First Report on the Genetic Diversity of Populations of Gossypium barbadense L. and Gossypium hirsutun L. in the Amazonian Native Communities, Cusco-Peru
Abstract
1. Introduction
2. Results
2.1. Distribution of G. barbadense L.
2.2. Distribution of G. barbadense L. var. Brasiliensis
2.3. Distribution of the Introduced Species G. hirsutum L.
2.4. Unidentified Samples of Gossypium spp.
2.5. Quantitative Characterization of the Collected Samples
2.6. Correlations of the Quantitative Descriptors Obtained among the Groups Derived from the Collected Samples
2.7. Qualitative Characterization of the Collected Samples
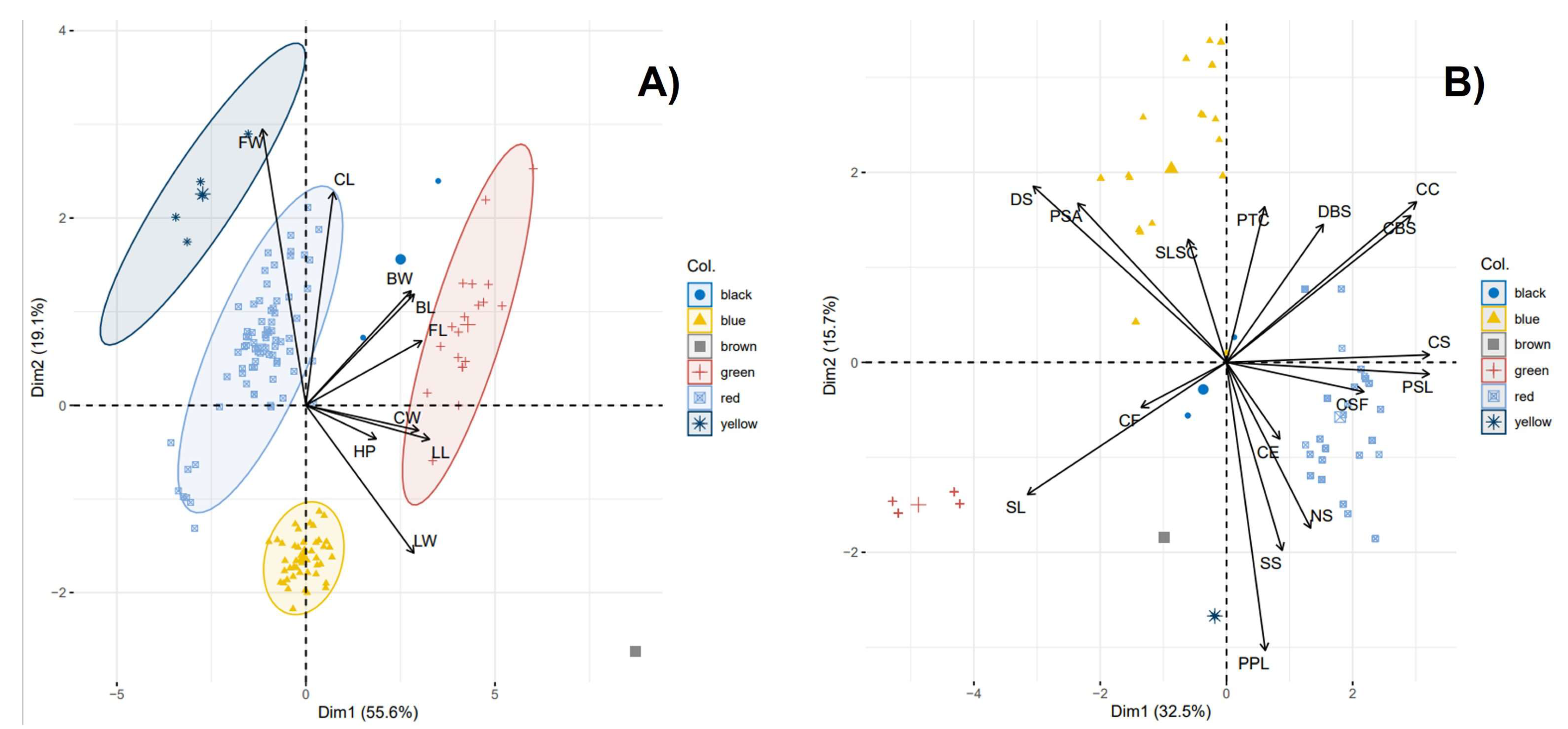
2.8. Principal Component Analysis (PCA) Using the Qualitative and Quantitative Descriptors
3. Discussion
4. Materials and Methods
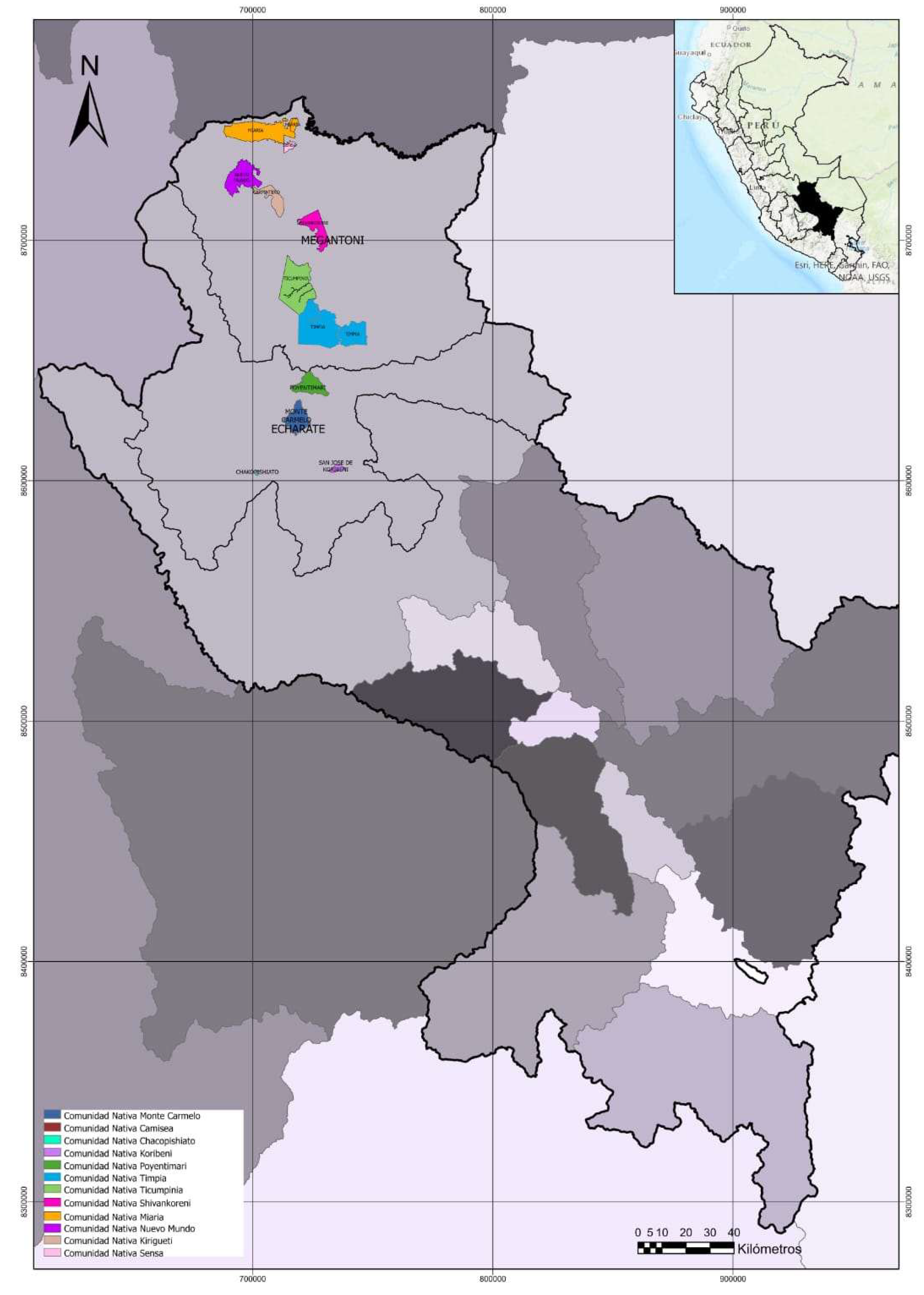
4.1. Study Site and Sampling Method
4.2. Qualitative Descriptors Evaluated
4.3. Quantitative Descriptors Assessed
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López, A.; López, S.; Gil, E.; Caicedo, E.; Mendoza, E. Characterization of fruits; seeds and fibers of Gossypium barbadense “algodón Pardo”. Sciéndo 2018, 21, 301–304. [Google Scholar] [CrossRef]
- MINAM—Ministerio del Ambiente del Perú. Línea de Base de la Diversidad del Algodón Peruano con Fines de Bioseguridad; Government of Peru: Lima, Peru, 2020; p. 83.
- MINAM—Ministerio del Ambiente del Perú. Colecta, Elaboración de Mapas de Distribución y Estudio Socioeconómico de la Diversidad del Algodón Nativo; Government of Peru: Lima, Peru, 2014; p. 83.
- Rojas, I.; Cuzquen, C.; Delgado, G. In vitro clonal propagation rooting of native cotton (Gossypium barbadense) cuttings. Rev. Acta Agron. 2014, 62, 312–320. [Google Scholar]
- Yuan, D.; Grover, C.E.; Hu, G.; Pan, M.; Miller, E.R.; Conover, J.L.; Hunt, S.P.; Udall, J.A.; Wendel, J.F. Parallel and Intertwining Threads of Domestication in Allopolyploid Cotton. Adv. Sci. 2021, 8, 2003634. [Google Scholar] [CrossRef] [PubMed]
- Lazo, J. Articles. Instituto Peruano del Algodón. 1990. Available online: https://www.ipaperu.org/descarga/ARTICULOS.pdf (accessed on 12 April 2018).
- Brubaker, C.L.; Wendel, J.F. Reevaluating the origin of domesticated cotton (Gossypium hirsutum, Malvaceae) using nuclear restriction fragment length polymorphisms (RFLPs). Am. J. Bot. 1994, 81, 1309–1326. [Google Scholar] [CrossRef]
- Stewart, J.M.; Oosterhuis, D.; Heitholt, J.J.; Mauney, J.R. Physiology of Cotton; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009; p. 562. [Google Scholar]
- Briuolo, A.L.W. Diversidad Genética y Conservación de Gossypium hirsutum Silvestre y Cultivado en México. Ph.D. Thesis, Universidad Nacional Autónoma de México, México City, México, 2013; p. 114. [Google Scholar]
- D’eeckenbrugge, G.C.; Lacape, J.M. Distribution and differentiation of wild, feral, and cultivated populations of perennial upland cotton (Gossypium hirsutum L.) in Mesoamerica and the Caribbean. PLoS ONE 2014, 9, e107458. [Google Scholar] [CrossRef]
- FAOSTAT. Available online: http://faostat.fao.org/ (accessed on 23 September 2020).
- Westengen, O.T.; Huaman, Z.; Heun, M. Genetic diversity and geographic pattern in early South American cotton domestication. Theor. Appl. Genet. 2005, 110, 392–402. [Google Scholar] [CrossRef]
- Chanco, M.; León, B.; Sánchez, I. Malvaceae endémicas del Perú. Rev. Peru. Biol. 2006, 13, 413s–425s. [Google Scholar] [CrossRef]
- López, S.; Gil, A. Phenology of Gossypium raimondii Ulbrich “native cotton” of green fiber. Sci. Agropecu. 2017, 8, 267–271. [Google Scholar] [CrossRef]
- López Medina, S.E.; Mostacero León, J.; Quijano Jara, C.H.; Gil Rivero, A.E.; Rabanal Che León, M. Caracterización Del Fruto; Semilla Y Fibra De algodón Silvestre (Gossypium raimondii). Cienc. Tecnol. Agropecu. 2019, 21, 1–8. [Google Scholar] [CrossRef]
- Smith, C.W.; Cothren, J.T. Cotton: Origin, History, Technology, and Production, 4th ed.; John Wiley & Sons: Hoboken, NJ, USA, 1999; 840p. [Google Scholar]
- Gross, B.L.; Strasburg, J.L. Cotton domestication: Dramatic changes in a single cell. BMC Biol. 2010, 8, 137. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Yu, S. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Huang, X. Pangenome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef]
- Li, Y.; Suontama, M.; Burdon, R.D.; Dungey, H.S. Genotype by environment interactions in forest tree breeding: Review of methodology and perspectives on research and application. Tree Genet. Genomes 2017, 13, 118. [Google Scholar] [CrossRef]
- Peixoto, M.A.; Malikouski, R.G.; Nascimento, E.F.D.; Schuster, A.; Farias, F.J.C.; Carvalho, L.P.; Teodoro, P.E.; Bhering, L.L. Genotype plus genotype by-environment interaction biplot and genetic diversity analyses on multi-environment trials data of yield and technological traits of cotton cultivars. Ciência Rural 2022, 52, e20201054. [Google Scholar] [CrossRef]
- Hinze, L.L.; Gazave, E.; Gore, M.A.; Fang, D.D.; Scheffler, B.E.; Yu, J.Z.; Jones, D.C.; Frelichowski, J.; Percy, R.G. Genetic diversity of the two commercial tetraploid cotton species in the Gossypium diversity reference set. J. Hered. 2016, 107, 274–286. [Google Scholar] [CrossRef]
- Wang, C.; Tang, Y.; Chen, J. Plant phenological synchrony increases under rapid within-spring warming. Sci. Rep. 2016, 6, 25460. [Google Scholar] [CrossRef]
- Lazo, J. Evolución del algodón Gossypium barbadense L. en el Perú y en el continente. Mundo Text. 2012, 115, 20–23. [Google Scholar]
- Ozyigit, I.I. In vitro shoot development from three different nodes of cotton (Gossypium hirsutum L.). Not. Bot. Horti Agrobot. 2009, 37, 74–78. [Google Scholar] [CrossRef]
- Ahmed, H.A.; Hajyzadeh, M.; Barpete, S.; Ozcan, S. In vitro plant regeneration of Iraqi cotton (Gossypium hirsutum L.) cultivars through embryonic axis. J. Biotechnol. Res. Cent. 2014, 8, 90–94. [Google Scholar] [CrossRef]
- Delgado-Paredes, G.E.; Vásquez-Díaz, C.; Esquerre-Ibáñez, B.; Huamán-Mera, A.; Rojas-Idrogo, C. Germplasm collection, in vitro clonal propagation, seed viability and vulnerability of ancient peruvian cotton (Gossypium barbadense L.). Pak. J. Bot. 2021, 53, 1259–1270. [Google Scholar] [CrossRef]
- Li, L.; Zhang, C.; Huang, J.; Liu, Q.; Wei, H.; Wang, H.; Yu, S. Genomic analyses reveal the genetic basis of early maturity and identification of loci and candidate genes in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 2021, 19, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F.; Grover, C.E. Taxonomy and evolution of the cotton genus; Gossypium. In Cotton, 2nd ed.; Fang, D.D., Percy, R.G., Eds.; American Society of Agronomy Inc.: Madison, WI, USA, 2015; pp. 25–44. [Google Scholar]
- MINAM—Ministerio del Ambiente del Perú. Elaboración del Mapa, Análisis Socioeconómico, y de Organismos y Microorganismos de aire y Suelo. Lineamientos para la Conservación de la Diversidad Genética de la Especie. Informe Final de la Consultoría; Ministerio del Ambiente—Dirección General de Diversidad Biológica: Lima, Peru, 2017; p. 95.
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef] [PubMed]
- Otzen, T.; Manterola, C. Sampling Techniques on a Population Study. Int. J. Morphol. 2017, 35, 227–232. [Google Scholar] [CrossRef]
- Manco Céspedes, E.I.; Chanamé Upay, J.; Arévalo Garazatúa, G.M.; Mamani Huarachi, W.V.; Hinostroza García, L.D.R.; Garay Duran, N.H.; Lindo Seminario, D.E.; Vasquez Oroya, J.; García Serquén, A.L. Descriptores para Algodón Peruano (Gossypium barbadense L.); Instituto Nacional de Innovación Agraria: La Molina, Peru, 2022.
- Bhering, L.L. Rbio: A Tool for Biometric and Statistical Analysis Using the R Platform. Crop Breed. Appl. Biotechnol. 2017, 17, 187–190. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]





| District | Community | G. barbadense L. | G. barbadense L. var. brasilensis | G. hirsutum L. | Unknown Groups | Total General | Altitude (m) | ||
|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | |||||||
| Megantoni | Camisea | 3 | 3 | 400 | |||||
| Kirigueti | 6 | 4 | 10 | 371–374 | |||||
| Miaria | 1 | 2 | 3 | 334–336 | |||||
| Nuevo Mundo | 6 | 4 | 10 | 364 | |||||
| Sensa | 4 | 2 | 6 | 425–426 | |||||
| Shivankoreni | 7 | 10 | 17 | 397–405 | |||||
| Ticumpinia | 1 | 2 | 3 | 377–382 | |||||
| Timpia | 10 | 5 | 15 | 421–447 | |||||
| Echarati | Chacopishiato | 16 | 20 | 36 | 1086–1137 | ||||
| Koribeni | 6 | 3 | 1 | 10 | 677–746 | ||||
| Monte Carmelo | 3 | 3 | 6 | 428 | |||||
| Poyentimari | 13 | 12 | 3 | 28 | 497–506 | ||||
| Total | 44 | 75 | 19 | 3 | 1 | 5 | 147 | ||
| Descriptors | F Value | Significance | CV (%) |
|---|---|---|---|
| Flower-length (FL, cm) | 73.57 | <0.001 | 2.29 |
| Flower-width (FW, cm) | 146.992 | <0.001 | 4.58 |
| Bracts-length (BL, cm) | 44.08 | <0.001 | 3.14 |
| Bracts-width (BW, cm) | 375.79 | <0.001 | 3.76 |
| Capsule-length (CL, cm) | 53.94 | <0.001 | 3.34 |
| Capsule-width (CW, cm) | 54.77 | <0.001 | 5.07 |
| Height-plant (HP, m) | 12.40 | <0.001 | 3.6 |
| Leaves-length (LL, cm) | 2011.48 | <0.001 | 1.81 |
| Leaves-width (LW, cm) | 2960.74 | <0.001 | 1.28 |
| Gossypium Described | FL | FW | BL | BW | CL | CW | HP | LL | LW | |
|---|---|---|---|---|---|---|---|---|---|---|
| G. barbadense L. | 7.25 c | 3.89 d | 5.86 c | 4.45 e | 5.92 d | 3.04 b | 3.34 bc | 15.18 d | 18.81 c | |
| G. barbadense L. var. brasiliensis | 7.27 c | 4.89 ab | 5.93 c | 4.69 d | 6.04 c | 2.83 c | 3.31 cd | 13.84 e | 15.83 d | |
| G. hirsutum L. | 7.88 b | 4.44 c | 6.46 b | 6.18 b | 6.42 b | 3.40 a | 3.48 b | 19.46 b | 20.14 b | |
| Gossypium unknown | G1 | 7.72 b | 4.61 bc | 6.33 b | 6.98 a | 6.32 bc | 2.99 bc | 3.11 d | 18.48 c | 18.71 c |
| G2 | 8.50 a | 3.48 d | 6.94 a | 5.74 c | 5.84 cd | 3.56 a | 3.58 ab | 25.48 a | 29.80 a | |
| G3 | 6.90 d | 5.12 a | 5.74 c | 3.88 f | 7.30 a | 2.74 c | 3.18 cd | 12.74 f | 15.54 d | |
| Total | 7.35 | 4.52 | 5.98 | 4.84 | 6.10 | 3.33 | 2.97 | 15.11 | 17.42 | |
| Gossypium Described | CE 1 | PSL | CSF | CF | PSA | DS | NS | CS | |
| G. barbadense L. | 10 | 2 | 4 | 3 | 1 | 1 | 2 | 1 | |
| G. barbadense L. var. brasiliensis | 16 | 1 | 4 | 4 | 2 | 1 | 2 | 1 | |
| G. hirsutum L. | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | |
| Gossypium unknown | G1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| G2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| G3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Total | 19 | 2 | 5 | 5 | 2 | 2 | 4 | 4 | |
| Gossypium Described | SS | DB | CB | SLSC | CC | PTC | SL | PPL | |
| G. barbadense L. | 2 | 3 | 2 | 2 | 1 | 2 | 1 | 1 | |
| G. barbadense L. var. brasiliensis | 2 | 4 | 2 | 3 | 1 | 2 | 1 | 1 | |
| G. hirsutum L. | 2 | 1 | 2 | 2 | 1 | 2 | 1 | 1 | |
| Gossypium unknown | G1 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 1 |
| G2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| G3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Total | 3 | 4 | 6 | 3 | 4 | 2 | 2 | 4 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Aranibar, L.; Yucra, F.E.Y.; Aranibar, C.G.M.; Sáenz, M.C.; Gonzales, H.H.S.; Aguilera, J.G.; Álvarez, J.L.L.; Zuffo, A.M.; Steiner, F.; Ratke, R.F.; et al. First Report on the Genetic Diversity of Populations of Gossypium barbadense L. and Gossypium hirsutun L. in the Amazonian Native Communities, Cusco-Peru. Plants 2023, 12, 865. https://doi.org/10.3390/plants12040865
Morales-Aranibar L, Yucra FEY, Aranibar CGM, Sáenz MC, Gonzales HHS, Aguilera JG, Álvarez JLL, Zuffo AM, Steiner F, Ratke RF, et al. First Report on the Genetic Diversity of Populations of Gossypium barbadense L. and Gossypium hirsutun L. in the Amazonian Native Communities, Cusco-Peru. Plants. 2023; 12(4):865. https://doi.org/10.3390/plants12040865
Chicago/Turabian StyleMorales-Aranibar, Luis, Francisca Elena Yucra Yucra, Carlos Genaro Morales Aranibar, Manuel Canto Sáenz, Hebert Hernán Soto Gonzales, Jorge González Aguilera, Juan Luis Lazo Álvarez, Alan Mario Zuffo, Fabio Steiner, Rafael Felippe Ratke, and et al. 2023. "First Report on the Genetic Diversity of Populations of Gossypium barbadense L. and Gossypium hirsutun L. in the Amazonian Native Communities, Cusco-Peru" Plants 12, no. 4: 865. https://doi.org/10.3390/plants12040865
APA StyleMorales-Aranibar, L., Yucra, F. E. Y., Aranibar, C. G. M., Sáenz, M. C., Gonzales, H. H. S., Aguilera, J. G., Álvarez, J. L. L., Zuffo, A. M., Steiner, F., Ratke, R. F., & Teodoro, P. E. (2023). First Report on the Genetic Diversity of Populations of Gossypium barbadense L. and Gossypium hirsutun L. in the Amazonian Native Communities, Cusco-Peru. Plants, 12(4), 865. https://doi.org/10.3390/plants12040865













