Genetic Diversity and Population Structure of Common Bean (Phaseolus vulgaris L.) Landraces in the Lazio Region of Italy
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material
2.2. Morpho-Phenotypic Seed Analysis
2.3. Growth Habit
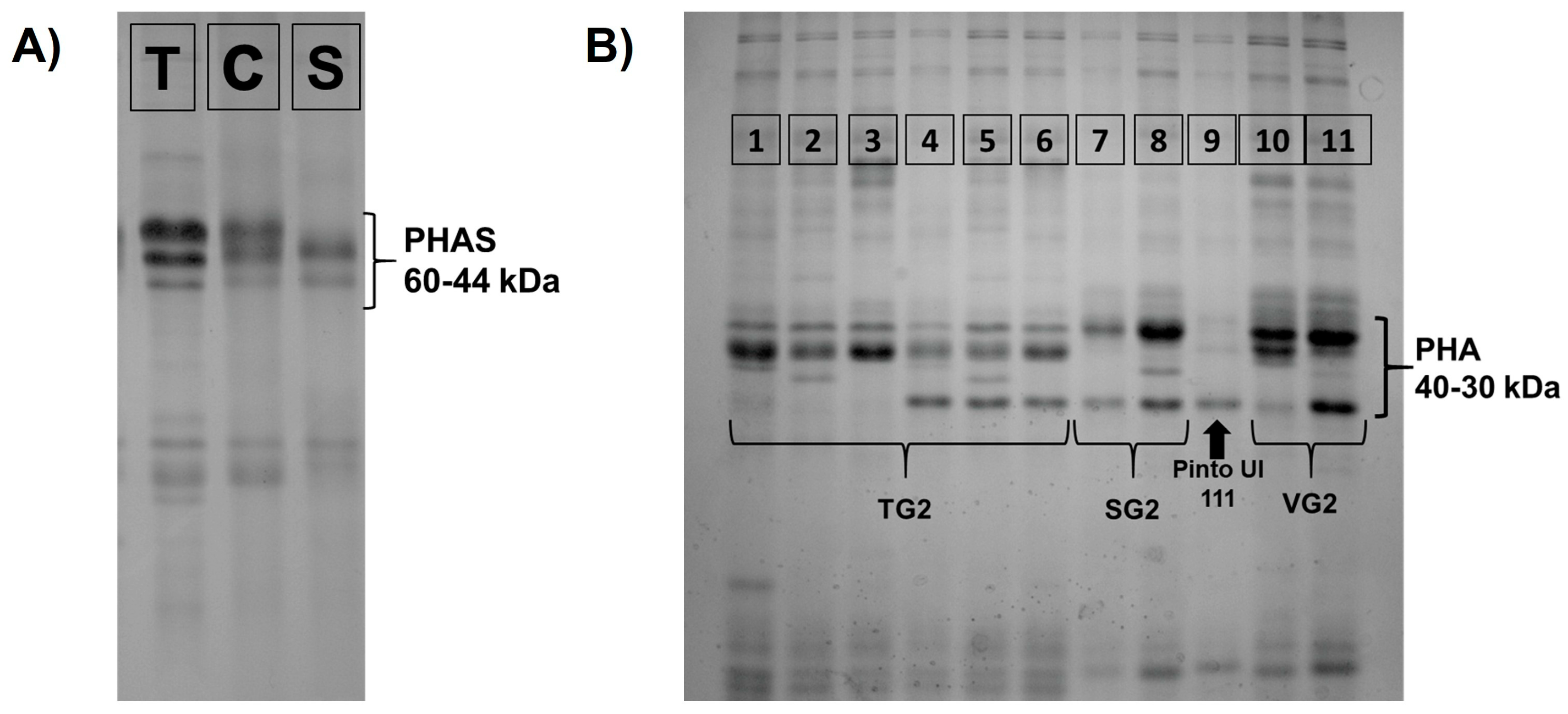
2.4. Phaseolin (PHAS) and Phytohemagglutinin (PHA) Analysis
2.5. DNA Isolation and Microsatellite Analysis
2.6. Data Analyses
3. Results
3.1. Seed Traits and Growth Habit
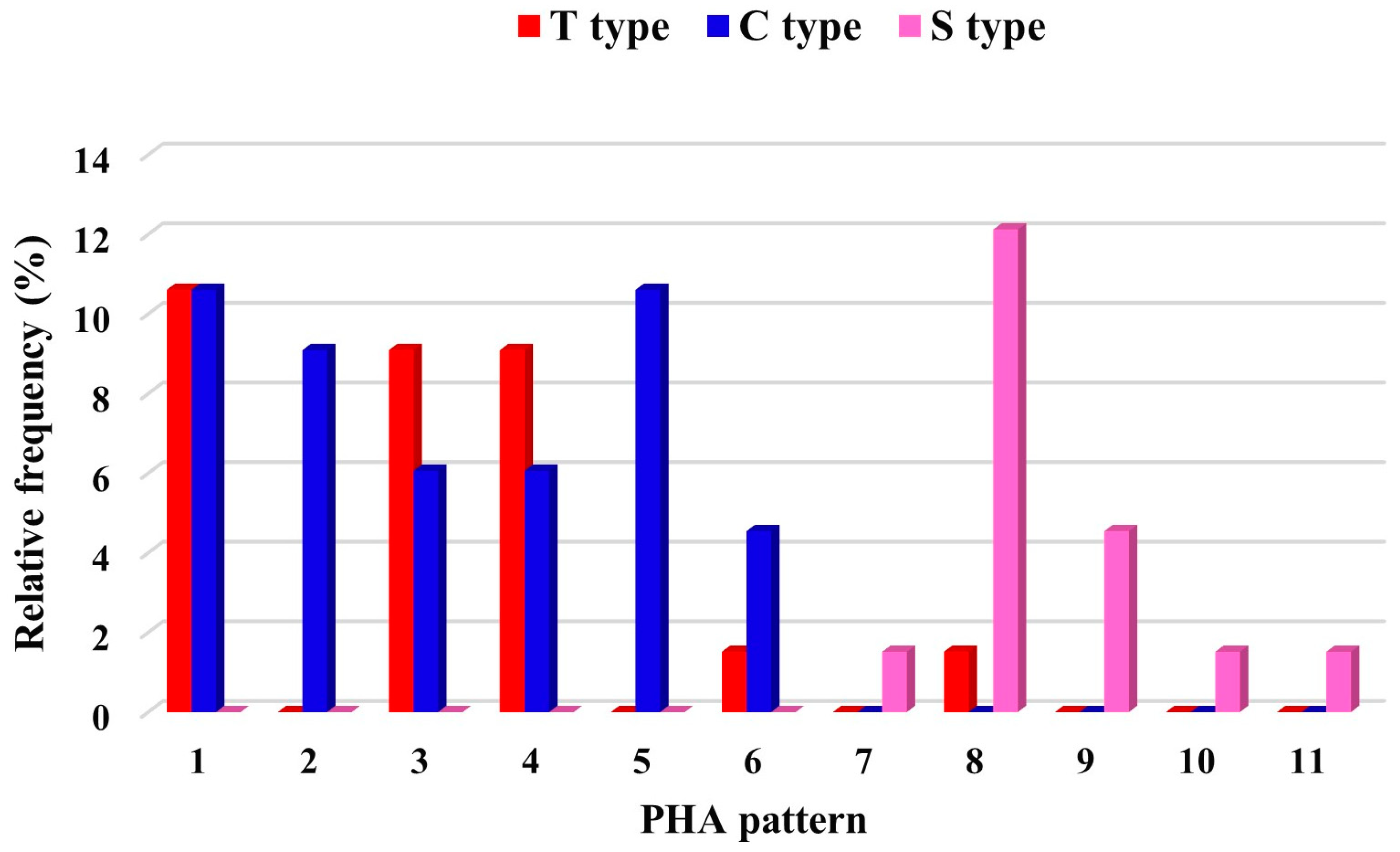
3.2. Phaseolin (PHAS) and Phytohemagglutinin (PHA) Variability
3.3. Relationship between PHAS and PHA Patterns, and PHAS Pattern and Morphological Data
3.4. Genetic Analysis by SSR Markers
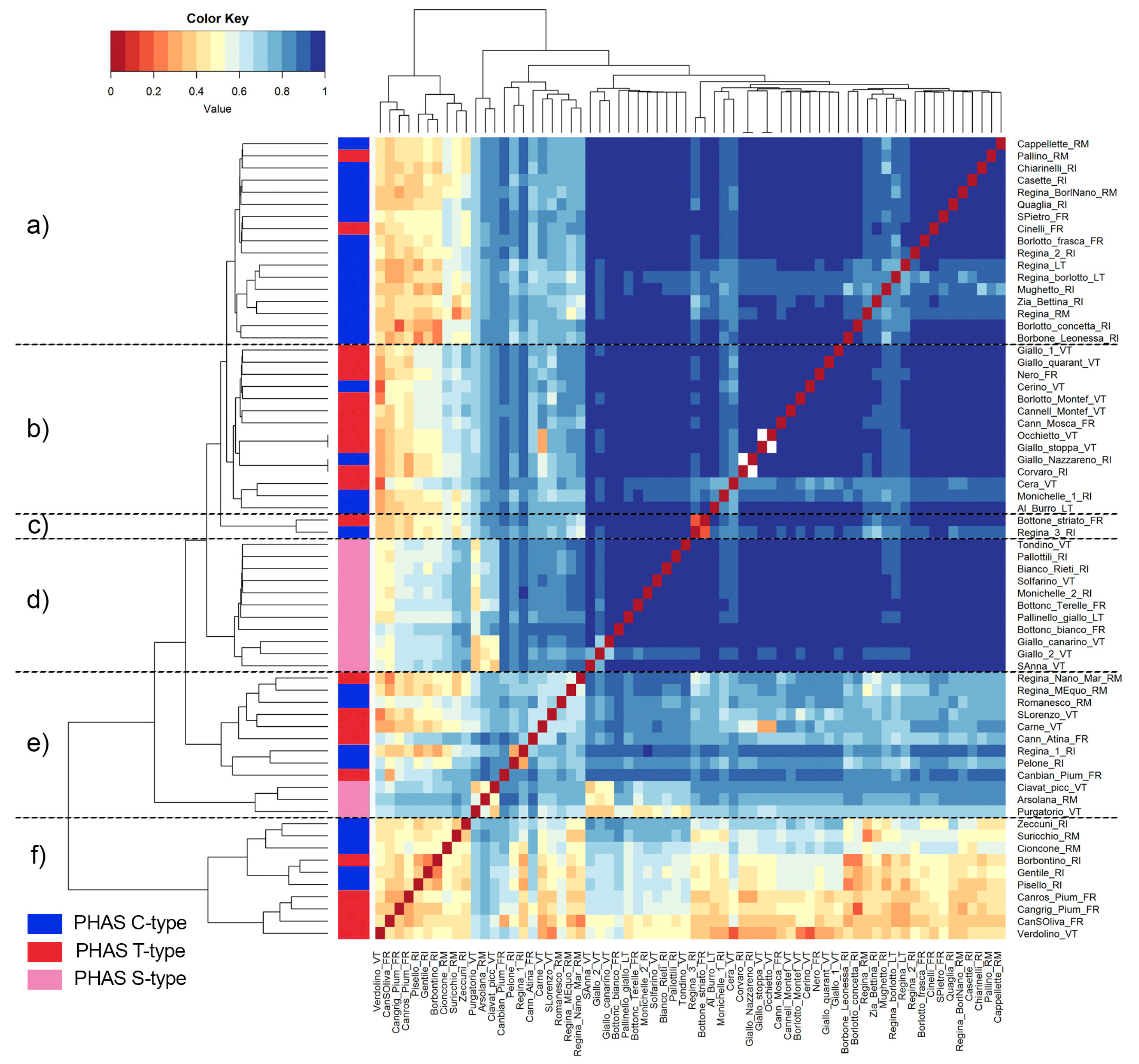
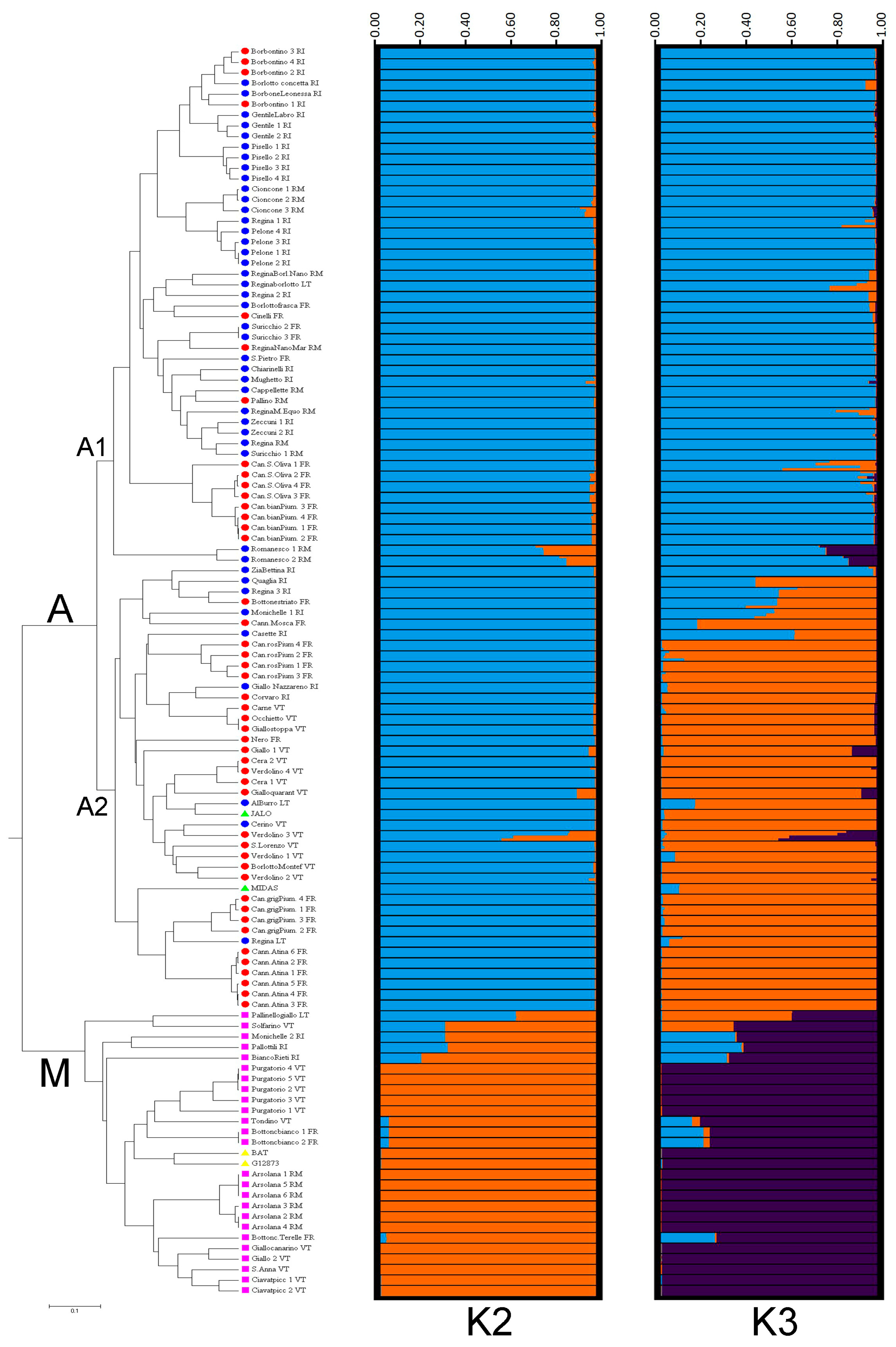
3.4.1. Genetic Diversity and Relationships among Landraces
3.4.2. Genetic Structure of Landraces
4. Discussion
4.1. Seed Morphological and Biochemical Traits of Lazio Germplasm
4.2. Molecular Characterization of Lazio Germplasm
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reynoso-Camacho, R.; Ramos-Gomez, M.; Loarca-Pina, G. Bioactive components in common beans (Phaseolus vulgaris L.). Adv. Agric. Food Biotechnol. 2006, 10, 217–236. [Google Scholar]
- Lioi, L.; Piergiovanni, A.R. European common bean. In Genetic and Genomic Resources of Grain Legume Improvement; Singh, M., Upadhyaya, H.D., Bisht, S., Eds.; Elsevier Inc.: London, UK, 2017; pp. 11–40. [Google Scholar] [CrossRef]
- Câmara, C.R.S.; Urrea, C.A.; Schlegel, V. Pinto beans (Phaseolus vulgaris L.) as a functional food: Implications on human health. Agriculture 2013, 3, 90–111. [Google Scholar] [CrossRef]
- Petry, N.; Boy, E.; Wirth, J.; Hurrell, R. The potential of the common bean (Phaseolus vulgaris) as a vehicle for iron biofortification. A review. Nutrients 2015, 7, 1144–1173. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 10 June 2022).
- ISTAT (Italian National Institute of Statistics) 2009. Available online: http://dati.istat.it/Index.aspx?DataSetCode=DCSP_COLTIVAZ&Lang=en (accessed on 29 August 2022).
- Rossi, M.; Bitocchi, E.; Bellucci, E.; Nanni, L.; Rau, D.; Attene, G.; Papa, R. Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L. Evol. Appl. 2009, 2, 504–522. [Google Scholar] [CrossRef] [PubMed]
- Bitocchi, E.; Nanni, L.; Bellucci, E.; Rossi, M.; Giardini, A.; Spagnoletti Zeuli, P.S.; Logozzo, G.; Stougaard, J.; McClean, P.; Attene, G.; et al. Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc. Natl. Acad. Sci. USA 2012, 109, E788–E796. [Google Scholar] [CrossRef]
- Bitocchi, E.; Rau, D.; Bellucci, E.; Rodriguez, M.; Murgia, M.L.; Gioia, T.; Santo, D.; Nanni, L.; Attene, G.; Papa, R. Beans (Phaseolus ssp.) as a model for understanding crop evolution. Front. Plant Sci. 2017, 8, 722. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J. On the origin of the common bean (Phaseolus vulgaris L.). Am. J. Plant Sci. 2013, 4, 1998–2000. [Google Scholar] [CrossRef]
- Mamidi, S.; Rossi, M.; Moghaddam, S.M.; Annam, D.; Lee, R.; Papa, R.; McClean, P. Demographic factors shaped diversity in the two gene pools of wild common bean Phaseolus vulgaris L. Heredity 2013, 110, 267–276. [Google Scholar] [CrossRef]
- Bitocchi, E.; Bellucci, E.; Giardini, A.; Rau, D.; Rodriguez, M.; Biagetti, E.; Santilocchi, R.; Spagnoletti Zeuli, P.; Gioia, T.; Logozzo, G.; et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2013, 197, 300–313. [Google Scholar] [CrossRef]
- Debouck, D.G.; Toro, O.; Paredes, O.M.; Johnson, W.C.; Gepts, P. Genetic diversity and ecological distribution of Phaseolus vulgaris in northwestern South America. Econ. Bot. 1993, 47, 408–423. [Google Scholar] [CrossRef]
- Kami, J.; Velásquez, V.B.; Debouck, D.G.; Gepts, P. Identification of presumed ancestral DNA sequences of phaseolin in Phaseolus vulgaris. Proc. Natl. Acad. Sci. USA 1995, 92, 1101–1104. [Google Scholar] [CrossRef]
- Gepts, P.; Debouck, D. Origin, domestication, and evolution of the common bean (Phaseolus vulgaris L.). In Common Beans: Research for Crop Improvement; van Schoonhoven, A., Voysest, O., Eds.; C.A.B. Int.: Wallingford, UK; CIAT: Cali, Colombia, 1991; pp. 7–53. [Google Scholar]
- Gepts, P. A Middle American and an Andean common bean gene pool. In Genetic Resources of Phaseolus Beans. Current Plant Science and Biotechnology in Agriculture; Gepts, P., Ed.; Springer: Dordrecht, The Netherlands, 1988; p. 6. [Google Scholar] [CrossRef]
- Singh, S.P.; Nodari, R.; Gepts, P. Genetic diversity in cultivated common bean: I. allozymes. Crop. Sci. 1991, 31, 19–23. [Google Scholar] [CrossRef]
- Šustar-Vozlič, J.; Maras, M.; Javornik, B.; Meglič, V. Genetic diversity and origin of slovene common bean (Phaseolus vulgaris L.) germplasm as revealed by AFLP markers and phaseolin analysis. J. Am. Soc. Hortic. Sci. 2006, 131, 242–249. [Google Scholar] [CrossRef]
- Desiderio, F.; Bitocchi, E.; Bellucci, E.; Rau, D.; Rodriguez, M.; Attene, G.; Papa, R.; Nanni, L. Chloroplast microsatellite diversity in Phaseolus vulgaris. Front. Plant Sci. 2013, 3, 312. [Google Scholar] [CrossRef] [PubMed]
- Blair, M.W.; Iriarte, G.; Beebe, S. QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean 9 wild common bean (Phaseolus vulgaris L.) cross. Theor. Appl. Genet. 2006, 112, 1149–1163. [Google Scholar] [CrossRef]
- Kwak, M.; Gepts, P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor. Appl. Genet. 2009, 118, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.; Rau, D.; Bitocchi, E.; Bellucci, E.; Biagetti, E.; Carboni, A.; Gepts, P.; Nanni, L.; Papa, R.; Attene, G. Landscape genetics, adaptive diversity, and population structure in Phaseolus vulgaris. New Phytol. 2016, 209, 1781–1794. [Google Scholar] [CrossRef]
- Lioi, L.; Zuluaga, L.D.; Pavan, S.; Sonnante, G. Genotyping-by-Sequencing Reveals Molecular Genetic Diversity in Italian Common Bean Landraces. Diversity 2019, 11, 154. [Google Scholar] [CrossRef]
- Angioi, S.A.; Rau, D.; Attene, G.; Nanni, L.; Bellucci, E.; Logozzo, G.; Negri, V.; Spagnoletti Zeuli, P.L.; Papa, R. Beans in Europe: Origin and structure of the European landraces of Phaseolus vulgaris L. Theor. Appl. Genet. 2010, 121, 829–843. [Google Scholar] [CrossRef]
- Blair, M.W.; Díaz, L.M.; Buendía, H.F.; Duque, M.C. Genetic diversity, seed size associations and population structure of a core collection of common beans (Phaseolus vulgaris L.). Theor. Appl. Genet. 2009, 119, 955–972. [Google Scholar] [CrossRef]
- Zhang, X.; Blair, M.W.; Wang, S. Genetic diversity in Chinese common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeats markers. Theor. Appl. Genet. 2008, 117, 629–640. [Google Scholar] [CrossRef]
- Raggi, L.; Tiranti, B.; Negri, V. Italian common bean landraces: Diversity and population structure. Genet. Resour. Crop Evol. 2013, 60, 1515–1530. [Google Scholar] [CrossRef]
- Maras, M.; Pipan, B.; Sustar-Vozlic, J.; Todorović, V.; Ðurić, G.; Vasić, M.; Kratovalieva, S.; Ibusoska, A.; Agi’c, R.; Matotan, Z.; et al. Examination of genetic diversity of common bean from the Western Balkans. J. Am. Soc. Hort. Sci. 2015, 140, 308–316. [Google Scholar] [CrossRef]
- Carović-Stanko, K.; Liber, Z.; Vidak, M.; Barešić, A.; Grdiša, M.; Lazarević, B.; Šatović, Z. Genetic diversity of Croatian common bean landraces. Front. Plant Sci. 2017, 8, 604. [Google Scholar] [CrossRef]
- Leitão, S.T.; Dinis, M.; Veloso, M.M.; Šatović, Z.; Vaz Patto, M.C. Establishing the bases for introducing the unexplored portuguese common bean germplasm into the breeding world. Front. Plant Sci. 2017, 8, 1296. [Google Scholar] [CrossRef]
- Šajgalík, M.; Ondreičková, K.; Hauptvogel, P.; Mihálik, D.; Glasa, M.; Kraic, J. Higher effectiveness of new common bean (Phaseolus vulgaris L.) germplasm acquisition by collecting expeditions associated with molecular analyses. Sustainability 2019, 11, 5270. [Google Scholar] [CrossRef]
- Gepts, P.; Kmiecik, K.; Pereira, P.; Bliss, F.A. Dissemination pathways of common bean (Phaseolus vulgaris, Fabaceae) deduced from phaseolin electrophoresis variability. I. The Americas. Econ. Bot. 1988, 42, 86–104. [Google Scholar] [CrossRef]
- Papa, R.; Nanni, L.; Sicard, D.; Rau, D.; Attene, G. The evolution of genetic diversity in Phaseolus vulgaris L. In Darwin’s Harvest: New Approaches to the Origins, Evolution and Conservation of Crops. Darwin’s Harvest; Motley, T.J., Zerega, N., Cross, H., Eds.; Columbia University Press: New York, NY, USA, 2006; pp. 121–143. [Google Scholar] [CrossRef]
- Gioia, T.; Logozzo, G.; Attene, G.; Bellucci, E.; Benedettelli, S.; Negri, V.; Papa, R.; Spagnoletti Zeuli, P. Evidence for introduction bottleneck and extensive inter-gene pool (Mesoamerica x Andes) hybridization in the European common bean (Phaseolus vulgaris L.) germplasm. PLoS ONE 2013, 8, e75974. [Google Scholar] [CrossRef]
- Negri, V.; Maxted, N.; Vetelainen, M. European landrace conservation: An introduction. In European Landraces: On-Farm Conservation, Management and Use; Vetelainen, M., Negri, V., Maxted, N., Eds.; Bioversity International: Rome, Italy, 2009; pp. 1–22. [Google Scholar]
- Esquinas-Alcazar, J. Protecting crop genetic diversity for food security: Political, ethical and technical challenges. Nat. Rev. Genet. 2005, 6, 946–953. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Lioi, L. Italian common bean landraces: History, genetic diversity and seed quality. A review. Diversity 2010, 2, 837–862. [Google Scholar] [CrossRef]
- Perrino, P.; Hammer, K.; Hanelt, P. Collection of landraces of cultivated plants in South Italy 1983. Die Kult. 1984, 32, 207–216. [Google Scholar] [CrossRef]
- Angioi, S.A.; Rau, D.; Rodriguez, M.; Logozzo, G.; Desiderio, F.; Papa, R.; Attene, G. Nuclear and chloroplast microsatellite diversity in Phaseolus vulgaris L. from Sardinia (Italy). Mol. Breed. 2009, 23, 413–429. [Google Scholar] [CrossRef]
- Scarano, D.; Rubio, F.; Ruiz, J.J.; Rao, R.; Giandomenico, C. Morphological and genetic diversity among and within common bean (Phaseolus vulgaris L.) landraces from the Campania region (Southern Italy). Sci. Hortic. 2014, 180, 72–78. [Google Scholar] [CrossRef]
- De Luca, D.; Cennamo, P.; del Guacchio, E.; di Novella, R.; Caputo, P. Conservation and genetic characterization of common bean landraces from Cilento region (southern Italy): High differentiation in spite of low genetic diversity. Genetica 2018, 146, 29–44. [Google Scholar] [CrossRef]
- Mercati, F.; Leone, M.; Lupini, A.; Sorgona, A.; Bacchi, M.; Abenavoli, M.R.; Sunseri, F. Genetic diversity and population structure of a common bean (Phaseolus vulgaris L.) collection from Calabria (Italy). Genet. Resour. Crop. Evol. 2013, 60, 839–852. [Google Scholar] [CrossRef]
- Lioi, L.; Nuzzi, A.; Campion, B.; Piergiovanni, A. Assessment of genetic variation in common bean (Phaseolus vulgaris L.) from Nebrodi mountains (Sicily, Italy). Genet. Resour. Crop Evol. 2012, 59, 455–464. [Google Scholar] [CrossRef]
- Fiore, M.C.; Raimondo, F.M.; Mercati, F.; Diganci, I.; Sunseri, F.; Scialabba, A. Preserving biodiversity in marginal rural areas: Assessment of morphological and genetic variability of a Sicilian common bean germplasm collection. Plants 2020, 9, 989. [Google Scholar] [CrossRef]
- International Board for Plant Genetic Resources (IBPGR). Phaseolus Vulgaris Descriptors; IBPGR: Rome, Italy, 1982. Available online: https://cgspace.cgiar.org/handle/10568/45742 (accessed on 29 August 2022).
- Chiorato, A.F.; Carbonell, S.A.M.; Benchimol, L.L.; Chiavegato, M.B.; Dias, L.A.S.; Colombo, C.A. Genetic diversity in common bean accessions evaluated by means of morpho-agronomical and RAPD data. Sci. Agric. 2007, 64, 256–262. [Google Scholar] [CrossRef]
- Singh, S.P. A key for identification of different growth habits of Phaseolus vulgaris L. Ann. Rep. Bean Improv. Coop. 1982, 25, 92–95. [Google Scholar]
- Limongelli, G.; Laghetti, G.; Perrino, P.; Piergiovanni, A.R. Variation of seed storage proteins in landraces of common bean (Phaseolus vulgaris L.) from Basilicata, southern Italy. Euphytica 1996, 92, 393–399. [Google Scholar] [CrossRef]
- Bollini, R.; Chrispeels, M.J. Characterization and subcellular localization of vicilin and phytohemagglutinin, the two major reserve proteins of Phaseolus vulgaris L. Planta 1978, 142, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Park, S.J.; Poysa, V.; Gepts, P. Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). J. Hered. 2000, 91, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Nanni, L.; Bitocchi, E.; Bellucci, E.; Rossi, M.; Rau, D.; Attene, G.; Gepts, P.; Papa, R. Nucleotide diversity of a genomic sequence similar to SHATTERPROOF (PvSHP1) in domesticated and wild common bean (Phaseolus vulgaris L.). Theor Appl Genet 2011, 123, 1341–1357. [Google Scholar] [CrossRef] [PubMed]
- Gaitán-Solís, E.; Duque, M.C.; Edwards, K.J.; Tohme, J. Microsatellite repeats in common bean (Phaseolus vulgaris L.): Isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop. Sci. 2002, 42, 2128–2136. [Google Scholar] [CrossRef]
- Mercati, F.; Catarcione, G.; Paolacci, A.R.; Abenavoli, M.R.; Sunseri, F.; Ciaffi, M. Genetic diversity and population structure of an Italian landrace of runner bean (Phaseolus coccineus L.): Inferences for its safeguard and on-farm conservation. Genetica 2015, 143, 473–485. [Google Scholar] [CrossRef]
- Cramer, H. Mathematical Methods of Statistics; Princeton University Press: Princeton, NJ, USA, 1999. [Google Scholar]
- Weber, J.L. Informativeness of human (dC-dA)n-(dGdT)n polymorphisms. Genomics 1990, 7, 524–530. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. Powermarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx6: Genetic analysis in excel. population genetic software for teaching and research. Mol Ecol Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT: A program to estimate and test gene diversities and fixation indices (Version 2.9.3.2). Institute of Ecology, Lausanne, Switzerland. Website 2002. Available online: https://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 29 August 2022).
- Nei, M. The theory and estimation of genetic distance. In Genetic Structure of Populations; Morton, N.E., Ed.; University Press of Hawaii: Honolulu, HI, USA, 1973; pp. 45–54. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wilgenbusch, J.C.; Swofford, D. Inferring evolutionary trees with PAUP*. Curr. Protoc Bioinform. 2003, 6, 4. [Google Scholar] [CrossRef]
- Wright, S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Goudet, J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Brown, J.W.S.; Osborn, T.C.; Bliss, F.A.; Hall, T.C. Bean lectins. Part 1: Relationships between agglutinating activity and electrophoretic variation in the lectin-containing G2/albumin seed proteins of French bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 1982, 62, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Bollini, R.; Genga, A.; Vitale, A. Lectin and lectin-like proteins in bean seeds. In Lectins; Biology, biochemistry, clinical biochemistry; Sigma Chemical Company: St. Louis, MO, USA, 1988; Volume 6, pp. 237–241. [Google Scholar]
- Lioi, L. Electrophoretic variation and geographical distribution of the seed protein phytohemagglutinin in cultivated Phaseolus vulgaris L. J. Genet. Breed. 1991, 45, 97–102. [Google Scholar]
- van Treuren, R.; van Hintum, T.J. Marker-assisted reduction of redundancy in germplasm collections: Genetic and economic aspects. Acta Hortic. 2003, 623, 139–149. [Google Scholar] [CrossRef]
- Jarvis, D.; Brown, A.H.D.; Cuong, P.H.; Panduro, C.L.; Moreno, L.L.; Gyawali, S.; Tanto, T.; Sawadogo, M.; Mar, I.; Sadiki, M.; et al. A global perspective of the richness and evenness of traditional crop-variety diversity maintained by farming communities. PNAS 2008, 105, 5326–5331. [Google Scholar] [CrossRef] [PubMed]
- Rana, J.C.; Sharma, T.R.; Tyagi, R.K.; Chahota, R.K.; Gautam, N.K.; Mohar, S.; Sharma, P.N.; Ojha, S.N. Characterisation of 4274 accessions of common bean (Phaseolus vulgaris L.) germplasm conserved in the Indian gene bank for phenological, morphological and agricultural traits. Euphytica 2015, 205, 441–457. [Google Scholar] [CrossRef]
- Perseguini, J.M.K.G.; Silva, G.M.B.; Rosa, J.R.B.F.; Gazaffi, R.; Marçal, J.F.; Carbonell, S.A.M.; Chiorat, A.F.; Zucchi, M.I.; Garcia, A.A.F.; Benchimol-Reis, V. Developing a common bean core collection suitable for association mapping studies. Genet. Mol. Biol. 2015, 38, 67–78. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Cerbino, D.; Brandi, M. The common bean populations from Basilicata (southern Italy). An evaluation of their variation. Genet. Resour. Crop Evol. 2000, 47, 489–495. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Taranto, G.; Pignone, D. Diversity among common bean populations from the Abruzzo region (Central Italy): A preliminary inquiry. Genet. Resour. Crop Evol. 2000, 47, 467–470. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Negri, V.; Tiranti, B. Effectiveness of in situ and ex situ conservation of crop diversity. What a Phaseolus vulgaris L. landrace case study can tell us. Genetica 2010, 138, 985–998. [Google Scholar] [CrossRef]
- Lakis, G.; Ousmane, A.M.; Sanoussi, D.; Habibou, A.; Badamassi, M.; Lamy, F.; Jika, N.; Sidikou, R.; Adam, T.; Sarr, A.; et al. Evolutionary dynamics of cycle length in pearl millet: The role of farmer’s practices and gene flow. Genetica 2011, 139, 1367–1380. [Google Scholar] [CrossRef] [PubMed]
- Zeven, A.C.; Waninge, T.; Van Hintum, T.; Singh, S.P. Phenotypic variation in a core collection of common bean (Phaseolus vulgaris L.) in the Netherlands. Euphytica 1999, 109, 93–106. [Google Scholar] [CrossRef]
- Pressoir, G.; Berthaud, J. Population structure and strong divergent selection shape phenotypic diversification in maize landraces. Heredity 2004, 92, 95–101. [Google Scholar] [CrossRef]
- Pusadee, T.; Jamjod, S.; Chiang, Y.C.; Rerkasem, B.; Schaal, B.A. Genetic structure and isolation by distance in a landrace of Thai rice. Proc. Natl. Acad. Sci. USA 2009, 106, 13880–13885. [Google Scholar] [CrossRef] [PubMed]
- Bellucci, E.; Nanni, L.; Biagetti, E.; Bitocchi, E.; Giardini, A.; Rau, D. Common bean origin, evolution and spread from America. In Phaseolus: A New World Gift to Mankind Why Common Beans Are so Common? Rubiales, D., Ed.; International Legume Society: Córdoba, Spain, 2014; pp. 12–16. [Google Scholar]
- Asfaw, A.; Blair, M.W.; Almekinders, C. Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from the East African highlands. Theor. Appl. Genet. 2009, 120, 1–12. [Google Scholar] [CrossRef]
- Angioi, S.A.; Rau, D.; Nanni, L.; Bellucci, E.; Papa, R.; Attene, G. The genetic make–up of the European landraces of the common bean. Plant Genet. Res. 2011, 9, 197–201. [Google Scholar] [CrossRef]
- Okii, D.; Tukamuhabwa, P.; Kami, J.; Namayanja, A.; Paparu, P.; Ugen, M.; Gepts, P. The genetic diversity and population structure of common bean (Phaseolus vulgaris L.) germplasm in Uganda. Afr. J. Biotechnol. 2014, 13, 2935–2949. [Google Scholar] [CrossRef]
- Burle, M.L.; Fonseca, J.R.; Kami, J.A.; Gepts, P. Microsatellite diversity and genetic structure among common bean (Phaseolus vulgaris L.) landraces in Brazil, a secondary center of diversity. Theor. Appl. Genet. 2010, 121, 801–813. [Google Scholar] [CrossRef]
- Santalla, M.; Rodiño, P.; De Ron, A.M. Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for the common bean. Theor. Appl. Genet. 2002, 104, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.; Toro, O.; Debouck, D.G.; Gepts, P. Multiple origins of the determinate growth habit in domesticated common bean (Phaseolus vulgaris). Ann. Bot. 2012, 110, 1573–1580. [Google Scholar] [CrossRef] [PubMed]

| Cluster/Phaseolin Type | N | N Lan | N Ind | N Det | Na | Ne | Npa | AR | Ho | He |
|---|---|---|---|---|---|---|---|---|---|---|
| M/PHAS type S (Mesoamerican) | 84 | 10 | 10 | 0 | 3.500 | 2.212 | 10 | 3.500 | 0.010 | 0.382 |
| A/PHAS type T or C (Andean) | 346 | 51 | 26 | 25 | 4.830 | 2.593 | 38 | 4.660 | 0.016 | 0.466 |
| p-value Test Kruskal–Wallis | 0.199 | 0.266 | 0.174 | |||||||
| M/PHAS type S (Mesoamerican) | 68 | 7 | 7 | 0 | 3.000 | 1.969 | 9 | 3.000 | 0.028 | 0.337 |
| A1/PHAS type C (Andean) | 128 | 20 | 18 | 2 | 3.833 | 2.087 | 13 | 3.802 | 0.018 | 0.382 |
| A2/PHAS type T (Andean) | 118 | 15 | 1 | 14 | 3.667 | 2.226 | 7 | 3.625 | 0.023 | 0.422 |
| Wilcoxon Test p-value | ||||||||||
| M/A1 | 0.320 | 0.220 | 0.622 | |||||||
| M/A2 | 0.398 | 0.530 | 0.283 | |||||||
| A1/A2 | 0.931 | 0.490 | 0.452 |
| Analysis | Source | df | SS | MS | Est. Var. | % | Φ-Statistic | P(Φ) |
|---|---|---|---|---|---|---|---|---|
| (A) | Among groups | 1 | 1518.301 | 1518.301 | 11.149 | 50 | 0.501 | <0.001 |
| Within groups | 428 | 4756.799 | 11.114 | 11.114 | 50 | |||
| Total | 429 | 6275.100 | 22.263 | 100 | ||||
| (B) | Among groups | 2 | 1861.247 | 930.623 | 9.091 | 50 | 0.503 | <0.001 |
| Within groups | 311 | 2796.690 | 8.993 | 8.993 | 50 | |||
| Total | 313 | 4657.936 | 18.084 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catarcione, G.; Paolacci, A.R.; Alicandri, E.; Gramiccia, E.; Taviani, P.; Rea, R.; Costanza, M.T.; De Lorenzis, G.; Puccio, G.; Mercati, F.; et al. Genetic Diversity and Population Structure of Common Bean (Phaseolus vulgaris L.) Landraces in the Lazio Region of Italy. Plants 2023, 12, 744. https://doi.org/10.3390/plants12040744
Catarcione G, Paolacci AR, Alicandri E, Gramiccia E, Taviani P, Rea R, Costanza MT, De Lorenzis G, Puccio G, Mercati F, et al. Genetic Diversity and Population Structure of Common Bean (Phaseolus vulgaris L.) Landraces in the Lazio Region of Italy. Plants. 2023; 12(4):744. https://doi.org/10.3390/plants12040744
Chicago/Turabian StyleCatarcione, Giulio, Anna Rita Paolacci, Enrica Alicandri, Elena Gramiccia, Paola Taviani, Roberto Rea, Maria Teresa Costanza, Gabriella De Lorenzis, Guglielmo Puccio, Francesco Mercati, and et al. 2023. "Genetic Diversity and Population Structure of Common Bean (Phaseolus vulgaris L.) Landraces in the Lazio Region of Italy" Plants 12, no. 4: 744. https://doi.org/10.3390/plants12040744
APA StyleCatarcione, G., Paolacci, A. R., Alicandri, E., Gramiccia, E., Taviani, P., Rea, R., Costanza, M. T., De Lorenzis, G., Puccio, G., Mercati, F., & Ciaffi, M. (2023). Genetic Diversity and Population Structure of Common Bean (Phaseolus vulgaris L.) Landraces in the Lazio Region of Italy. Plants, 12(4), 744. https://doi.org/10.3390/plants12040744










