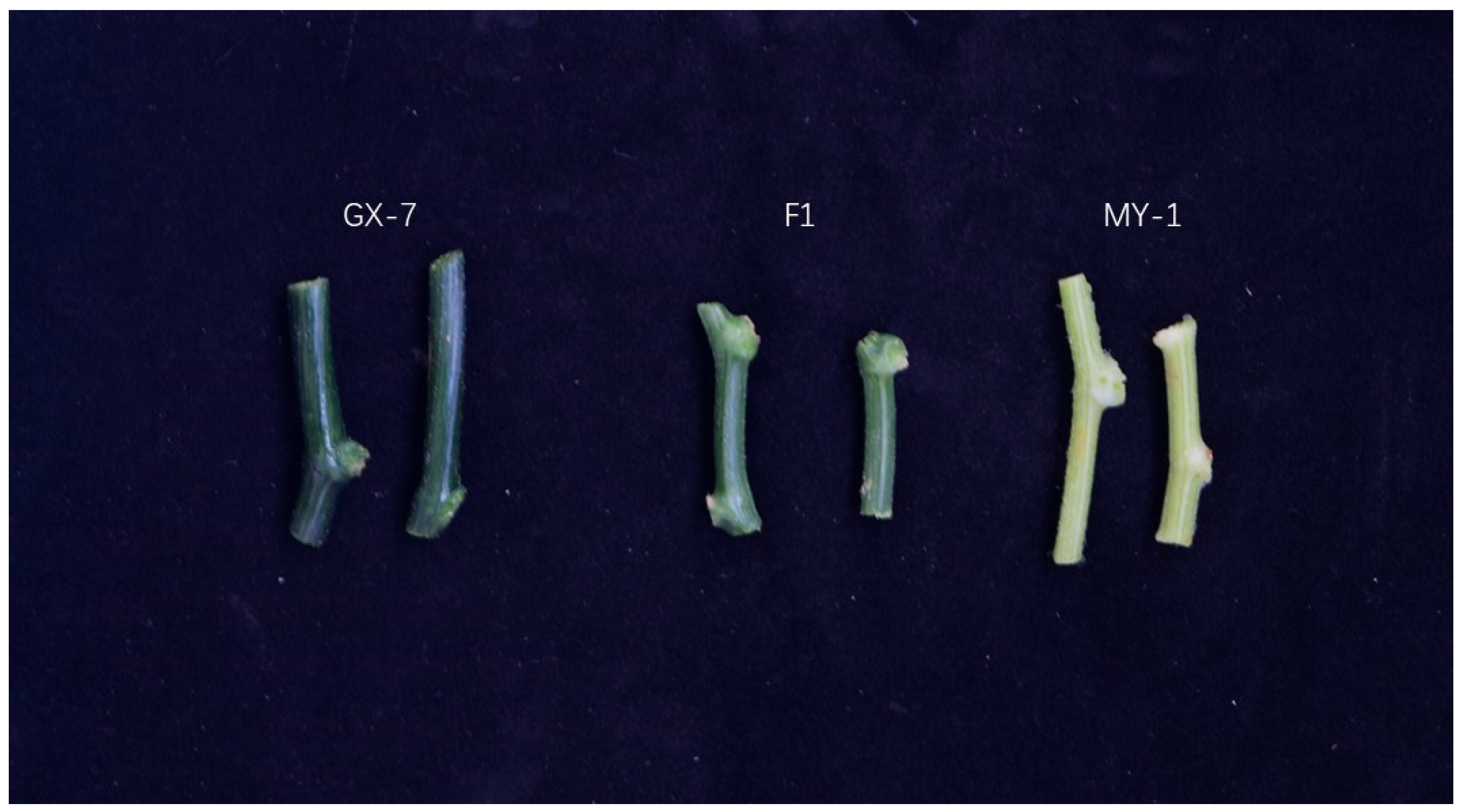
2.1. Phenotypic and Genetic Analyses of the Stem Color
2.2. Determination of Pigment Content
2.3. QTL Mapping and Candidate Gene Identification
2.4. qRT-PCR Analysis
2.5. InDel Marker-Assisted Breeding
3. Discussion
Molecular Mechanism Regulating the Stem Color of Wax Gourds
4. Materials and Methods
4.1. Plant Material and Phenotypic Evaluation
4.2. Extraction and Determination of Chlorophyll and Carotenoid Contents
4.3. DNA Extraction
4.4. QTL Mapping and InDel Marker Analysis
4.5. RNA Extraction and Candidate Gene Prediction Analysis
4.6. qRT-PCR Analysis of Candidate Genes
4.7. Molecular Marker-Assisted Breeding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, W.; Wang, P.; Huang, X.; Su, L.; Lv, H.; Gou, J.; Cheng, Z.; Ma, L.; Yu, W.; Liu, Z. Fine Mapping and Functional Analysis of Major Regulatory Genes of Soluble Solids Content in Wax Gourd (Benincasa hispida). Int. J. Mol. Sci. 2022, 23, 6999. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wang, P.; Liu, T.; Nong, L.; Cheng, Z.; Su, L.; Bai, W.; Deng, Y.; Chen, Z.; Liu, Z. Fine mapping of the major gene BhHLS1 controlling seed size in wax gourd (Benincasa hispida). Front. Plant Sci. 2023, 14, 1266796. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Yan, J.; He, C.; Liu, W.; Xie, D.; Jiang, B. Genome-Wide Identification of the SAUR Gene Family in Wax Gourd (Benincasa hispida) and Functional Characterization of BhSAUR60 during Fruit Development. Int. J. Mol. Sci. 2022, 23, 14021. [Google Scholar] [CrossRef] [PubMed]
- Oren, E.; Tzuri, G.; Vexler, L.; Dafna, A.; Meir, A.; Faigenboim, A.; Kenigswald, M.; Portnoy, V.; Schaffer, A.A.; Levi, A.; et al. The multi-allelic APRR2 gene is associated with fruit pigment accumulation in melon and watermelon. J. Exp. Bot. 2019, 70, 3781–3794. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Zhong, J.; Cheng, J.; Wang, Y.; Hu, K. Map-based cloning of the APRR2 gene controlling green stigma in bitter gourd (Momordica charantia). Front. Plant Sci. 2023, 14, 1128926. [Google Scholar] [CrossRef]
- Huo, Y.; Zhang, G.; Yu, W.; Liu, Z.; Shen, M.; Zhao, R.; Hu, S.; Zheng, X.; Wang, P.; Yang, Y. Forward genetic studies reveal LsAPRR2 as a key gene in regulating the green color of pericarp in bottle gourd (Lagenaria siceraria). Front. Plant Sci. 2023, 14, 1130669. [Google Scholar]
- Zhu, L.; Wang, Y.; Zhang, Z.; Hu, D.; Wang, Z.; Hu, J.; Ma, C.; Yang, L.; Sun, S.; Li, Y. Chromosomal fragment deletion in APRR2-repeated locus modulates the dark stem color in Cucurbita pepo. Theor. Appl. Genet. 2022, 135, 4277–4288. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liu, Z.; Cheng, Z.; Gou, J.; Chen, J.; Yu, W.; Wang, P. Identification and Application of BhAPRR2 Controlling Peel Colour in Wax Gourd (Benincasa hispida). Front. Plant Sci. 2021, 12, 716772. [Google Scholar]
- Huang, X.; Wu, W.; Su, L.; Lv, H.; Cheng, Z.; Yang, W.; Nong, L.; Liu, T.; Chen, Y.; Wang, P.; et al. Development and Application of InDel Markers Linked to Fruit-Shape and Peel-Colour Genes in Wax Gourd. Genes 2022, 13, 1567. [Google Scholar] [CrossRef] [PubMed]
- Bang, W.Y.; Jeong, I.S.; Kim, D.W.; Im, C.H.; Ji, C.; Hwang, S.M.; Kim, S.W.; Son, Y.S.; Jeong, J.; Shiina, T.; et al. Role of Arabidopsis CHL27 Protein for Photosynthesis, Chloroplast Development and Gene Expression Profiling. Plant Cell Physiol. 2008, 49, 1350–1363. [Google Scholar] [PubMed]
- Stenbaek, A.; Jensen, P.E. Redox Regulation of Chlorophyll Biosynthesis. Phytochemistry 2010, 71, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.-H.; Li, X.; Zhang, X.-X.; Zhang, H.; Zhao, X.-Y. Mutation Mechanism of Leaf Color in Plants: A Review. Forests 2020, 11, 851. [Google Scholar] [CrossRef]
- Fu, M.; Cheng, S.; Xu, F.; Chen, Z.; Liu, Z.; Zhang, W.; Zheng, J.; Wang, L. Advance in mechanism of plant leaf colour mutation. Not. Bot. Horti Agrobot. Cluj-Napoca 2021, 49, 12071. [Google Scholar] [CrossRef]
- Yang, S.; Wang, X.; Yan, W.; Zhang, Y.; Song, P.; Guo, Y.; Xie, K.; Hu, J.; Hou, J.; Wu, Y.; et al. Melon yellow-green plant (Cmygp) encodes a Golden2-like transcription factor regulating chlorophyll synthesis and chloroplast development. Theor. Appl. Genet. 2023, 136, 66. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.G.; Liang, Q.W.; Su, Y.; Huang, C.; Mo, B.X.; Yu, Y.; Xiao, L.T. Auxin inhibits chlorophyll accumulation through ARF7-IAA14-mediated repression of chlorophyll biosynthesis genes in Arabidopsis. Front. Plant Sci. 2023, 14, 1172059. [Google Scholar]
- Cheng, M.; Meng, F.; Mo, F.; Qi, H.; Wang, P.; Chen, X.; Liu, J.; Ghanizadeh, H.; Zhang, H.; Wang, A. Slym1 control the color etiolation of leaves by facilitating the decomposition of chlorophyll in tomato. Plant Sci. 2022, 324, 111457. [Google Scholar] [PubMed]
- Wu, M.; Xu, X.; Hu, X.; Liu, Y.; Cao, H.; Chan, H.; Gong, Z.; Yuan, Y.; Luo, Y.; Feng, B.; et al. SlMYB72 Regulates the Metabolism of Chlorophylls, Carotenoids, and Flavonoids in Tomato Fruit1. Plant Physiol. 2020, 183, 854–868. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Moylan, E.C.; Langdale, J.A. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008, 56, 432–444. [Google Scholar] [CrossRef]
- Nadakuduti, S.S.; Holdsworth, W.L.; Klein, C.L.; Barry, C.S. KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 2014, 78, 1022–1033. [Google Scholar] [PubMed]
- Jiang, L.; Fu, Y.; Tian, X.; Ma, Y.; Chen, F.; Wang, G. The Anthurium APRR2-like Gene Promotes Photosynthetic Pigment Accumulation in Response to Salt Stress. Trop. Plant Biol. 2022, 15, 12–21. [Google Scholar]
- Shen, S.; Yuan, J.; Xu, Y.; Ma, B.; Chen, X. Biological Function and Molecular Mechanism of the Transcription Factor GLKs in Plants: A Review. Chin. J. Biotechnol. 2022, 8, 2700–2712. [Google Scholar]
- Yan, F.; Gao, Y.; Pang, X.; Xu, X.; Zhu, N.; Chan, H.; Hu, G.; Wu, M.; Yuan, Y.; Li, H.; et al. BEL1-LIKE HOMEODOMAIN4 regulates chlorophyll accumulation, chloroplast development, and cell wall metabolism in tomato fruit. J. Exp. Bot. 2020, 71, 5549–5561. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Wang, Z.; Yang, P. Review: The effect of light on the key pigment compounds of photosensitive etiolated tea plant. Bot. Stud. 2021, 62, 21. [Google Scholar] [CrossRef] [PubMed]
- You, Q.; Li, H.; Wu, J.; Li, T.; Wang, Y.; Sun, G.; Li, Z.; Sun, B. Mapping and validation of the epistatic D and P genes controlling anthocyanin biosynthesis in the peel of eggplant (Solanum melongena L.) fruit. Hortic. Res. 2023, 10, uhac268. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Tian, M.; An, G.; Zhang, W.; Chen, J.; Yan, C. Rapid identification of the purple stem (Ps) gene of Chinese kale (Brassica oleracea var. alboglabra) in a segregation distortion population by bulked segregant analysis and RNA sequencing. Mol. Breed. 2017, 37, 153. [Google Scholar]
- Shmakov, N.A.; Vasiliev, G.V.; Shatskaya, N.V.; Doroshkov, A.V.; Gordeeva, E.I.; Afonnikov, D.A.; Khlestkina, E.K. Identification of nuclear genes controlling chlorophyll synthesis in barley by RNA-seq. BMC Plant Biol. 2016, 16, 245. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, H.; Yang, Y.; Li, M.; Zhang, Y.; Liu, J.; Dong, J.; Li, J.; Butelli, E.; Xue, Z.; et al. The control of red colour by a family of MYB transcription factors in octoploid strawberry (Fragaria × ananassa) fruits. Plant Biotechnol. J. 2020, 18, 1169–1184. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, R.; Zeng, X.-Y.; Lee, S.; Ye, L.-H.; Tian, S.-L.; Zhang, Y.-J.; Busch, W.; Zhou, W.-B.; Zhu, X.-G.; et al. GLK transcription factors accompany ELONGATED HYPOCOTYL5 to orchestrate light-induced seedling development in Arabidopsis. Plant Physiol. 2024, 194, 2400–2421. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, R.; Abe, S.; Marugami, M.; Yamagami, A.; Akema, R.; Ohashi, T.; Nishida, K.; Nosaki, S.; Miyakawa, T.; Tanokura, M.; et al. BPG4 regulates chloroplast development and homeostasis by suppressing GLK transcription factors and involving light and brassinosteroid signaling. Nat. Commun. 2024, 15, 370. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Bradley, G.; Pyke, K.; Ball, G.; Lu, C.; Fray, R.; Marshall, A.; Jayasuta, S.; Baxter, C.; van Wijk, R.; et al. Network Inference Analysis Identifies an APRR2-Like Gene Linked to Pigment Accumulation in Tomato and Pepper Fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Nong, L.; Wang, P.; Yang, W.; Liu, T.; Su, L.; Cheng, Z.; Bai, W.; Deng, Y.; Chen, Z.; Liu, Z. Analysis of BhAPRR2 allele variation, chlorophyll content, and chloroplast structure of different peel colour varieties of wax gourd (Benincasa hispida) and development of molecular markers. Euphytica 2023, 219, 107. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian Waves of Expression of the APRR1/TOC1 Family of Pseudo-Response Regulators in Arabidopsis thaliana: Insight into the Plant Circadian Clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Nakamichi, N.; Matsushika, A.; Fujimori, T.; Yamashino, T.; Mizuno, T. Molecular Dissection of the Promoter of the Light-Induced and Circadian-Controlled APRR9 Gene Encoding a Clock-Associated Component of Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2005, 69, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCENCE1 Gene Encodes a Novel Phospholipase A1 Catalyzing the Initial Step of Jasmonic Acid Biosynthesis, Which Synchronizes Pollen Maturation, Anther Dehiscence, and Flower Opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Principles and Techniques of Plant Physiological and Biochemical Experiments; Higher Education Press: Beijing, China, 2000. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Plant Type | Total No. | Green (Light Green) | White | X2 (9:7) | p |
|---|---|---|---|---|---|
| GX-71 | 10 | 10 | - | - | - |
| MY-1 | 10 | - | 10 | - | - |
| F1 population | 10 | 10 | - | - | - |
| F2 population | 786 | 449 | 337 | 0.244 | 0.19 |
| Gene ID | Nonsynonymous Mutations in Coding Sequences | Physical Location | Gene Annotation |
|---|---|---|---|
| Bch05G003860 | Yes | Chr5:13207272–13212788 (+) | protein NRT1/ PTR FAMILY 7.3-like |
| Bch05G003870 | Yes | Chr5:13213544–13235133 (−) | Pyruvate kinase isozyme G |
| Bch05G003880 | Yes | Chr5:13274698–13276057 (+) | - |
| Bch05G003890 | No | Chr5:13276096–13281871 (+) | Protein ABCI7, chloroplastic |
| Bch05G003900 | Yes | Chr5:13341180–13346696 (−) | Probable ribose-phosphate Pyrophosphokinase 1-like |
| Bch05G003910 | Yes | Chr5:13367114–13372139 (−) | Probable tobamovirus multiplication protein 2B isoform X1 |
| Bch05G003920 | Yes | Chr5:13438283–13441573 (−) | Probable peroxidase 20 isoform X1 |
| Bch05G003930 | No | Chr5:13441626–13447889 (−) | Tobamovirus multiplication protein 2A-like |
| Bch05G003940 | Yes | Chr5:13484454–13484678 (−) | - |
| Bch05G003950 | Yes | Chr5:13491669–13499644 (−) | Two-component response Regulator-like protein APRR2 |
| Bch05G003960 | Yes | Chr5:13676056–13689823 (−) | MSC domain-containing protein |
| Bch05G003970 | No | Chr5:13805666–13806021 (+) | Vacuolar protein sorting-associated protein 55 homolog |
| Bch05G003980 | No | Chr5:13810161–13810902 (−) | Uncharacterized protein LOC111455443 |
| Bch05G003990 | Yes | Chr5:13825779–13838208 (−) | Transcription factor MAMYB |
| Gene ID | Nonsynonymous Mutations in Coding Sequences | Physical Location | Gene Annotation |
|---|---|---|---|
| Bch12G020370 | Yes | Chr12:75155250–75160607 (−) | Probable H/ACA ribonucleoprotein complex subunit 1-like |
| Bch12G020380 | No | Chr12:75215164–75221452 (−) | RGG repeats nuclear RNA binding protein A-like isoform X2 |
| Bch12G020390 | No | Chr12:75225625–75297961 (−) | Probable glycine—tRNA ligase, chloroplastic/mitochondrial 2 isoform X1 |
| Bch12G020400 | No | Chr12:75332840–75335150 (−) | Phospholipase A(1) DAD1, chloroplast-like |
| Bch12G020410 | No | Chr12:75409841–75418358 (+) | Carotenoid cleavage dioxygenase 7, chloroplastic |
| Bch12G020420 | No | Chr12:75418509–75419660 (+) | Probable major pollen allergen Ole e 6-like |
| Bch12G020430 | No | Chr12:75498816–75499589 (+) | Probable major pollen allergen Ole e 6-like |
| Bch12G020440 | No | Chr12:75510818–75511148 (−) | - |
| Bch12G020450 | No | Chr12:75522841–75523897 (+) | Probable major pollen allergen Ole e 6-like |
| Bch12G020460 | No | Chr12:75529773–75530669 (+) | Probable major pollen allergen Ole e 6-like |
| Bch12G020470 | No | Chr12:75548538–75551483 (−) | - |
| Bch12G020480 | No | Chr12:75557730–75559794 (−) | Pentatricopeptide repeat-containing protein At4g01570 |
| Bch12G020490 | No | Chr12:75564027–75565069 (−) | Probable uncharacterized protein LOC103498828 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Wang, P.; Bai, W.; Deng, Y.; Cheng, Z.; Su, L.; Nong, L.; Liu, T.; Yang, W.; Yang, X.; et al. Quantitative Trait Loci Sequencing and Genetic Mapping Reveal Two Main Regulatory Genes for Stem Color in Wax Gourds. Plants 2024, 13, 1804. https://doi.org/10.3390/plants13131804
Chen Z, Wang P, Bai W, Deng Y, Cheng Z, Su L, Nong L, Liu T, Yang W, Yang X, et al. Quantitative Trait Loci Sequencing and Genetic Mapping Reveal Two Main Regulatory Genes for Stem Color in Wax Gourds. Plants. 2024; 13(13):1804. https://doi.org/10.3390/plants13131804
Chicago/Turabian StyleChen, Zhihao, Peng Wang, Wenhui Bai, Yan Deng, Zhikui Cheng, Liwen Su, Lifeng Nong, Ting Liu, Wenrui Yang, Xiping Yang, and et al. 2024. "Quantitative Trait Loci Sequencing and Genetic Mapping Reveal Two Main Regulatory Genes for Stem Color in Wax Gourds" Plants 13, no. 13: 1804. https://doi.org/10.3390/plants13131804
APA StyleChen, Z., Wang, P., Bai, W., Deng, Y., Cheng, Z., Su, L., Nong, L., Liu, T., Yang, W., Yang, X., & Liu, Z. (2024). Quantitative Trait Loci Sequencing and Genetic Mapping Reveal Two Main Regulatory Genes for Stem Color in Wax Gourds. Plants, 13(13), 1804. https://doi.org/10.3390/plants13131804






