Abstract
Pineapple is one of the most economically important fruits in tropical countries, particularly in Thailand. Canned pineapple is currently Thailand’s main exported commodity to many countries, including the United States, Russia, Germany, Poland, and Japan. Fungal diseases are considered a permanent threat to fruits in the pre- and post-harvest stages, leading to considerable economic losses. Fungal disease is one of the primary causes of massive yield losses in pineapples around the world. Colletotrichum species are the most common fungal pathogens affecting different tropical fruits. Although there are many reports regarding Colletotrichum species associated with pineapple, they do not have molecular data to confirm species identification. However, the occurrence of Colletotrichum species on pineapple has not been reported in Thailand so far. In this study, we isolated and identified Colletotrichum fructicola on pineapple in northern Thailand and have proven its pathogenicity to the host. This is the first report of the occurrence of Colletotrichum in pineapple, based on morpho-molecular approaches.
1. Introduction
Pineapple (Ananas comosus (L.) Merr.) is one of the edible and nutritious fruits of the Bromeliaceae, grown in tropical and subtropical countries [1]. Asia, South Central America, and Africa are the world’s leading areas producing this fruit [2]. Pineapple is the second largest tropical crop in the world [2] and the third most consumed fruit worldwide [3]. Brazil, China, Costa Rica, India, the Philippines, and Thailand are the top pineapple-producing countries [1]. Pineapple could be eaten as a fresh fruit or selected as a basic raw ingredient used in the confectionery industry [4,5,6]. The fruit contains immense nutrients and is abundant in vitamins A, C, B1, and B6 [7,8,9]. It also has proteins, carbohydrates, fiber, copper, manganese, and several minerals [5,10].
In Thailand, pineapple products have been regarded as economic commodities for export [11]. Thailand is currently the second-largest supplier of processed pineapple fruit in the world. Production areas for pineapple in Thailand have been divided into northern, northeastern, central, and southern parts [12]. Most cultivated areas are in Prachuap Khiri Khan, Rayong, Ratchaburi, and Chonburi provinces [11]. Pineapple is classified into five groups based on the morphology of the leaf and fruit, namely Abacaxi, Cayenne, Maipure or Perolera, Queen, and Spanish [13]. Among these, Cayenne, Queen, and Spanish are cultivated in Thailand [14]. Nang Lae district in Chiang Rai province is the most pineapple-cultivating area, and Nang Lae (Cayenne group) and Phu Lae (Queen group) are the most common varieties in northern Thailand (http://www.doa.go.th/; https://www.saio.co.th/; accessed on 12 December 2022). The pineapple-growing area in Thailand is around 72,656 hectares, with an annual production of 1,680,884 metric tons. The total canned pineapples exported from Thailand were 290,524 metric tons in 2020, valued at about 345 million US dollars [15].
Diseases are the key elements of significant yield losses in pineapples across the world [1]. Among fungal pathogens, Colletotrichum species are the most important fungi responsible for the diseases of tropical and sub-tropical fruits [16]. Many Colletotrichum species have been reported from different hosts in Thailand such as C. aenigma [17], C. aeschynomenes [18], C. artocarpicola [19], C. asianum [20,21,22,23], C. boninense [18,24], C. brevisporum [16,25,26,27], C. chiangraiense [24,28], C. cordylinicola [21,29,30], C. endophytica [31,32,33,34], C. orchidearum [24,25], C. orchidophilum [24], C. plurivorum [35], and C. siamense [16,21,24,36,37], majority belonging to gloeosporioides species complex. Colletotrichum fructicola is one of the most invasive species and has been reported as the causal agent of anthracnose, leaf spots and bitter rots in more than 90 plant species [38]. It was originally isolated from Coffea arabica in northern Thailand [39]. It has also been reported from Capsicum annuum [24], Carica papaya [20,24], Cymbopogon citratus [16,24], Dendrobium sp. [24], Dimocarpus longan [20,24,29], Freycinetia sp. [24], Pandanus sp. [24], Pennisetum purpureum [16,24] in Thailand. Mealybug wilt-associated virus, bacterial heart rot, fruit collapse, butt rot, fruitlet core rot, black rot, yeasty, and fusariosis are the main diseases of pineapple, discussed by Sapak et al., (2021). Despite the importance of pineapple in Thailand, studies for the isolation and identification of fungal pathogens associated with pineapple have not been conducted in Thailand.
2. Results
2.1. Morphological Studies
Following 7–14 days of incubation, morphological features including culture (color and growth rate) and microscopic features (conidiogenous cells and conidial measurements, appressoria measurements) were recorded for P76 (MFLU 22-0302) and P76-3 (MFLU 22-0303). The two strains, P76 and P76-3 were isolated from rotting pineapple fruit and leaf dieback, respectively. Morphological comparisons of P76 and P76-3 were performed on 14-day-old cultures grown on PDA at 25 °C ± 2 °C. There were minor differences in size of conidia and conidiogenous cells and also in appressoria shape which are very common in a specific species within the C. gloeosporioides species complex. Finally, these two isolates were identified as C. fructicola based on morpho-molecular evidence.
2.2. Phylogenetic Analyses
The five-locus (ITS, ACT, GAPDH, CHS-1, and TUB2) phylogenetic analysis included 73 reference isolates [40,41,42]. The phylogenetic tree consisted of 71 ingroup and 2 outgroup taxa (Colletotrichum truncatum, CBS 151.35 and C. acidae, MFLUCC 17-2659). The data matrix contained a total of 1670 characters, of which 269 were parsimony-uninformative and 384 were parsimony-informative. The most parsimonious tree (Tree Length (TL) = 1321, Consistency Index (CI) = 0.673, Retention Index (RI) = 0.824, Rescaled Consistency Index (RC) = 0.554, Homoplasy Index (HI) = 0.327) was presented (Figure 1). The ML, MP and BYPP trees were identical in topology. The best-scoring RAxML tree with final optimization showed a likelihood value of −9834.854352. The dataset comprised 754 distinct alignment patterns, with 5.31% of characters being gaps or undetermined. Estimated base frequencies were as follows: A = 0.229261, C = 0.299900, G = 0.241493, T = 0.229347, with substitution rates AC = 1.133302, AG = 2.907494, AT = 1.290422, CG = 0.902726, CT = 4.957643, GT = 1.000000. The gamma distribution shape parameter is 0.422238 and the tree length is 0.963648. Based on the phylogenetic analysis, strains P76 and P76-3 clustered with C. fructicola, showing 86/77/0.99 ML, MP, and BYPP values, respectively (Figure 1). The base pair differences between these two strains and the ex-type of C. fructicola (ICMP 18581) were shown (Table 1).
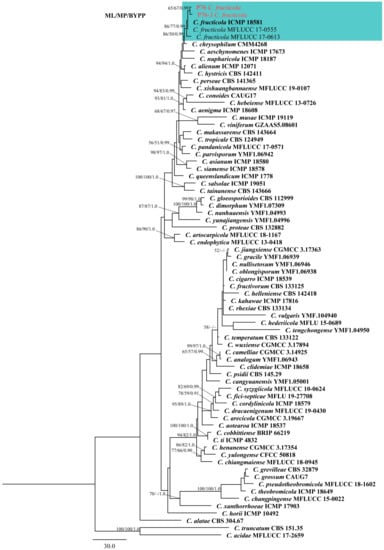
Figure 1.
Maximum parsimony tree of the Colletotrichum gloeosporioides species complex generated by analysis of combined ITS, ACT, CHS-1, GAPDH, and TUB2 sequence data. The tree was rooted with Colletotrichum truncatum (CBS 151.35) and Colletotrichum acidae (MFLUCC 17.2659). Maximum likelihood and maximum parsimony bootstrap values ≥ 50% and bayesian posterior probabilities ≥ 0.90 are shown near the nodes, respectively. Type strains are in bold and the newly generated isolates are in red.

Table 1.
Base pair differences between C. fructicola (ICMP 18581) and two newly isolated strains.
2.3. Taxonomy

Figure 2.
Colletotrichum fructicola P76 (MFLU 22-0302); (a,b). Rotted pineapple fruit; (c). Acervuli on the fruit; (d). Setae; (e). Conidiophores; (f). Conidiogenous cells and conidial attachment; (g). Conidia; (h). Appressoria; (i). Upper and reverse view of colony on PDA. Scale bars: (c) = 200 µm, (d) = 20 µm, (e–h) = 10 µm.
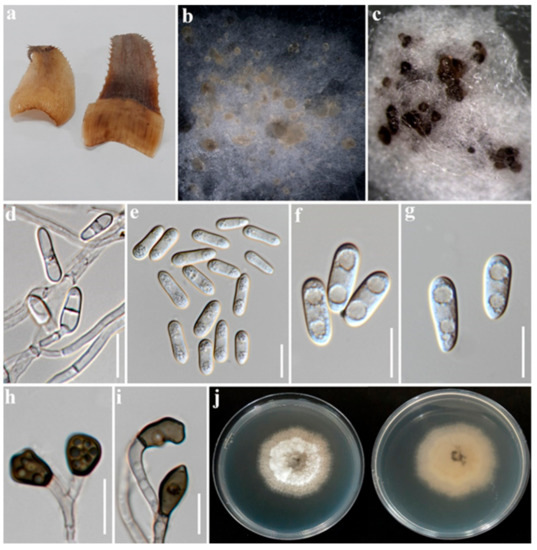
Figure 3.
Colletotrichum fructicola P76-3 (MFLUCC 22-0182); (a). Symptomatic leaves; (b). Conidial masses on PDA; (c). Acervuli on PDA; (d). Conidial attachment; (e–g). Conidia; (h,i). Appressoria; (j). Upper and reverse view of colony on PDA. Scale bars: (d–i) 10 µm.
Index fungorum number: IF 515409; Faces of Fungi number: FoF 06767.
Associated with pineapple fruit rot and leaf dieback. Sexual morph: Not observed. Asexual morph: Vegetative hyphae hyaline, smooth-walled, septate, branched. Conidiomata acervular, dark brown, bearing conidial mass, and setae. Setae brown to dark brown, smooth-walled, 2–4 septate, 38–83 μm long ( = 59.5 μm, n = 6), base cylindrical, 3–5 μm diam. ( = 4.5 µm, n = 6), tip acute or obtuse. Conidiophores rarely observed, hyaline, septate, branched, cylindrical to inflated. Conidiogenous cells hyaline, cylindrical or clavate, 12–25 × 3–4.5 μm ( = 18 × 3.5 µm, n = 20). Conidia hyaline, aseptate, smooth-walled, cylindrical, rounded at apex, sub-acute at base, guttulate, 12.5–19 × 4.5–6 μm ( = 16 × 5 µm, n = 30).
Culture characteristics: Colonies on PDA 65–85 mm in diam. after 7 days at 28 °C, velvety, circular, undulate; surface pale grey in center and white in margin, becoming grey with age; reverse same color. Colonies on OA 60–71 mm in diam. after 7 days, cottony, slightly raised, entire; surface white to whitish grey; reverse same color. Appressoria produced on slide culture, brown to dark brown, irregular in shape, undulate, 7–9 × 4.5–6 μm ( = 7.8 × 5 µm, n = 15), producing on hyphae and conidia.
Material examined: Thailand, Chiang Rai Province, Mueang Chiang Rai District, Ban Du Sub-district. On pineapple rotting fruit, 27 June 2022, Alireza Armand, P76 (MFLU 22-0302), living culture, MFLUCC 22-0181. On pineapple leaf dieback, 27 June 2022, Alireza Armand, P76-3 (MFLU 22-0303), living culture, MFLUCC 22-0182.
Notes: The species within the gloeosporioides species complex are mainly distinguished by producing cylindrical conidia with rounded ends, tapering slightly towards the base [16]. The strain P76 was isolated directly from rotting pineapple, whereas P76-3 was obtained by tissue isolation from a fresh leaf with tip dieback symptoms. Based on the phylogenetic tree (Figure 1), isolates P76 (MFLUCC 22-0182) and P76-3 (MFLU 22-0303) clustered with C. fructicola strains with 86/77/0.99 ML, MP, and BYPP values, respectively. Morphologically, P76 and P76-3 are similar. However, P76 produced slightly larger conidia than P76-3 (13–19 × 4.5–6 μm in P76 vs. 12.5–17.5 × 4–6 μm in P76-3). The conidial shape was slightly different, as P76 produced conidia with obtuse ends, whereas P76-3 mostly produced conidia with rounded ends. However, morphological comparison with the ex-type of C. fructicola revealed no significant differences between the type strain and our isolates (P76, P76-3) [39].
2.4. Pathogenicity Assay
Pathogenicity test results related to strain P76 showed that this strain can cause disease on both wounded and non-wounded host leaves. The wounded leaves inoculated with P76 showed dieback symptoms 4 days after the inoculation, whereas those of the non-wounded leaves showed symptoms 6 days after the inoculation. However, the symptoms continued to spread in both wounded and non-wounded leaves after 10 days. After 11 days, aerial mycelia started to grow on the surface of the symptomatic area in both wounded and non-wounded leaves. (Figure 4, P76/W, P76/NW).
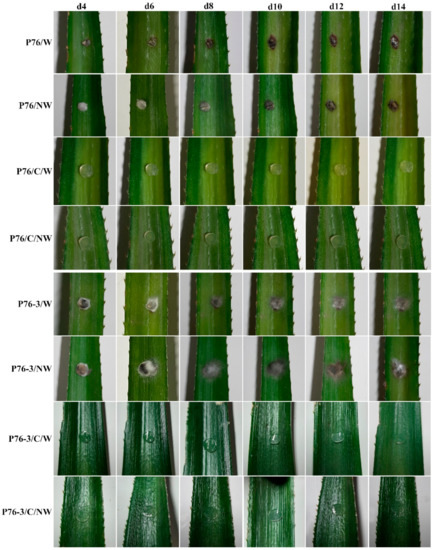
Figure 4.
Pathogenicity testing on pineapple leaves. Symptoms (dieback) on days 4 (d4), 6 (d6), 8 (d8), 10 (d10), 12 (d12), and 14 (d14) after inoculation are shown. C: Control, W: Wounded, NW: Non-wounded.
Six days after the inoculation, symptoms on the injured leaves treated with P76-3 were only present at the border of the mycelial plugs (Figure 4, P76-3/W). The non-wounded leaves inoculated with P76-3 remained asymptomatic during the test (Figure 4, P76-3/NW) along with the negative control treatments. The re-isolated fungi were identified as C. fructicola according to morphological characteristics.
3. Discussion
In this study, the diseased leaves showing dieback symptoms and rotting fruits of pineapple were collected in Chiang Rai province, northern Thailand. Based on a direct isolation from the fruiting bodies (Figure 2) and an indirect isolation of infected leaves via tissue culture (Figure 3), we obtained two Colletotrichum isolates. Jayawardena et al., (2021) recommended using a polyphasic approach to identify Colletotrichum species [43]. We used both morphological examination and multi-loci molecular analysis for species-level identification. The phylogenetic analysis of a combined dataset of ITS, ACT, CHS-1, GAPDH, and TUB2 showed that the two isolates of Colletotrichum associated with pineapple belong to C. fructicola (Figure 1). Morphological studies also confirmed the phylogenetic results. Colletotrichum species have a wide host range and geographical distribution worldwide [16,44]. However, the occurrence of Colletotrichum species on pineapple has not so far been reported in Thailand. In the USDA host fungal database, there are 12 records of Colletotrichum species on Ananas sp. [45]. Colletotrichum ananas was reported in India [46], while C. truncatum was reported to cause leaf-tip dieback in Malaysia [45]. Colletotrichum gloeosporioides has been recorded to cause anthracnose on pineapple in China and the United States [45]. Colletotrichum gloeosporioides was also recorded from Ananas spp. in Brazil on pineapple leaves. Colletotrichum sp. was identified in Cuba, India, Korea, Panama, and the West Indies [47]. However, these species were identified based only on morphology.
In recent decades, identification based on morphology has led to the misidentification of fungal pathogens [43]. In plant pathology, correct identification of fungal species is a fundamental step that links information concerning biology, host range, distribution, pathogenicity and food security [43,48], indicating the importance of species identification to barricade future afflictions provoked by these pathogens [49]. Moreover, emerging pathogens have also been increasing threats during the last decade [24]. Colletotrichum, being a complex genus, shares overlapping morphological characteristics among species. Colletotrichum fructicola belongs to the gloeosporioides species complex, which comprises fruit rots and post-harvest pathogens. Since this species complex is the most confusing within the Colletotrichum genus, morphological identification alone cannot be trusted to identify the species correctly. It follows that the use of a polyphasic approach in plant pathology is crucial for the precise identification and naming of fungi, which will advance the management and control of both recognized and newly emerging diseases. [43]. Furthermore, a polyphasic approach using molecular analysis is an effective tool to identify cryptic species and estimate species diversity [49].
Among Colletotrichum species in the gloeosporioides species complex, C. fructicola has a very broad host range, isolated from more than eight plant families as endophytes and plant pathogens [48]. It has been reported from America, Asia, Africa, Europe, and Oceania [38]. In this study, we isolated and illustrated C. fructicola as the first report of Colletotrichum species associated with pineapple in Thailand, based on molecular and morphological analyses. Furthermore, the pathogenicity tests proved that the isolates are pathogens to pineapple (Figure 4).
The results of the present study can be useful for pathologists in understanding the fungal pathogen diversity associated with pineapple, disease management and quarantine purposes. Many new Colletotrichum species have been introduced in 2022 (https://www.mycobank.org/; accessed on 8 December 2022), and there are potentially many novel species of Colletotrichum that remained undiscovered [49]. Additionally, many studies reported new hosts for existing Colletotrichum species [50,51,52,53,54,55,56,57,58], broadening their host and geographical ranges. Therefore, more investigations on the isolation and identification of Colletotrichum associated with pineapple can improve our knowledge about fungal diversity and host range and will potentially lead to the discovery of novel species of Colletotrichum.
4. Materials and Methods
4.1. Sample Collection, Examination and Isolation
In order to isolate the fungal pathogens associated with pineapple plants, pineapple leaves with diebacks and leaf spots, and rotting fruits were considered. In total, 10 symptomatic leaves and 12 rotting pineapple fruits were collected during June–July 2022 from organic farms in Ban Du and Nang Lae sub-districts, Mueang Chiang Rai district, Chiang Rai province, northern Thailand. The samples were kept in plastic bags labelled with the collection date, collection site and host name before being transported to the laboratory for further examination. The fruiting bodies on natural substrates were observed and photographed using a stereomicroscope (OLYMPUS SZX16; Tokyo, Japan). Morphological features were observed using a LEICA-EZ4 stereomicroscope and photographed with an optical microscope equipped with a Nikon DS-Ri2 camera. The photo plates were made by the Adobe Photoshop v.21.1.2 software, and the scales were measured by the Tarosoft (R) Image Frame Work software.
Direct isolation and indirect isolation (tissue isolation) were used to obtain cultures [59]. Further, 30 mm2 leaf fragments were cut from the margins of lesions for tissue isolation, and were sterilized by submerging in 70% ethanol for 2 min, 10% sodium hypochlorite solution for 60 s, followed by three times rinsing in sterile distilled water for 60 s [32]. Following the procedures outlined by Senanayake et al., (2020), single-spore isolation and hyphal tip isolation were done to purify the isolates. Finally, the pure cultures were deposited in the Mae Fah Luang University Culture Collection (MFLUCC), Chiang Rai, Thailand. Specimens were deposited in the herbarium of the Center of Excellence in Fungal Research (CEFR), Mae Fah Luang University (MFLU).
4.2. DNA Extraction and PCR Amplification
Using a DNA Extraction Kit (Omega Biotek) in accordance with the manufacturer’s instructions, genomic DNA was extracted from fresh mycelia cultured on potato dextrose agar (PDA) for 14 days. The internal transcribed spacer (ITS), actin (ACT), chitin synthase (CHS-1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-tubulin (TUB2) were amplified using primers ITS5/ITS4, ACT-512F/ACT-783R, CHS-79F/CHS-345R, GDF/GDR, and BT-2Fd/BT-4Rd, respectively (Table 2). The polymerase chain reaction was carried out in a total volume of 25 µL, including 12.5 µL of 2 × Power Taq PCR Master Mix, 1 µL of each primer (20 µM), 1 µL genomic DNA, and 9.5 µL of deionized water. The PCR procedure was done under the following conditions: Initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation for 30 s at 95 °C; annealing at 53 °C for 60 s (ITS), 55 °C for 50 s (ACT), 58 °C for 30 s (CHS-1); 58 °C for 50 s (GAPDH), 58 °C for 90 s (TUB2); extension at 72 °C for 60 s; and the final extension at 72 °C for 10 min. PCR amplification was performed in an eppendorf thermal cycler (Master Cycler X50s). PCR products were sequenced by the SolGent company, Republic of Korea.

Table 2.
Primers used in this study.
4.3. Phylogenetic Analyses
Sequences for the selected strains were obtained from GenBank (Table 3), according to blast-searching and related publications [40,41,42]. Multiple sequence alignments for ITS, ACT, CHS-1, GAPDH, and TUB2 were constructed using MAFFT v.7.11 on the web server (https://mafft.cbrc.jp/alignment/server, accessed on 12 February 2023) with the default settings [64]. BioEdit v.7.0.9.0 was used for adjusting the sequences [65] and TrimAl software was used to trim aligned sequences automatically using the gappyout command. Maximum parsimony (MP) analysis was done using PAUP XSEDE [66]. Maximum likelihood (ML) analysis was performed on XSEDE with the GTR + Gamma model and 1000 replications using RAxML-HPC2 The Bayesian posterior probabilities analysis (BYPP) was carried out using a Markov Chain Monte Carlo (MCMC) algorithm using MrBayes on XSEDE [67]. In order to choose the best-fit evolutionary models for each dataset, jModeltest 2.1.10 and the Akaike Information Criterion (AIC) were employed on the CIPRES platform. Four MCMC chains were run from random trees for 1,000,000 generations and sampled every 100th generation. The first 25% of the generated trees were ignored as burn-in and the remaining trees were used for analyzing the posterior probabilities. Gaps were considered missing data, and ambiguously aligned parts were eliminated. The phylogenetic trees were visualized in FigTree v.1.4.0 [68], and were further edited in Adobe Illustrator CC 22.0.0 (Adobe Systems, San Jose, CA, USA).

Table 3.
Taxa and their GenBank accession numbers used in the phylogenetic analysis.
4.4. Pathogenicity Assay
Koch’s postulates were applied according to the procedures demonstrated by Bhunjun et al., (2021) to confirm the pathogenicity of our isolates [69]. Three replicates of detached leaves from an organic farm were considered for both wounded and non-wounded assays using mycelial plug incubation because of the lack of culture sporulation. The pineapple leaves were surface sterilized by washing them in 70% ethanol for 2 min, then in 2% sodium hypochlorite for 2 min, followed by three washes with sterile distilled water and laminar air drying. We chose a pineapple leaf instead of fruit to accurately assess symptoms and fungal spread through the host. Mycelial plugs were obtained from the fresh colonies grown on PDA (10-day colonies). Control inoculations were performed using uninoculated PDA plugs. In a moist chamber at 28 °C and 80% relative humidity, the inoculated and control leaves were incubated. Koch’s postulates were confirmed by re-isolating the fungus from the infected leaves. The re-isolated fungus was identified based on cultural and morphological features.
Author Contributions
Conceptualization, R.S.J.; Methodology, A.A.; Formal analysis, A.A. and R.S.J.; Investigation, A.A.; Resources, R.S.J.; Data curation, A.A.; Writing—original draft, A.A.; Writing—review & editing, R.S.J. and K.D.H.; Supervision, R.S.J.; Project administration, R.S.J.; Funding acquisition, R.S.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Research Council of Thailand (NRCT) grant entitled “Biodiversity, taxonomy, phylogeny and evolution of Colletotrichum in northern Thailand” (grant no. NRCT-TRG010).
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: https://www.ncbi.nlm.nih.gov (accessed on 12 November 2022).
Acknowledgments
We would like to thank KC. Mallikarathna and S. Wangsafoo for providing the organic pineapple leaves to carry out the pathogenicity tests. Alireza Armand would like to thank Arttapon Walker for his kind help in obtaining local information.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sapak, Z.; Mohd Faisol Mahadeven, A.N.; Nurul Farhana, M.H.; Norsahira, S.; Mohd Zafri, A.W. A review of common diseases of pineapple: The causal pathogens, disease symptoms, and available control measures. Food Res. 2021, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Altendorf, S. Major Tropical Fruits Market Review 2018. Food and Agriculture Organization of the United NationS (FAO). Available online: http://www.fao.org/3/ca5692en/ca5692en.pdf (accessed on 5 December 2022).
- Baruwa, O.I. Profitability and constraints of pineapple production in Osun State, Nigeria. J. Hortic. Res. 2013, 21, 59–64. [Google Scholar] [CrossRef]
- Hossain, M.F.; Akhtar, S.; Anwar, M. Nutritional value and medicinal benefits of pineapple. Int. J. Nutr. Food Sci. 2015, 4, 84–88. [Google Scholar] [CrossRef]
- Sodjinou, M.K.; Assouma-Imorou, A.; Olounlade, A.O. Technical efficiency of pineapple production and challenges in Southern Benin. Afr. J. Agric. Res. 2022, 18, 522–534. [Google Scholar]
- Umi, H.N.; Tricahya, R.A.; Farid, A.M. Performance analysis of drip and sprinkler irrigation on pineapple cultivation. IOP Conf. Ser. Earth Environ. Sci. 2020, 451, 012034. [Google Scholar]
- Joy, P.P. Benefits and Uses of Pineapple; Pineapple Research Station (Kerala Agricultural University): Vazhakulam, India, 2010. [Google Scholar]
- Kader, A.; Hossain, F.M.; Islam, M.M.; Kabir, G.; Sarkar, S.K.; Absar, N.A. Comparative analysis on the nutritional contents of two varieties of pineapple of Chittagong region. Chittagong Univ. J. Biol. Sci. 2010, 5, 105–112. [Google Scholar] [CrossRef]
- Williams, P.A.; Crespo, O.; Atkinson, C.J.; Essegbey, G.O. Impact of climate variability on pineapple production in Ghana. Agric. Food Secur. 2017, 6, 26. [Google Scholar] [CrossRef]
- Hasan, S.; Hasan, S.S.; Saha, S.; Islam, M.R. Identify problems and suggest possible solutions for safe pineapple production in Madhupur tract. Eur. J. Agric. Food Sci. 2022, 4, 68–74. [Google Scholar] [CrossRef]
- Apipatpapha, T.; Ongkunaruk, P.; Chollakup, R. Pineapple leaf fiber supply chain analysis for the sustainability of community enterprise: A case study in Thailand. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2022; Volume 1074, p. 012032. [Google Scholar]
- Hemung, B.O.; Sompholkrang, M.; Wongchai, A.; Chanshotikul, N.; Ueasin, N. A study of the potential of by-products from pineapple processing in Thailand. Int. J. Health Sci. 2022, 6, 12605–12615. [Google Scholar] [CrossRef]
- Py, C.; Lacoevilhe, J.J.; Teisson, C. Pineapple, Cultivation and Uses. Techniques Agricoles et Productions Tropicales; P. Maisonneuve Et Larose: Paris, France, 1987; p. 205. ISBN1 2-7068-0844-6. ISBN2 92-9028-058-1. [Google Scholar]
- Popluechai, S.; Onto, S.; Eungwanichayapant, P.D. Relationships between some Thai cultivars of pineapple (Ananas comosus) revealed by RAPD analysis. Songklanakarin J. Sci. Technol. 2007, 29, 1491–1497. [Google Scholar]
- International Trade Centre Trade Map: List of Exporters for Product. Pineapples in 2020 Trade Map. Available online: https://www.trademap.org (accessed on 10 December 2022).
- Jayawardena, R.S.; Hyde, K.D.; Damm, U.; Cai, L.; Liu, M.; Li, X.H.; Zhang, W.; Zhao, W.S.; Yan, J.Y. Notes on currently accepted species of Colletotrichum. Mycosphere 2016, 7, 1192–1260. [Google Scholar] [CrossRef]
- Meetum, P.; Leksomboon, C.; Kanjanamaneesathian, M. First report of Colletotrichum aenigma and C. siamense, the causal agents of anthracnose disease of dragon fruit in Thailand. J. Plant Pathol. 2015, 97, 402. [Google Scholar]
- Sangpueak, R.; Phansak, P.; Buensanteai, N. Morphological and molecular identification of Colletotrichum species associated with cassava anthracnose in Thailand. J. Phytopathol. 2018, 166, 129–142. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Jayawardena, R.S.; Wei, D.P.; Huanraluek, N.; Abeywickrama, P.D.; Jeewon, R.; Monkai, J.; Hyde, K.D. Multigene phylogenetic characterissation of Colletotrichum artocarpicola sp. nov. from Artocarpus heterophyllus in northern Thailand. Phytotaxa 2019, 418, 273–286. [Google Scholar] [CrossRef]
- Phoulivong, S.; McKenzie, E.H.C.; Hyde, K.D. Cross infection of Colletotrichum species; a case study with tropical fruits. Curr. Res. Environ. Appl. Mycol. 2012, 2, 99–111. [Google Scholar] [CrossRef]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef]
- Wang, Q.H.; Fan, K.; Li, D.W.; Han, C.M.; Qu, Y.Y.; Qi, Y.K.; Wu, X.Q. Identification, virulence and fungicide sensitivity of Colletotrichum gloeosporioides s.s. responsible for walnut anthracnose disease in China. Plant Dis. 2020, 104, 1358–1368. [Google Scholar] [CrossRef]
- Noor, N.M.; Zakaria, L. Identification and characterization of Colletotrichum spp. associated with chili anthracnose in peninsular Malaysia. Eur. J. Plant Pathol. 2018, 151, 961–973. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Chen, J.; Dissanayake, A.J.; Doilom, M.; Hongsanan, S.; Jayawardena, R.S.; Jeewon, R.; Perera, R.H.; Thongbai, B.; et al. Thailand’s amazing diversity: Up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 2018, 93, 215–239. [Google Scholar] [CrossRef]
- Damm, U.; Sato, T.; Alizadeh, A.; Groenewald, J.Z.; Crous, P.W. The Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Stud. Mycol. 2019, 92, 1–46. [Google Scholar] [CrossRef]
- Chai, A.L.; Zhao, Q.; Li, B.J.; Sinsiri, W. First report of anthracnose caused by Colletotrichum brevisporum on gac (Momordica cochinchinensis) in Thailand. Plant Dis. 2018, 102, 2378. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.W.; Liu, F.; Duan, W.J.; Cai, L. Colletotrichum aracearum and C. camelliae-japonicae, two holomorphic new species from China and Japan. Mycosphere 2016, 7, 1111–1123. [Google Scholar] [CrossRef]
- Ma, X.; Nontachaiyapoom, S.; Jayawardena, R.S.; Hyde, K.D.; Gentekaki, E.; Zhou, S.; Qian, Y.; Wen, T.; Kang, J. Endophytic Colletotrichum species from Dendrobium spp. in China and northern Thailand. MycoKeys 2018, 43, 23–57. [Google Scholar] [CrossRef] [PubMed]
- Phoulivong, S.; Cai, L.; Parinn, N.; Chen, H.; Abd-Elsalam, K.A.; Chukeatirote, E.; Hyde, K.D. A new species of Colletotrichum from Cordyline fruticosa and Eugenia javanica causing anthracnose disease. Mycotaxon 2010, 114, 247–257. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Huang, J.K.; Jin, B.C.; Yan, J.Y.; Li, X.H.; Hyde, K.D.; Bahkali, A.H.; Yin, S.L.; Zhang, G.Z. An account of Colletotrichum species associated with strawberry anthracnose in China based on morphology and molecular data. Mycosphere 2016, 7, 1147–1163. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Udayanga, D.; Cai, L.; Chukeatirote, E.; Hyde, K.D. Endophytic Colletotrichum from tropical grasses with a new species Colletotrichum endophytica. Fungal Divers. 2013, 61, 107–115. [Google Scholar] [CrossRef]
- Liu, F.; Weir, B.S.; Damm, U.; Crous, P.W.; Wang, Y.; Liu, B.; Wang, M.; Zhang, M.; Cai, L. Unravelling Colletotrichum species associated with Camellia: Employing ApMat and GS loci to resolve species in the C. gloeosporioides complex. Persoonia 2015, 35, 63–86. [Google Scholar] [CrossRef]
- Sharma, G.; Shenoy, B.D. Colletotrichum fructicola and C. siamense are involved in chili anthracnose in India. Arch. Phytopathol. 2014, 47, 1179–1194. [Google Scholar] [CrossRef]
- Diao, Y.Z.; Zhang, C.; Liu, F.; Wang, W.Z.; Liu, L.; Cai, L.; Liu, X.L. Colletotrichum species causing anthracnose disease of chili in China. Persoonia 2017, 38, 20–37. [Google Scholar] [CrossRef]
- De Silva, D.D.; Groenewald, J.Z.; Crous, P.W.; Ades, P.K.; Nasruddin, A.; Mongkolporn, O.; Taylor, P.W.J. Identification, prevalence and pathogenicity of Colletotrichum species causing anthracnose of Capsicum annuum in Asia. IMA Fungus 2019, 10, 8. [Google Scholar] [CrossRef]
- Suwannarat, S.; Steinkellner, S.; Songkumarn, P.; Sangchote, S. Diversity of Colletotrichum spp. isolated from chili pepper fruit exhibiting symptoms of anthracnose in Thailand. Mycol. Progr. 2017, 16, 677–686. [Google Scholar] [CrossRef]
- Sharma, G.; Maymon, M.; Freeman, S. Epidemiology, pathology and identification of Colletotrichum including a novel species associated with avocado (Persea americana) anthracnose in Israel. Sci. Rep. 2017, 7, 15839. [Google Scholar] [CrossRef]
- Bragard, C.; Dehnen-Schmutz, K.; Di Serio, F.; Gonthier, P.; Jacques, M.A.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P. Pest categorisation of Colletotrichum Fruct. EFSA J. 2021, 19, e06803. [Google Scholar]
- Prihastuti, H.; Cai, L.; Chen, H.; McKenzie, E.H.; Hyde, K.D. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 2009, 39, 89–109. [Google Scholar]
- Jayawardena, R.S.; Hyde, K.D.; Chen, Y.J.; Papp, V.; Palla, B.; Papp, D.; Bhunjun, C.S.; Hurdeal, V.G.; Senwanna, C.; Manawasinghe, I.S.; et al. One stop shop IV: Taxonomic update with molecular phylogeny for important phytopathogenic genera. Fungal Divers. 2020, 103, 87–218. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Bhunjun, C.S.; Hyde, K.D.; Gentekaki, E.; Itthayakorn, P. Colletotrichum: Lifestyles, biology, morpho-species, species complexes and accepted species. Mycosphere 2021, 12, 519–669. [Google Scholar] [CrossRef]
- Liu, F.; Ma, Z.Y.; Hou, L.W.; Diao, Y.Z.; Wu, W.P.; Damm, U.; Song, S.; Cai, L. Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud. Mycol. 2022, 101, 1–56. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; de Farias, A.R.; Bhunjun, C.S.; Ferdinandez, H.S.; Manamgoda, D.S.; Udayanga, D.; Herath, I.S.; Thambugala, K.M.; Manawasinghe, I.S.; et al. What is a species in fungal plant pathogens? Fungal Divers. 2021, 109, 239–266. [Google Scholar] [CrossRef]
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum-current status and future directions. Stud. Mycol. 2012, 73, 181–213. [Google Scholar] [CrossRef]
- Farr, D.F.; Rossman, A.Y. Fungal Databases; Systematic Mycology Microbiology Laboratory, USDA-ARS: Beltsville, MD, USA, 2022; Volume 12. [Google Scholar]
- Garud, A.B. An anthracnose disease of pineapple in India. Plant Dis. Report. 1968, 52, 436–437. [Google Scholar]
- Johnston, A. A supplement to a host list of plant diseases in Malaya. Mycol. Pap. 1960, 77, 1–30. [Google Scholar]
- Chen, Y.; Fu, D.; Wang, W.; Gleason, M.L.; Zhang, R.; Liang, X.; Sun, G. Diversity of Colletotrichum species causing apple bitter rot and Glomerella leaf spot in China. J. Fungi 2022, 8, 740. [Google Scholar] [CrossRef] [PubMed]
- Bhunjun, C.S.; Niskanen, T.; Suwannarach, N.; Wannathes, N.; Chen, Y.J.; McKenzie, E.H.; Maharachchikumbura, S.S.; Buyck, B.; Zhao, C.L.; Fan, Y.G.; et al. The numbers of fungi: Are the most speciose genera truly diverse? Fungal Divers. 2022, 114, 387–462. [Google Scholar] [CrossRef]
- Castro, J.F.; Millas, P.; Cisterna-Oyarce, V.; Carrasco, J.; Santelices, C.; Muñoz-Reyes, V.; Guerra, M.; Barra-Bucarei, L.; France, A. First report of Colletotrichum fioriniae causing anthracnose fruit rot on Vaccinium corymbosum in Chile. Plant Dis. 2022. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, H. First report of Colletotrichum aeschynomenes causing anthracnose on Camellia oleifera in China. For. Pathol. 2022, 52, e12770. [Google Scholar] [CrossRef]
- Nigar, Q.; Cadle-Davidson, L.; Gadoury, D.M.; Hassan, M.U. First report of Colletotrichum fioriniae causing grapevine anthracnose in New York. Plant Dis. 2022, 107, 223. [Google Scholar] [CrossRef]
- Pacheco-Esteva, M.C.; Soto-Castro, D.; Vásquez-López, A.; Lima, N.B.; Tovar-Pedraza, J.M. First report of Colletotrichum chrysophilum causing papaya anthracnose in Mexico. Plant Dis. 2022, 106, 3213. [Google Scholar] [CrossRef]
- Sun, W.; Lei, T.; Yuan, H.; Chen, S. First report of anthracnose caused by Colletotrichum kahawae and Colletotrichum horri on tea-oil tree in China. Plant Dis. 2022. [Google Scholar] [CrossRef]
- Wang, Y.; Jia, W.; Zhu, X.; Mi, Z.; Wang, E.; Chang, G.; Yan, S.; Wang, X.; Kong, D.; He, S. First report of Colletotrichum aenigma causing anthracnose of tree peony in China. Plant Dis. 2022. [Google Scholar] [CrossRef]
- Yu, J.; Li, Q.; Zhang, W.; Bai, P.; Huo, J.; Gao, W.; Liu, Y. First report of Colletotrichum truncatum causing anthracnose of green foxtail (Setaria viridis) in China. Plant Dis. 2022. [Google Scholar] [CrossRef]
- Zhang, N.; Yuan, S.; Zhu, Y.; Wang, B.; Xu, Y.; Yang, W. First report of anthracnose on chili Pepper caused by Colletotrichum sojae in Hebei Province, China. Plant Dis. 2022. [Google Scholar] [CrossRef]
- Zhu, Z.; Dong, Z.; Mo, R.; Zhang, C.; Zuo, Y.; Yu, C.; Hu, X. First report of Colletotrichum aenigma causing anthracnose on mulberry leaves in China. Plant Dis. 2023. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Templeton, M.D.; Rikkerink, E.H.; Solon, S.L.; Crowhurst, R.N. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene 1992, 122, 225–230. [Google Scholar] [CrossRef]
- Woudenberg, J.H.; Aveskamp, M.M.; De Gruyter, J.; Spiers, A.G.; Crous, P.W. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Pers. Mol. Phylogeny Evol. Fungi 2009, 22, 56–62. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Swofford, D.L.; Sullivan, J. Phylogeny inference based on parsimony and other methods using PAUP. In The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny, Cáp; Cambridge University Press: Cambridge, UK, 2003; Volume 7, pp. 160–206. [Google Scholar]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J. FigTree Version 1.4.0.2012. Available online: http://Tree.bio.ed.ac.uk/software/figtree (accessed on 12 February 2017).
- Bhunjun, C.S.; Phillips, A.J.; Jayawardena, R.S.; Promputtha, I.; Hyde, K.D. Importance of molecular data to identify fungal plant pathogens and guidelines for pathogenicity testing based on Koch’s Postulates. Pathogens 2021, 10, 1096. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).