Morphological, Molecular, and Nutritional Characterisation of the Globe Artichoke Landrace “Carciofo Ortano”
Abstract
1. Introduction
2. Materials and Methods
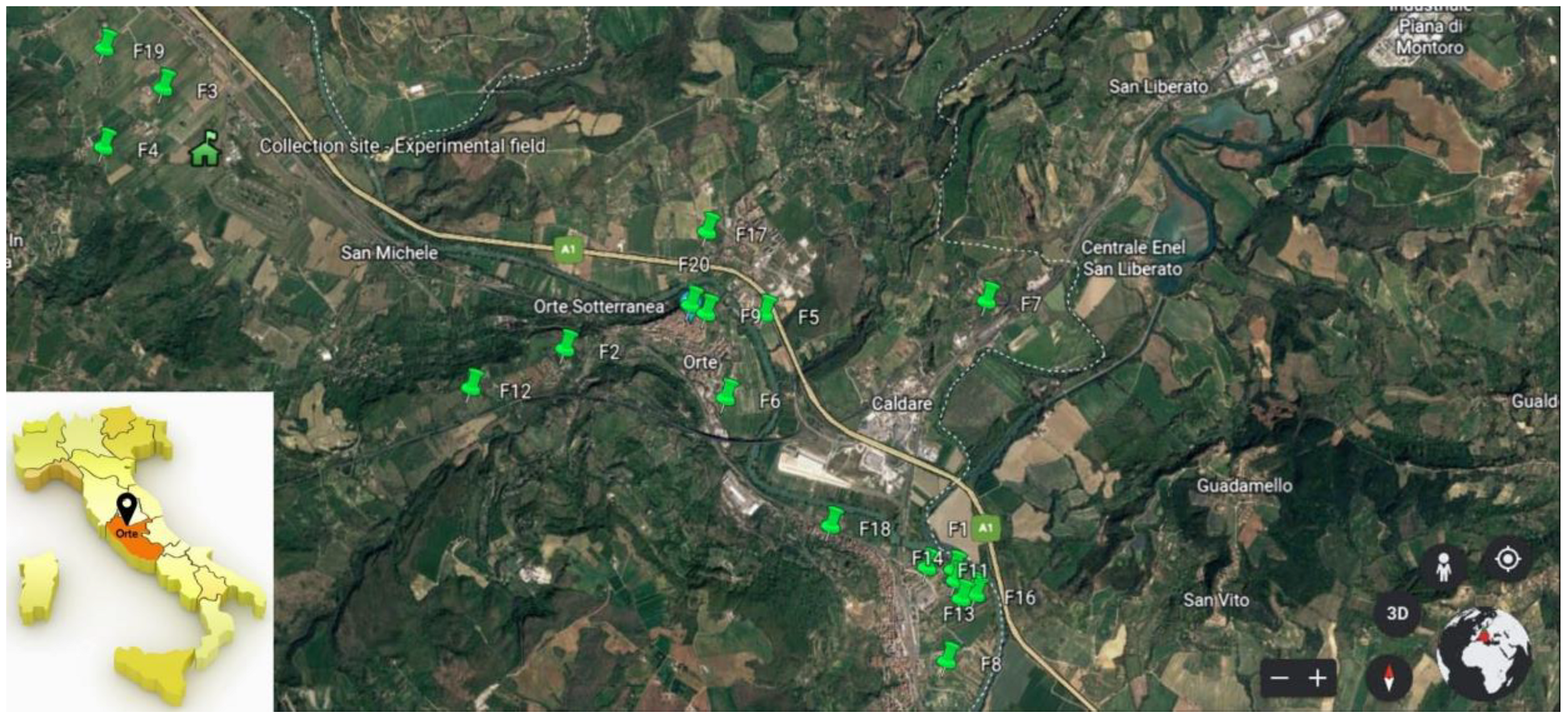
2.1. Identification and Survey of Smallholdings and Family Gardens Growing the Landrace “Carciofo Ortano”
2.2. Molecular Characterisation of the “Carciofo Ortano” Landrace
2.2.1. Plant Material
2.2.2. DNA Extraction and Molecular Markers (SSR and ISSR) Analysis
2.3. Planting of Collection Experimental Field for Morphological and Nutritional Analyses
2.4. Morphological Analysis
2.5. Nutritional and Chemical Analysis of Primary Flower Heads
2.5.1. Sampling and Preparation of Plant Material
2.5.2. Nutritional Value and Mineral Composition Analysis
2.6. Data Analysis
2.6.1. Molecular Data
2.6.2. Morphological Data
2.6.3. Nutritional and Chemical Data
3. Results and Discussion
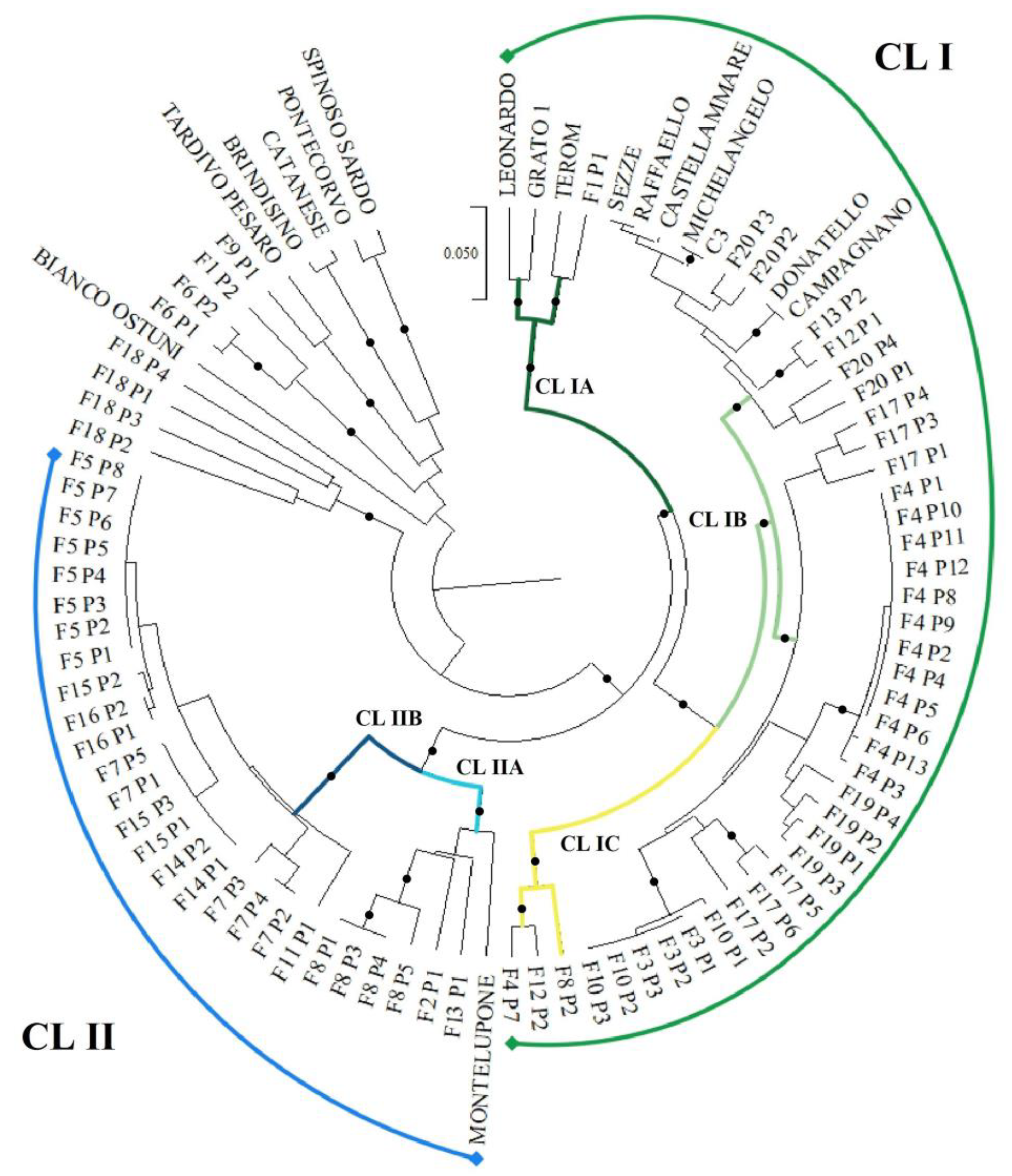
3.1. Genetic Analysis by SSR and ISSR Markers
3.1.1. Genetic Diversity
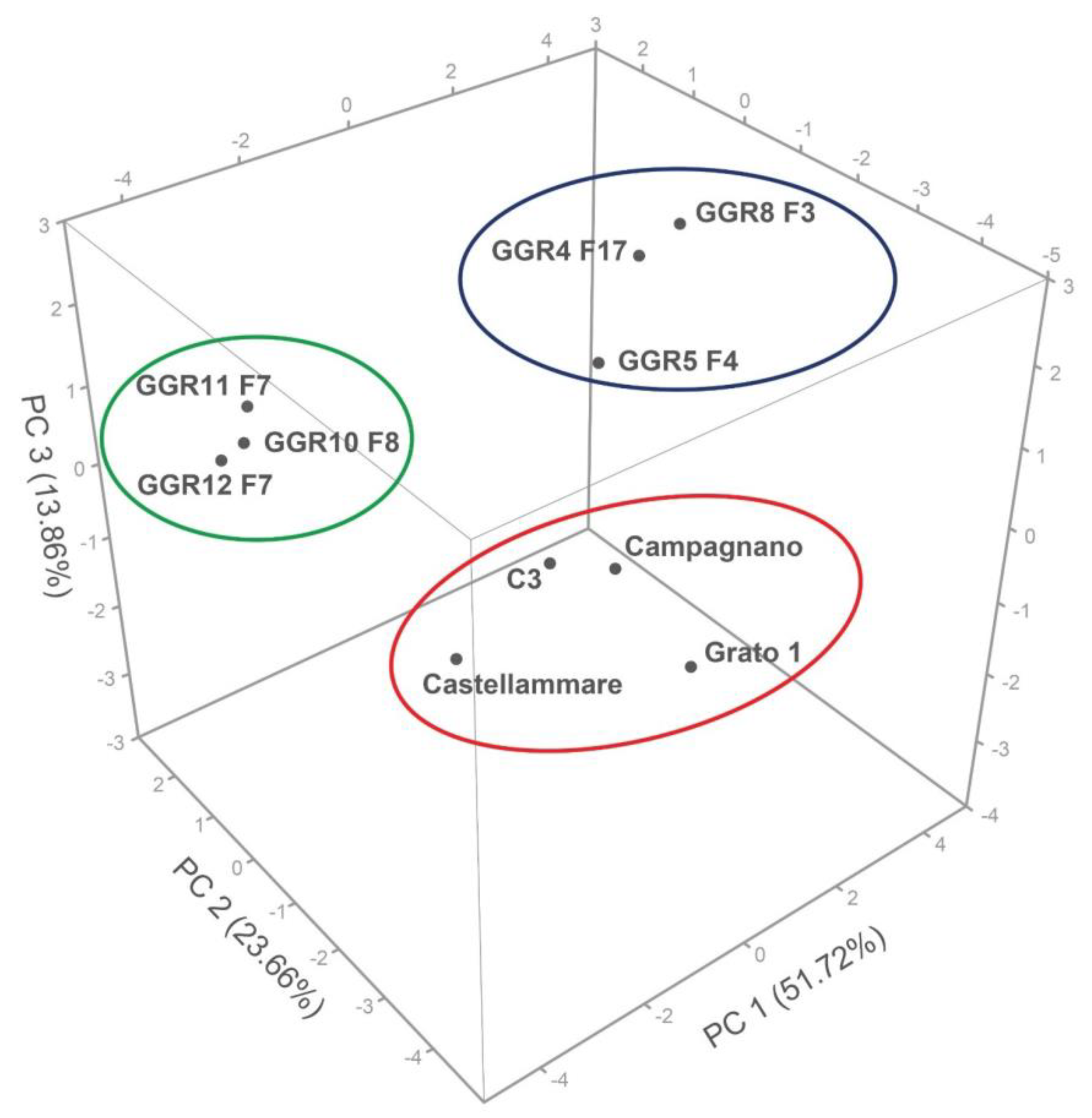
3.1.2. Genetic Relationship among Genotypes
3.1.3. Genetic Structure of the “Carciofo Ortano” Landrace
3.2. Setting up of the Collection Experimental Field
3.3. Morphological Characterisation
3.4. Nutritional and Chemical Characterisation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bianco, V.V. Carciofo (Cynara scolymus). In Orticoltura; Orticoltura Bianco, V.V., Rimpini, F., Eds.; Patron Editore: Bologna, Italy, 1990; pp. 209–251. [Google Scholar]
- Sonnante, G.; Pignone, D.; Hammer, K. The domestication of artichoke and cardoon: From roman times to the genomic age. Ann. Bot. 2007, 100, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Gatto, A.; De Paola, D.; Bagnoli, F.; Vendramin, G.G.; Sonnante, G. Population structure of Cynara cardunculus complex and the origin of the conspecific crops artichoke and cardoon. Ann. Bot. 2013, 112, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Pandino, G.; Lombardo, S.; Mauromicale, G. Chemical and morphological characteristics of new clones and commercial varieties of globe artichoke (Cynara cardunculus Var scolymus). Plant Foods Hum. Nutr. 2011, 66, 291–297. [Google Scholar] [CrossRef] [PubMed]
- De Falco, B.; Incerti, G.; Amato, M.; Lanzotti, V. Artichoke: Botanical, agronomical, phytochemical, and pharmacological overview. Phytochem. Rev. 2015, 14, 993–1018. [Google Scholar] [CrossRef]
- Lattanzio, V.; Kroon, P.A.; Linsalata, V.; Cardinali, A. Globe artichoke: A functional food and source of nutraceutical ingredients. J. Funct. Foods 2009, 1, 131–144. [Google Scholar] [CrossRef]
- Petropoulos, S.A.; Pereira, C.; Ntatsi, G.; Danalatos, N.; Barros, L.; Ferreira, I.C.F.R. Nutritional value and chemical composition of Greek artichoke genotypes. Food Chem. 2018, 267, 296–302. [Google Scholar] [CrossRef]
- Gostin, A.I.; Waisundara, V.Y. Edible flowers as functional food: A review on artichoke (Cynara Cardunculus L.). Trends Food Sci. Technol. 2019, 86, 381–391. [Google Scholar] [CrossRef]
- Ciancolini, A.; Rey, N.A.; Pagnotta, M.A.; Crinò, P. Characterization of italian spring globe artichoke germplasm: Morphological and molecular profiles. Euphytica 2012, 186, 433–443. [Google Scholar] [CrossRef]
- Rouphael, Y.; Bernardi, J.; Cardarelli, M.; Bernardo, L.; Kane, D.; Colla, G.; Lucini, L. Phenolic compounds and sesquiterpene lactones profile in leaves of nineteen artichoke cultivars. J. Agric. Food Chem. 2016, 64, 8540–8548. [Google Scholar] [CrossRef]
- Ierna, A.; Mauro, R.P.; Mauromicale, G. Biomass, grain and energy yield in Cynara cardunculus L. as affected by fertilization, genotype and harvest time. Biomass Bioenergy 2012, 36, 404–410. [Google Scholar] [CrossRef]
- Ledda, L.; Deligios, P.A.; Farci, R.; Sulas, L. Biomass supply for energetic purposes from some Cardueae species grown in Mediterranean farming systems. Ind. Crops Prod. 2013, 47, 218–226. [Google Scholar] [CrossRef]
- FAOSTAT Data. 2013. Available online: http://www.fao.org/faostat/en/#data (accessed on 4 April 2023).
- ISTAT (Italian National Institute of Statistics). 2009. Available online: http://dati.istat.it/Index.aspx?DataSetCode=DCSP_COLTIVAZ&Lang=en (accessed on 4 April 2023).
- Porceddu, E.; Dellacecca, V.; Blanco, V.V. Classificazione numerica di cultivar di carciofo. In Proceedings of the II International Congress of Artichoke, Bari, Italy, 17–21 October 1976; pp. 1105–1119. [Google Scholar]
- Pagnotta, M.A.; Noorani, A. Genetic Diversity assessment in European Cynara collections. In Genomics of Plant Genetic Resources; Tuberosa, R., Graner, A., Frison, E., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 559–584. [Google Scholar] [CrossRef]
- Pagnotta, M.A.; Fernández, J.A.; Sonnante, G.; Egea-Gilabert, C. Genetic diversity and accession structure in European Cynara cardunculus Collections. PLoS ONE 2017, 12, e0178770. [Google Scholar] [CrossRef]
- Bonasia, A.; Conversa, G.; Lazzizera, C.; Gambacorta, G.; Elia, A. Morphological and qualitative characterization of globe artichoke head from new seed-propagated cultivars. J. Sci. Food Agric. 2010, 90, 2689–2693. [Google Scholar] [CrossRef]
- Crinò, P.; Pagnotta, M.A. Phenotyping, genotyping, and selections within Italian local landraces of Romanesco globe artichoke. Diversity 2017, 9, 14. [Google Scholar] [CrossRef]
- Mauro, R.P.; Portis, E.; Lanteri, S.; Mauromicale, G. Genotypic and bio-agronomical characterization of an early Sicilian landrace of globe artichoke. Euphytica 2012, 186, 357–366. [Google Scholar] [CrossRef]
- Dosi, R.; Daniele, A.; Guida, V.; Ferrara, L.; Severino, V.; Di, A. Nutritional and metabolic profiling of the globe artichoke (Cynara scolymus L. “Capuanella” Heads) in province of Caserta, Italy. Aust. J. Crop. Sci. 2013, 7, 1927–1934. Available online: http://www.cropj.com/dimaro_7_12_2013_1927_1934.pdf (accessed on 22 February 2023).
- Portis, E.; Mauromicale, G.; Barchi, L.; Mauro, R.; Lanteri, S. Population structure and genetic variation in autochthonous globe artichoke germplasm from Sicily Island. Plant Sci. 2005, 168, 1591–1598. [Google Scholar] [CrossRef]
- Romani, A.; Pinelli, P.; Cantini, C.; Cimato, A.; Heimler, D. Characterization of Violetto Di Toscana, a typical italian variety of artichoke (Cynara scolymus L.). Food Chem. 2006, 95, 221–225. [Google Scholar] [CrossRef]
- Crinò, P.; Tavazza, R.; Rey Muñoz, N.A.; Trionfetti Nisini, P.; Saccardo, F.; Ancora, G.; Pagnotta, M.A. Recovery, morphological and molecular characterization of globe artichoke ‘Romanesco’ landraces. Genet. Resour. Crop. Evol. 2008, 55, 823–833. [Google Scholar] [CrossRef]
- Mauro, R.; Portis, E.; Acquadro, A.; Lombardo, S.; Mauromicale, G.; Lanteri, S. Genetic diversity of globe artichoke landraces from Sicilian small holdings: Implications for evolution and domestication of the species. Conserv. Genet. 2009, 10, 431–440. [Google Scholar] [CrossRef]
- Soressi, G. Available variability usable for breeding of globe artichoke [Cynara cardunculus L. var. scolymus L.]. Inf. Agrar. 2003, 22, 47. [Google Scholar]
- Cappelletti, R.; Mezzetti, B.; Balducci, F.; Diamanti, J.; Capocasa, F. Morphological, nutraceutical and chemical characterization of globe artichoke (Cynara cardunculus L. var. scolymus (L.) Fiori) landraces typically cultivated in Marche area. Acta Hortic. 2013, 983, 39–46. [Google Scholar] [CrossRef]
- Istituto Centrale di Statistica del Regno d’Italia. Catasto agrario 1929. Prov. Viterbo 1935, 61, 57.
- Noorani, A.; Rey, N.A.; Saccardo, F.; Pagnotta, M.A.; Dulloo, E.; Crinò, P. Diversity assessment of seven Italian globe artichoke varieties using agromorphological parameters. Acta Hortic. 2012, 942, 95–101. [Google Scholar] [CrossRef]
- Galieni, A.; Stagnari, F.; Pisante, M.; Platani, C.; Ficcadenti, N. Biochemical characterization of artichoke (Cynara cardunculus var scolymus L.) spring genotypes from Marche and Abruzzo regions (Central Italy). Adv. Hortic. Sci. 2018, 33, 23–32. [Google Scholar] [CrossRef]
- Available online: https://www.coribia.it/wp-content/uploads/2021/07/SCHEDA-CARCIOFO-VIOLETTO-CATANESE.pdf (accessed on 4 April 2023).
- Available online: https://www.politicheagricole.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/2059 (accessed on 4 April 2023).
- Available online: http://www.carciofosardodop.it/ (accessed on 4 April 2023).
- Available online: https://biodiversitapuglia.it/fbpost/bianco-di-ostuni-bianco-di-taranto-brindisino-centofoglie-di-rutigliano-carciofo-di-lucera-fran/ (accessed on 4 April 2023).
- Acquadro, A.; Portis, E.; Lanteri, S. Isolation of microsatellite loci in artichoke (Cynara cardunculus L. var. scolymus): Primer note. Mol. Ecol. Notes 2003, 3, 37–39. [Google Scholar] [CrossRef]
- Acquadro, A.; Portis, E.; Lee, D.; Donini, P.; Lanteri, S. Development and characterization of microsatellite markers in Cynara cardunculus L. Genome 2005, 48, 217–225. [Google Scholar] [CrossRef]
- Acquadro, A.; Portis, E.; Albertini, E.; Lanteri, S. M-AFLP-based protocol for microsatellite loci isolation in Cynara cardunculus L. (Asteraceae): Primer note. Mol. Ecol. Notes 2005, 5, 272–274. [Google Scholar] [CrossRef]
- Acquadro, A.; Lanteri, S.; Scaglione, D.; Arens, P.; Vosman, B.; Portis, E. Genetic mapping and annotation of genomic microsatellites isolated from globe artichoke. Appl. Genet. 2009, 118, 1573–1587. [Google Scholar] [CrossRef]
- Sonnante, G.; Carluccio, A.V.; De Paolis, A.; Pignone, D. Identification of artichoke SSR markers: Molecular variation and patterns of diversity in genetically cohesive taxa and wild allies. Genet. Resour. Crop. Evol. 2008, 55, 1029–1046. [Google Scholar] [CrossRef]
- Alicandri, E.; Vettraino, A.M.; Agrimi, M.; Ciaffi, M.; Kuzminsky, E. Molecular markers dataset to assess the genetic diversity of oriental plane trees from historical sites in Lazio (Central Italy). Data Brief. 2022, 42, 108100. [Google Scholar] [CrossRef]
- Ciaffi, M.; Alicandri, E.; Vettraino, A.M.; Paolacci, A.R.; Tamantini, M.; Tomao, A.; Agrimi, M.; Kuzminsky, E. Conservation of veteran trees within historical gardens (COVE): A case study applied to Platanus orientalis L. in central Italy. Urban For. Urban Green. 2018, 34, 336–347. [Google Scholar] [CrossRef]
- Association of Official Analytical Chemists. AOAC Official Methods of Analysis, 19th ed.; Association of Official Analytical Chemists: Washington, DC, USA, 2012. [Google Scholar]
- Lee, S.C.; Prosky, L. International survey on dietary fiber: Definition, analysis, and reference materials. J. AOAC Int. 1995, 78, 22–36. [Google Scholar] [CrossRef]
- El-Kheir, K.S.E.; Murwa, A.M. Chemical composition, minerals, protein fractionation, and anti-nutrition factors in leaf of hargel plant (Solenostemma Argel). Eur. J. Sci. Res. 2010, 43, 430–434. [Google Scholar]
- Steegmans, M.; Iliaens, S.; Hoebregs, H. Enzymatic, spectrophotometric determination of glucose, fructose, sucrose, and inulin/oligofructose in foods. J. AOAC Int. 2004, 87, 1200–1207. [Google Scholar] [CrossRef]
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- Weber, J.L. Informativeness of human (dC-dA)n-(dGdT)n polymorphisms. Genomics 1990, 7, 524–530. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx6: Genetic analysis in excel. population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Nei, M. The theory and estimation of genetic distance. In Genetic Structure of Populations; Morton, N.E., Ed.; University Press of Hawaii: Honolulu, HI, USA, 1973; pp. 45–54. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wilgenbusch, J.C.; Swofford, D. Inferring evolutionary trees with PAUP*. CP Bioinform. 2003, 1, 6-4. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Ciaffi, M.; Vettraino, A.M.; Alicandri, E.; Tomao, A.; Adducci, F.; Kuzminsky, E.; Agrimi, M. Dimensional and genetic characterization of the last oriental plane trees (Platanus orientalis L.) of historical sites in Lazio (central Italy). Urban For. Urban Green. 2022, 69, 127506. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- De Felice, B.; Borra, M.; Manfellotto, F.; Anna, S.; Biffali, E.; Guida, M. Assessment of genetic diversity between wild and cultivated artichokes using SSR markers. Genet. Resour. Crop. Evol. 2016, 63, 1363–1369. [Google Scholar] [CrossRef]
- Rau, D.; Attene, G.; Rodriguez, M.; Baghino, L.; Pisanu, A.B.; Sanna, D.; Acquadro, A.; Portis, E.; Comino, C. The population structure of a globe artichoke worldwide collection, as revealed by molecular and phenotypic analyzes. Front. Plant Sci. 2022, 13, 898740. [Google Scholar] [CrossRef]
- El Sohaimy, S.A. Chemical composition, antioxidant and antimicrobial potential of artichoke. Open Nutraceuticals J. 2009, 1, 15–20. [Google Scholar] [CrossRef]
- Negro, D.; Montesano, V.; Sonnante, G.; Rubino, P.; De Lisi, A.; Sarli, G. Fertilization Strategies on cultivars of globe artichoke: Effects on yield and quality performance. J. Plant Nutr. 2016, 39, 279–287. [Google Scholar] [CrossRef]
- Li, Y.O.; Komarek, A.R. Dietary fibre basics: Health, nutrition, analysis, and applications. Food Qual. Saf. 2017, 1, 47–59. [Google Scholar] [CrossRef]
- Jones, J.M. CODEX-aligned dietary fiber definitions help to bridge the ‘fiber gap’. Nutr. J. 2014, 13, 34. [Google Scholar] [CrossRef]
- Alimentarius CODEX. (CODEX) guidelines on nutrition labeling CAC/GL 2–1985 as last amended 2010. In Joint FAO/WHO Food Standards Programme, Secretariat of the CODEX Alimentarius Commission; FAO: Rome, Italy, 2010. [Google Scholar]
- Dai, F.J.; Chau, C.F. Classification and regulatory perspectives of dietary fiber. J. Food Drug Anal. 2017, 25, 37–42. [Google Scholar] [CrossRef]
- Ruiz-Cano, D.; Pérez-Llamas, F.; Frutos, M.J.; Arnao, M.B.; Espinosa, C.; López-Jiménez, J.Á.; Castillo, J.; Zamora, S. Chemical and functional properties of the different by-products of artichoke (Cynara Scolymus L.) from industrial canning processing. Food Chem. 2014, 160, 134–140. [Google Scholar] [CrossRef]
- Machado, M.T.C.; Eça, K.S.; Vieira, G.S.; Menegalli, F.C.; Martínez, J.; Hubinger, M.D. Prebiotic oligosaccharides from artichoke industrial waste: Evaluation of different extraction methods. Ind. Crops Prod. 2015, 76, 141–148. [Google Scholar] [CrossRef]
- Wan, X.; Guo, H.; Liang, Y.; Zhou, C.; Liu, Z.; Li, K.; Niu, F.; Zhai, X.; Wang, L. The physiological functions and pharmaceutical applications of inulin: A review. Carbohydr. Polym. 2020, 246, 116589. [Google Scholar] [CrossRef]
- Di Venere, D.; Linsalata, V.; Pace, B.; Bianca, V.; Perrino, P. Polyphenol and inulin content in a collection of artichoke. IV Int. Congr. Artichoke 2005, 681, 453–460. [Google Scholar] [CrossRef]
- Melilli, M.G.; Tringali, S.; Bognanni, R.; Argento, S.; Calderaro, P.; Raccuia, S.A. Nutritional Quality of globe artichoke [Cynara Cardunculus L. subsp. scolymus (L.) Hegi] head as affected by genotype and environment of cultivation. Acta Hortic. 2014, 1040, 187–192. [Google Scholar] [CrossRef]
- Pandino, G.; Lombardo, S.; Mauromicale, G. Mineral profile in globe artichoke as affected by genotype, head part and environment. J. Sci. Food Agric. 2011, 91, 302–308. [Google Scholar] [CrossRef]
- Nicoletto, C.; Santagata, S.; Tosini, F.; Sambo, P. Qualitative and healthy traits of different Italian typical artichoke genotypes. CyTA-J. Food 2013, 11, 108–113. [Google Scholar] [CrossRef]
- Rincón, L.; Pérez, C.; Pellicer, C.; Abadía, A.; Sáez, J. Nutrient uptake by artichoke. Acta Hort. 2007, 630, 287–292. [Google Scholar] [CrossRef]
- Cory, H.; Passarelli, S.; Szeto, J.; Tamez, M.; Mattei, J. The role of polyphenols in human health and food systems: A mini-review. Front. Nutr. 2018, 5, 87. [Google Scholar] [CrossRef]


| SSR Marker | Allele No | MAF | He | PIC | Ho | ||
| CDAT_02 | 7 | 0.505 | 0.635 | 0.577 | 0.374 | ||
| CELMS_08 | 7 | 0.703 | 0.485 | 0.465 | 0.978 | ||
| CDAT_04 | 8 | 0.495 | 0.658 | 0.609 | 0.429 | ||
| Cmal_06 | 4 | 0.879 | 0.221 | 0.212 | 0.934 | ||
| CELMS_05 | 9 | 0.505 | 0.637 | 0.581 | 0.912 | ||
| Cmal_08 | 4 | 0.912 | 0.165 | 0.161 | 0.967 | ||
| CDAT_01 | 8 | 0.505 | 0.646 | 0.594 | 0.989 | ||
| CELMS_40 | 10 | 0.527 | 0.650 | 0.609 | 0.912 | ||
| CsPal_03 | 8 | 0.527 | 0.620 | 0.563 | 0.89 | ||
| TOT | 65 | ||||||
| MEAN | 7.22 | 0.618 | 0.524 | 0.486 | 0.821 | ||
| ST. DEV. | 0.17 | 0.195 | 0.176 | 0.24 | |||
| ISSR Marker | TB | PB | % Pol | MAF | He | PIC | |
| ISSR_24 | 17 | 17 | 100 | 0.789 | 0.29 | 0.236 | |
| ISSR_28 | 11 | 7 | 64 | 0.979 | 0.04 | 0.039 | |
| UBC_840 | 15 | 15 | 100 | 0.870 | 0.200 | 0.170 | |
| UBC_848 | 10 | 10 | 100 | 0.699 | 0.363 | 0.282 | |
| UBC_855 | 12 | 12 | 100 | 0.803 | 0.265 | 0.213 | |
| UBC_857 | 11 | 11 | 100 | 0.783 | 0.292 | 0.235 | |
| TOT | 76 | 72 | |||||
| MEAN | 12.67 | 12 | 94 | 0.821 | 0.242 | 0.196 | |
| ST. DEV. | 0.087 | 0.102 | 0.078 | ||||
| SSR Marker | ||||||
| N | Ne | Npa | I | He | Ho | |
| Orte 1 | 37.00 | 1.78 | 4.00 | 0.54 | 0.39 | 0.78 |
| Orte 2 | 27.00 | 2.20 | 11.00 | 0.82 | 0.54 | 1.00 |
| p-value Kruskal–Wallis Test | <0.01 * | <0.01 * | <0.01 * | >0.01 | ||
| ISSR Marker | ||||||
| N | Ne | Npa | I | He | ||
| Orte 1 | 37.00 | 1.39 | 15.00 | 0.35 | 0.23 | |
| Orte 2 | 27.00 | 1.36 | 6.00 | 0.28 | 0.20 | |
| p-value Kruskal–Wallis Test | >0.01 | >0.01 | >0.01 |
| SSR Marker | |||||||
| df | SS | MS | Est. Var. | % | Φ-statistic | p(Φ) | |
| Among Pops | 1.00 | 71.408 | 71.408 | 2.596 | 80.55% | 0.905 | <0.001 |
| Within Pops | 62.00 | 38.889 | 0.627 | 0.627 | 19.45% | ||
| Total | 63.00 | 110.297 | 3.223 | 100% | |||
| ISSR Marker | |||||||
| df | SS | MS | Est. Var. | % | |||
| Among Pops | 1.00 | 106.87 | 106.87 | 3.23 | 34.94% | 0.349 | <0.001 |
| Within Pops | 62.00 | 372.93 | 6.01 | 6.01 | 65.06% | ||
| Total | 63.00 | 479.80 | 9.25 | 100.00% |
| PH | MSD | DHL | LCH | L/D Ratio | RECD | WCH | LH1 | L/D Ratio1 | WH1 | Y | THA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Campagnano | 66.833 b | 2.316 bc | 24.750 ab | 90.000 a | 0.947 b | 4.566 ab | 260.558 abc | 5.333 a | 1.011 b | 81.240 bc | 692.798 bc | 102.333 b |
| Castellammare | 52.833 d | 2.621 a | 20.450 c | 88.500 a | 0.938 b | 4.666 ab | 245.468 abc | 5.066 ab | 0.996 b | 71.316 c | 617.913 cd | 95.000 c |
| C3 | 58.666 cd | 2.666 a | 20.666 c | 96.833 a | 0.973 b | 4.983 a | 287.665 ab | 5.350 a | 0.983 b | 94.765 a | 798.718 a | 84.166 d |
| Grato 1 | 61.666 bc | 2.200 c | 22.916 bc | 96.000 a | 1.115 a | 4.000 b | 230.485 bc | 5.350 a | 1.173 a | 76.138 c | 597.956 d | 103.833 b |
| Orte1-GGR4 F17 | 73.000 a | 2.533 ab | 27.000 a | 98.166 a | 0.960 b | 5.150 a | 303.815 a | 5.783 a | 0.991 b | 91.836 ab | 768.151 a | 105.000 b |
| Orte1-GGR5 F4 | 67.000 b | 2.483 ab | 23.166 bc | 95.666 a | 0.953 b | 4.983 a | 304.283 a | 5.716 a | 1.028 b | 93.245 a | 785.028 a | 105.666 b |
| Orte1-GGR8 F3 | 74.333 a | 2.316 bc | 24.333 ab | 95.000 a | 0.955 b | 5.16 a | 285.355 ab | 5.366 a | 1.016 b | 90.566 ab | 724.255 ab | 103.333 b |
| Orte2-GGR12 F7 | 53.333 d | 2.383 bc | 20.000 c | 72.666 b | 0.805 c | 4.800 a | 231.573 bc | 4.300 c | 0.876 c | 72.033 c | 589.273 d | 118.000 a |
| Orte2-GGR11 F7 | 54.500 d | 2.403 bc | 20.333 c | 74.833 b | 0.790 c | 4.850 a | 238.103 bc | 4.533 bc | 0.865 c | 74.323 c | 608.093 d | 118.000 a |
| Orte2-GGR10 F8 | 54.000 d | 2.400 bc | 21.000 c | 74.000 b | 0.801 c | 4.716 a | 222.413 c | 4.416 bc | 0.885 c | 73.206 c | 588.620 d | 118.333 a |
| CV | 13.368 | 5.947 | 10.497 | 11.724 | 10.823 | 7.099 | 12.159 | 10.336 | 9.305 | 11.829 | 12.769 | 10.301 |
| p | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** |
| Moisture | Ash | Proteins | Lipids | Carbohydrates | THMWDF | IHMWDF | SHMWDF | Inulin | |
|---|---|---|---|---|---|---|---|---|---|
| Campagnano | 82.10 b | 1.51 a | 2.96 b | 0.23 a | 7.13 b | 6.08 b | 4.03 c | 2.05 bc | 3.30 b |
| Castellammare | 81.53 c | 1.45 ab | 3.25 a | 0.22 a | 7.30 b | 6.25 b | 4.43 b | 1.82 cd | 2.93 c |
| C3 | 83.38 a | 1.45 ab | 3.02 ab | 0.23 a | 6.17 c | 5.75 c | 4.1 c | 1.67 d | 2.92 c |
| Grato 1 | 80.88 d | 1.40 b | 3.18 ab | 0.25 a | 7.35 b | 6.93 a | 3.63 c | 2.70 a | 2.25 d |
| Orte1-GGR4 F17 | 82.96 a | 1.36 b | 3.06 ab | 0.22 a | 6.32 c | 6.07 b | 3.87 c | 2.20 b | 3.33 b |
| Orte1-GGR5 F4 | 83.33 a | 1.35 b | 3.00 ab | 0.23 a | 5.97 c | 6.12 b | 3.85 c | 2.27 b | 3.57 b |
| Orte2-GGR10 F8 | 80.50 de | 1.20 c | 3.03 ab | 0.22 a | 7.98 a | 7.07 a | 4.94 a | 2.13 b | 5.12 a |
| Orte2-GGR11 F7 | 80.25 e | 1.21 c | 3.00 ab | 0.23 a | 8.20 a | 7.10 a | 4.93 a | 2.17 b | 5.05 a |
| CV | 1.55 | 8.21 | 3.27 | 4.33 | 11.77 | 8.22 | 11.75 | 14.49 | 28.69 |
| p | *** | *** | * | ns | *** | *** | *** | *** | *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alicandri, E.; Paolacci, A.R.; Catarcione, G.; Del Lungo, A.; Iacoponi, V.; Pati, F.; Scarascia Mugnozza, G.; Ciaffi, M. Morphological, Molecular, and Nutritional Characterisation of the Globe Artichoke Landrace “Carciofo Ortano”. Plants 2023, 12, 1844. https://doi.org/10.3390/plants12091844
Alicandri E, Paolacci AR, Catarcione G, Del Lungo A, Iacoponi V, Pati F, Scarascia Mugnozza G, Ciaffi M. Morphological, Molecular, and Nutritional Characterisation of the Globe Artichoke Landrace “Carciofo Ortano”. Plants. 2023; 12(9):1844. https://doi.org/10.3390/plants12091844
Chicago/Turabian StyleAlicandri, Enrica, Anna Rita Paolacci, Giulio Catarcione, Alberto Del Lungo, Valentina Iacoponi, Francesco Pati, Giuseppe Scarascia Mugnozza, and Mario Ciaffi. 2023. "Morphological, Molecular, and Nutritional Characterisation of the Globe Artichoke Landrace “Carciofo Ortano”" Plants 12, no. 9: 1844. https://doi.org/10.3390/plants12091844
APA StyleAlicandri, E., Paolacci, A. R., Catarcione, G., Del Lungo, A., Iacoponi, V., Pati, F., Scarascia Mugnozza, G., & Ciaffi, M. (2023). Morphological, Molecular, and Nutritional Characterisation of the Globe Artichoke Landrace “Carciofo Ortano”. Plants, 12(9), 1844. https://doi.org/10.3390/plants12091844








