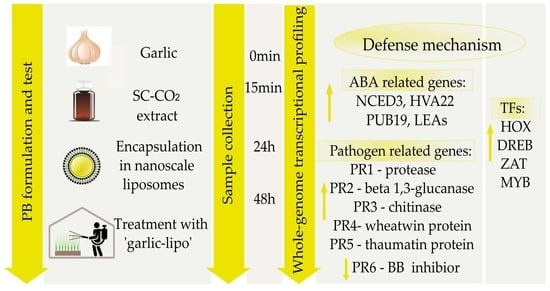
‘Garlic-lipo’4Plants: Liposome-Encapsulated Garlic Extract Stimulates ABA Pathway and PR Genes in Wheat (Triticum aestivum)
Abstract
1. Introduction
2. Materials and Methods
2.1. Garlic-Lipo Stability and Particle Size Measurements of Liposomes
2.2. Plant Materials and Treatment with ‘Garlic-Lipo’
2.3. Sequencing, Data Processing, Gene-Level Quantification, and DEG Determination
2.4. Time-Course Expression Analysis
2.5. Functional Annotation
2.6. Real-Time PCR Analysis
3. Results
3.1. Dynamic Light Scattering
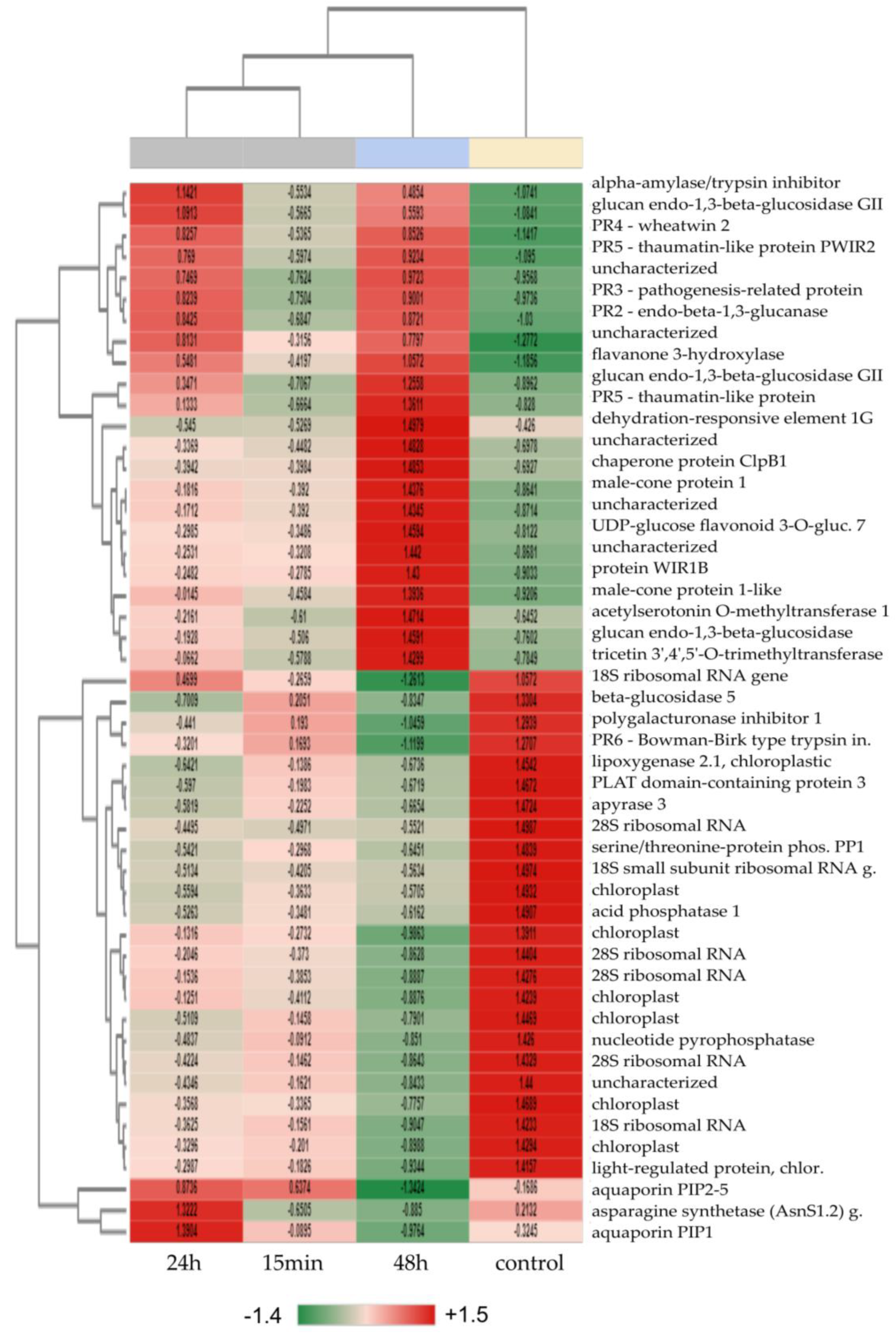
3.2. Pairwise Expression Analysis and Determination of Top 50 DEGs
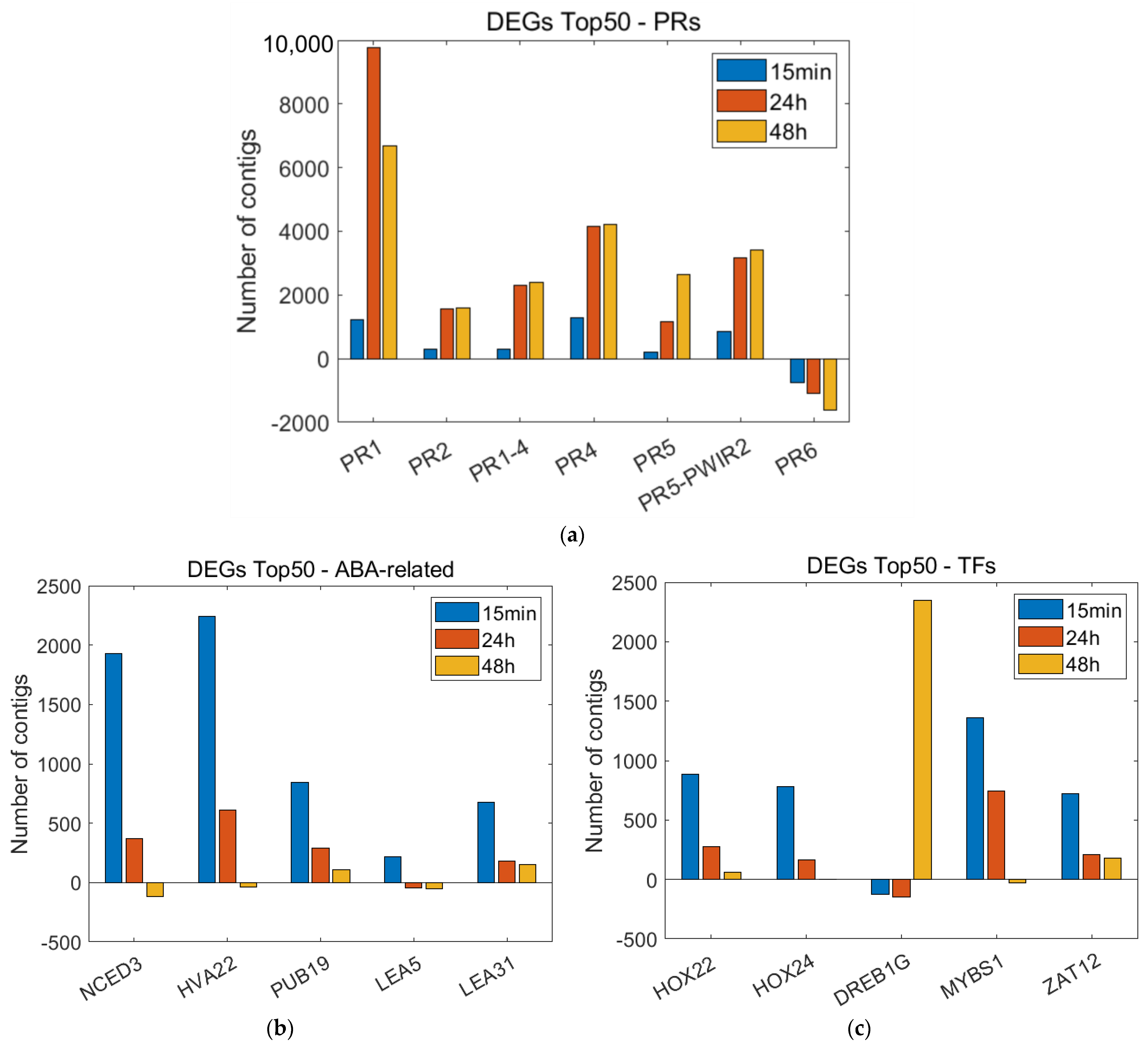
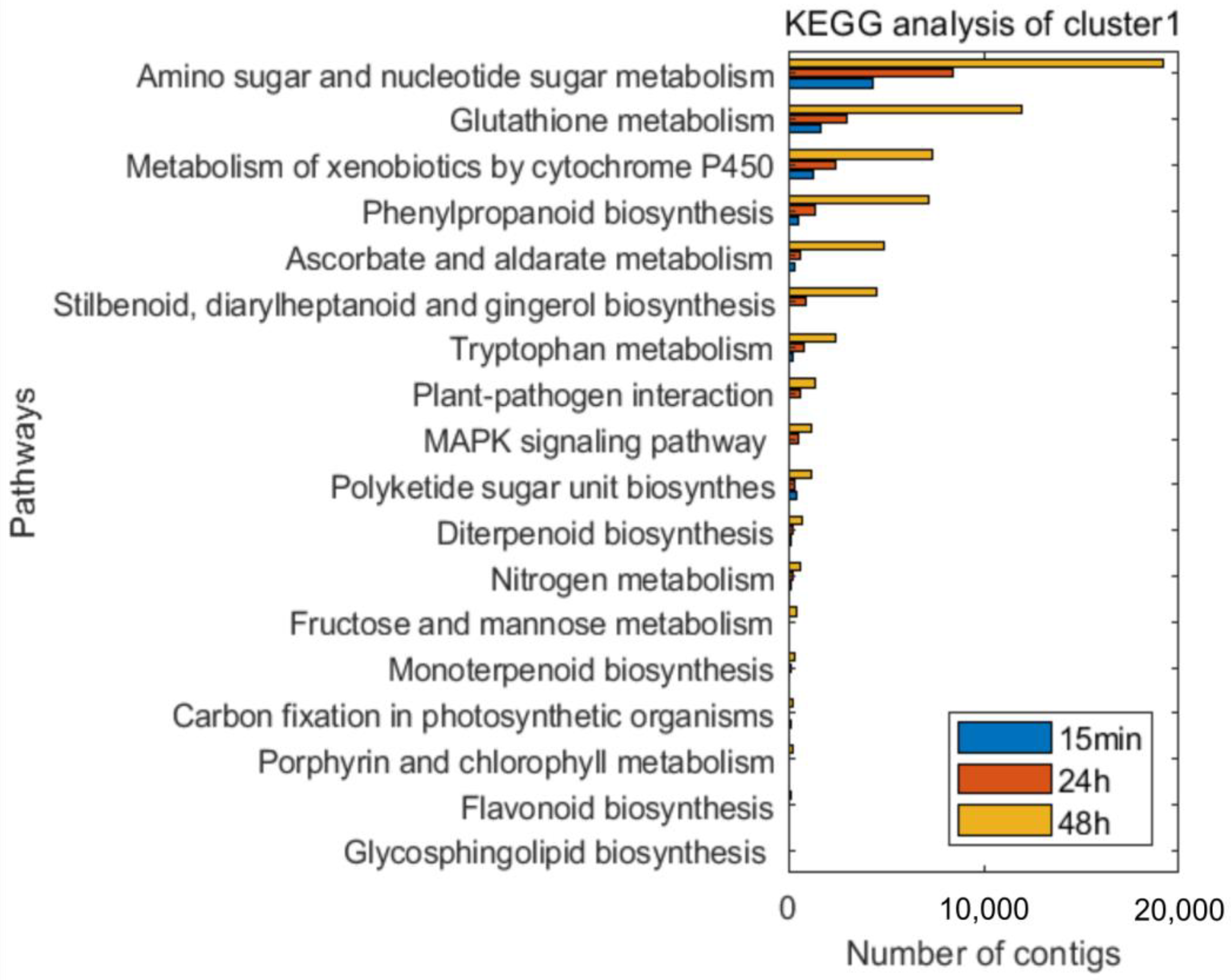
3.3. Time-Course Expression Analysis
3.4. Real-Time PCR Analysis
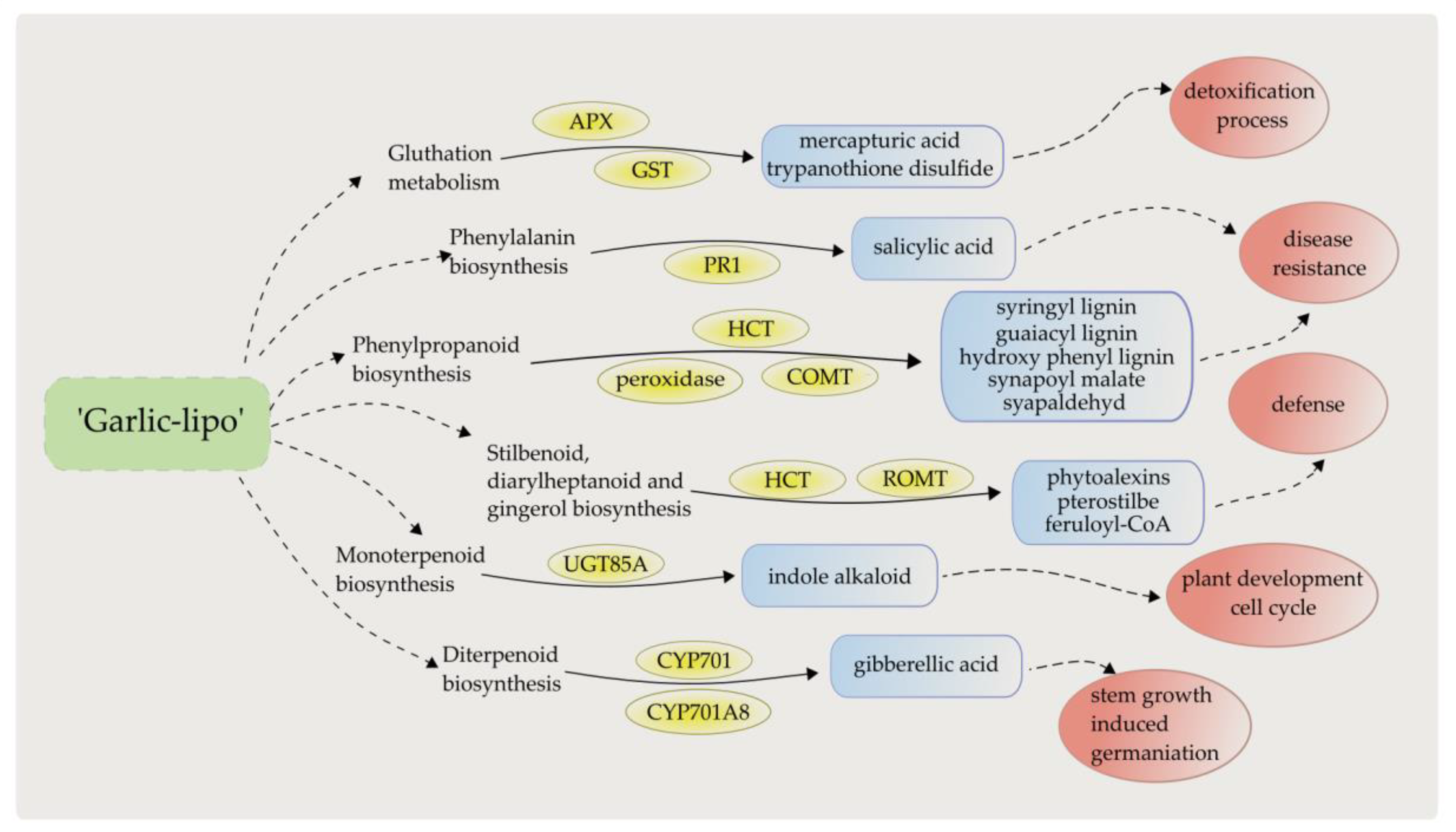
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lawson, L.D. Garlic: A review of its medicinal effects and indicated active compounds. Blood 1998, 179, 62. [Google Scholar] [CrossRef]
- Diretto, G.; Rubio-Moraga, A.; Argandoña, J.; Castillo, P.; Gómez-Gómez, L.; Ahrazem, O. Tissue-specific accumulation of sulfur compounds and saponins in different parts of garlic cloves from purple and white ecotypes. Molecules 2017, 22, 1359. [Google Scholar] [CrossRef]
- Divya, B.; Suman, B.; Venkataswamy, M.; Thyagaraju, K. A study on phytochemicals, functional groups and mineral composition of Allium sativum (garlic) cloves. Int. J. Curr. Pharm. Res. 2017, 9, 42–45. [Google Scholar] [CrossRef]
- Muir, J.G.; Shepherd, S.J.; Rosella, O.; Rose, R.; Barrett, J.S.; Gibson, P.R. Fructan and free fructose content of common Australian vegetables and fruit. J. Agric. Food Chem. 2007, 55, 6619–6627. [Google Scholar] [CrossRef] [PubMed]
- Miean, K.H.; Mohamed, S. Flavonoid (myricetin, quercetin, kaempferol, luteolin, and apigenin) content of edible tropical plants. J. Agric. Food Chem. 2001, 49, 3106–3112. [Google Scholar] [CrossRef]
- Lanzotti, V. The analysis of onion and garlic. J. Chromatogr. A 2006, 1112, 3–22. [Google Scholar] [CrossRef]
- Gorinstein, S.; Drzewiecki, J.; Leontowicz, H.; Leontowicz, M.; Najman, K.; Jastrzebski, Z.; Zachwieja, Z.; Barton, H.; Shtabsky, B.; Katrich, E. Comparison of the bioactive compounds and antioxidant potentials of fresh and cooked Polish, Ukrainian, and Israeli garlic. J. Agric. Food Chem. 2005, 53, 2726–2732. [Google Scholar] [CrossRef]
- Nencini, C.; Menchiari, A.; Franchi, G.G.; Micheli, L. In vitro antioxidant activity of aged extracts of some Italian Allium species. Plant Foods Hum. Nutr. 2011, 66, 11–16. [Google Scholar] [CrossRef]
- Perello, A.E.; Noll, U.; Slusarenko, A.J. In vitro efficacy of garlic extract to control fungal pathogens of wheat. J. Med. Plants Res. 2013, 7, 1809–1817. [Google Scholar] [CrossRef]
- Mougou, I.; Boughalleb-M’hamdi, N. Biocontrol of Pseudomonas syringae pv. syringae affecting citrus orchards in Tunisia by using indigenous Bacillus spp. and garlic extract. Egypt. J. Biol. Pest Control 2018, 28, 60. [Google Scholar] [CrossRef]
- Sarfraz, M.; Nasim, M.J.; Jacob, C.; Gruhlke, M.C. Efficacy of allicin against plant pathogenic fungi and unveiling the underlying mode of action employing yeast based chemogenetic profiling approach. Appl. Sci. 2020, 10, 2563. [Google Scholar] [CrossRef]
- Slusarenko, A.J.; Patel, A.; Portz, D. Control of plant diseases by natural products: Allicin from garlic as a case study. Sustain. Dis. Manag. A Eur. Context 2008, 121, 313–322. [Google Scholar] [CrossRef]
- Curtis, H.; Noll, U.; Störmann, J.; Slusarenko, A.J. Broad-spectrum activity of the volatile phytoanticipin allicin in extracts of garlic (Allium sativum L.) against plant pathogenic bacteria, fungi and Oomycetes. Physiol. Mol. Plant Pathol. 2004, 65, 79–89. [Google Scholar] [CrossRef]
- Godlewska, K.; Ronga, D.; Michalak, I. Plant extracts-importance in sustainable agriculture. Ital. J. Agron. 2021, 16. [Google Scholar] [CrossRef]
- Rouphael, Y.; Colla, G. Biostimulants in agriculture. Front. Plant Sci. 2020, 11, 40. [Google Scholar] [CrossRef]
- Du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Calvo, P.; Nelson, L.; Kloepper, J.W. Agricultural uses of plant biostimulants. Plant Soil 2014, 383, 3–41. [Google Scholar] [CrossRef]
- Hayat, S.; Ahmad, H.; Ali, M.; Hayat, K.; Khan, M.A.; Cheng, Z. Aqueous garlic extract as a plant biostimulant enhances physiology, improves crop quality and metabolite abundance, and primes the defense responses of receiver plants. Appl. Sci. 2018, 8, 1505. [Google Scholar] [CrossRef]
- Hayat, S.; Ahmad, H.; Ali, M.; Ren, K.; Cheng, Z. Aqueous garlic extract stimulates growth and antioxidant enzymes activity of tomato (Solanum lycopersicum). Sci. Hortic. 2018, 240, 139–146. [Google Scholar] [CrossRef]
- Hayat, S.; Cheng, Z.; Ahmad, H.; Ali, M.; Chen, X.; Wang, M. Garlic, from remedy to stimulant: Evaluation of antifungal potential reveals diversity in phytoalexin allicin content among garlic cultivars; allicin containing aqueous garlic extracts trigger antioxidants in cucumber. Front. Plant Sci. 2016, 7, 1235. [Google Scholar] [CrossRef]
- Olusanmi, M.; Amadi, J. Studies on the antimicrobial properties and phytochemical screening of garlic (Allium sativum) extracts. Ethnobot. Leafl. 2010, 2009, 10. [Google Scholar]
- Mohamed, M.H.; Badr, E.A.; Sadak, M.S.; Khedr, H.H. Effect of garlic extract, ascorbic acid and nicotinamide on growth, some biochemical aspects, yield and its components of three faba bean (Vicia faba L.) cultivars under sandy soil conditions. Bull. Natl. Res. Cent. 2020, 44, 100. [Google Scholar] [CrossRef]
- Elzaawely, A.A.; Ahmed, M.E.; Maswada, H.F.; Al-Araby, A.A.; Xuan, T.D. Growth traits, physiological parameters and hormonal status of snap bean (Phaseolus vulgaris L.) sprayed with garlic cloves extract. Arch. Agron. Soil Sci. 2018, 64, 1068–1082. [Google Scholar] [CrossRef]
- Arguello, J.; Ledesma, A.; Bottini, R. Hormonal regulation of dormancy in garlic (Allium sativum L.) cv Rosado Paraguayo. Agriscientia 1991, 8, 9–14. [Google Scholar] [CrossRef]
- Rohkin Shalom, S.; Gillett, D.; Zemach, H.; Kimhi, S.; Forer, I.; Zutahy, Y.; Tam, Y.; Teper-Bamnolker, P.; Kamenetsky, R.; Eshel, D. Storage temperature controls the timing of garlic bulb formation via shoot apical meristem termination. Planta 2015, 242, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Beckman, E.J. Supercritical and near-critical CO2 in green chemical synthesis and processing. J. Supercrit. Fluids 2004, 28, 121–191. [Google Scholar] [CrossRef]
- Brunner, G. Supercritical fluids: Technology and application to food processing. J. Food Eng. 2005, 67, 21–33. [Google Scholar] [CrossRef]
- Michalak, I.; Chojnacka, K.; Dmytryk, A.; Wilk, R.; Gramza, M.; Rój, E. Evaluation of supercritical extracts of algae as biostimulants of plant growth in field trials. Front. Plant Sci. 2016, 7, 1591. [Google Scholar] [CrossRef]
- Michalak, I.; Chojnacka, K.; Saeid, A. Plant growth biostimulants, dietary feed supplements and cosmetics formulated with supercritical CO2 algal extracts. Molecules 2017, 22, 66. [Google Scholar] [CrossRef]
- Hincapié, C.A.L.; López, G.E.P.; Torres, R.C. Comparison and characterization of garlic (Allium sativum L.) bulbs extracts and their effect on mortality and repellency of Tetranychus urticae Koch (Acari: Tetranychidae). Chil. J. Agric. Res. 2008, 68, 317–327. [Google Scholar] [CrossRef]
- Bravi, E.; Perretti, G.; Falconi, C.; Marconi, O.; Fantozzi, P. Antioxidant effects of supercritical fluid garlic extracts in sunflower oil. J. Sci. Food Agric. 2017, 97, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Cota-Ruiz, K.; Hernández-Viezcas, J.A.; Valdés, C.; Medina-Velo, I.A.; Turley, R.S.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Manganese nanoparticles control salinity-modulated molecular responses in Capsicum annuum L. through priming: A sustainable approach for agriculture. ACS Sustain. Chem. Eng. 2020, 8, 1427–1436. [Google Scholar] [CrossRef]
- Latef, A.A.H.A.; Alhmad, M.F.A.; Abdelfattah, K.E. The possible roles of priming with ZnO nanoparticles in mitigation of salinity stress in lupine (Lupinus termis) plants. J. Plant Growth Regul. 2017, 36, 60–70. [Google Scholar] [CrossRef]
- Dutta, P. Seed priming: New vistas and contemporary perspectives. In Advances in Seed Priming; Springer: Singapore, 2018; pp. 3–22. [Google Scholar]
- Karny, A.; Zinger, A.; Kajal, A.; Shainsky-Roitman, J.; Schroeder, A. Therapeutic nanoparticles penetrate leaves and deliver nutrients to agricultural crops. Sci. Rep. 2018, 8, 7589. [Google Scholar] [CrossRef] [PubMed]
- Hegedűs, G.; Kutasy, B.; Kiniczky, M.; Decsi, K.; Juhász, Á.; Nagy, Á.; Pallos, J.P.; Virág, E. Liposomal Formulation of Botanical Extracts may Enhance Yield Triggering PR Genes and Phenylpropanoid Pathway in Barley (Hordeum vulgare). Plants 2022, 11, 2969. [Google Scholar] [CrossRef]
- Decsi, K.; Kutasy, B.; Kiniczky, M.; Hegedűs, G.; Virág, E. RNA-seq datasets of field soybean cultures conditioned by Elice16Indures® biostimulator. Data Brief 2022, 42, 108182. [Google Scholar] [CrossRef]
- Decsi, K.; Kutasy, B.; Hegedűs, G.; Alföldi, Z.P.; Kálmán, N.; Nagy, Á.; Virág, E. Natural immunity stimulation using ELICE16INDURES® plant conditioner in field culture of soybean. Heliyon 2023, 9, e12907. [Google Scholar] [CrossRef] [PubMed]
- Kutasy, B.; Decsi, K.; Kiniczky, M.; Hegedűs, G.; Virág, E. Time-course gene expression profiling data of Triticum aestivum treated by supercritical CO2 garlic extract encapsulated in nanoscale liposomes. Data Brief 2022, 42, 108287. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Hegedűs, G.; Nagy, Á.; Decsi, K.; Kutasy, B.; Virág, E. Transcriptome datasets of β-Aminobutyric acid (BABA)-primed mono-and dicotyledonous plants, Hordeum vulgare and Arabidopsis thaliana. Data Brief 2022, 41, 107983. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Beckers, G.J.; Conrath, U. Priming for stress resistance: From the lab to the field. Curr. Opin. Plant Biol. 2007, 10, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Hanachi, A.; Bianchi, A.; Kahn, C.J.; Velot, E.; Arab-Tehrany, E.; Cakir-Kiefer, C.; Linder, M. Encapsulation of Salmon Peptides in Marine Liposomes: Physico-Chemical Properties, Antiradical Activities and Biocompatibility Assays. Mar. Drugs 2022, 20, 249. [Google Scholar] [CrossRef]
- Atif, M.J.; Amin, B.; Ghani, M.I.; Ali, M.; Liu, X.; Zhang, Y.; Cheng, Z. Allium sativum L.(Garlic) bulb enlargement as influenced by differential combinations of photoperiod and temperature. Food Chem. 2021, 338, 127991. [Google Scholar] [CrossRef] [PubMed]
- Travaglia, C.; Reinoso, H.; Cohen, A.; Luna, C.; Tommasino, E.; Castillo, C.; Bottini, R. Exogenous ABA increases yield in field-grown wheat with moderate water restriction. J. Plant Growth Regul. 2010, 29, 366–374. [Google Scholar] [CrossRef]
- Aroca, R.; del Mar Alguacil, M.; Vernieri, P.; Ruiz-Lozano, J.M. Plant responses to drought stress and exogenous ABA application are modulated differently by mycorrhization in tomato and an ABA-deficient mutant (sitiens). Microb. Ecol. 2008, 56, 704–719. [Google Scholar] [CrossRef] [PubMed]
- Chan, Z. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 2012, 100, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Hao, G.P.; Zhang, X.H.; Wang, Y.Q.; Wu, Z.Y.; Huang, C.L. Nucleotide variation in the NCED3 region of Arabidopsis thaliana and its association study with abscisic acid content under drought stress. J. Integr. Plant Biol. 2009, 51, 175–183. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, H.; You, S.; Xu, Y.; Ran, K.; Fan, S. Cloning, characterization and functional analysis of the role MhNCED3, a gene encoding 9-cis-epoxycarotenoid dioxygenase in Malus hupehensis Rehd., plays in plant tolerance to osmotic and Cd2+ stresses. Plant Soil 2014, 381, 143–160. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef]
- Shen, Q.; Chen, C.-N.; Brands, A.; Pan, S.-M.; Tuan-Hua, D.H. The stress-and abscisic acid-induced barley gene HVA22: Developmental regulation and homologues in diverse organisms. Plant Mol. Biol. 2001, 45, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Bergler, J.; Hoth, S. Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. 2011, 13, 725–730. [Google Scholar] [CrossRef]
- Seo, D.H.; Ahn, M.Y.; Park, K.Y.; Kim, E.Y.; Kim, W.T. The N-terminal UND motif of the Arabidopsis U-box E3 ligase PUB18 is critical for the negative regulation of ABA-mediated stomatal movement and determines its ubiquitination specificity for exocyst subunit Exo70B1. Plant Cell 2016, 28, 2952–2973. [Google Scholar] [CrossRef]
- Azevedo, C.; Santos-Rosa, M.J.; Shirasu, K. The U-box protein family in plants. Trends Plant Sci. 2001, 6, 354–358. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Wu, Y.-R.; Huang, X.-H.; Sun, J.; Xie, Q. AtPUB19, a U-box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol. Plant 2011, 4, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Staneloni, R.J.; Rodriguez-Batiller, M.J.; Casal, J.J. Abscisic acid, high-light, and oxidative stress down-regulate a photosynthetic gene via a promoter motif not involved in phytochrome-mediated transcriptional regulation. Mol. Plant 2008, 1, 75–83. [Google Scholar] [CrossRef]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Zarka, D.G.; Van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005, 41, 195–211. [Google Scholar] [CrossRef]
- González-Diéguez, D.O.; Orozco, C. Identificación de marcadores moleculares (SNP) y genes candidatos asociados a la tolerancia genética al complejo mancha de asfalto en variedades nativas de maíz (Zea mays L.). Cienc. Tecnol. Salud 2020, 7, 218–235. [Google Scholar] [CrossRef]
- Pasini, L.; Bergonti, M.; Fracasso, A.; Marocco, A.; Amaducci, S. Microarray analysis of differentially expressed mRNAs and miRNAs in young leaves of sorghum under dry-down conditions. J. Plant Physiol. 2014, 171, 537–548. [Google Scholar] [CrossRef]
- Chaudhary, C.; Sharma, N.; Khurana, P. Decoding the wheat awn transcriptome and overexpressing Ta Rca1β in rice for heat stress tolerance. Plant Mol. Biol. 2021, 105, 133–146. [Google Scholar] [CrossRef]
- Mohanty, B. Genomic architecture of promoters and transcriptional regulation of candidate genes in rice involved in tolerance to anaerobic germination. Curr. Plant Biol. 2022, 29, 100236. [Google Scholar] [CrossRef]
- Lu, C.-A.; Lin, C.-C.; Lee, K.-W.; Chen, J.-L.; Huang, L.-F.; Ho, S.-L.; Liu, H.-J.; Hsing, Y.-I.; Yu, S.-M. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 2007, 19, 2484–2499. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Moon, S.-J.; Min, M.K.; Kim, J.; Kim, D.Y.; Yoon, I.S.; Kwon, T.R.; Byun, M.O.; Kim, B.-G. Ectopic expression of OsDREB1G, a member of the OsDREB1 subfamily, confers cold stress tolerance in rice. Front. Plant Sci. 2019, 10, 297. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Medina, J.; Catalá, R.; Salinas, J. The CBFs: Three Arabidopsis transcription factors to cold acclimate. Plant Sci. 2011, 180, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Sun, J.; Gong, D.; Kong, Y. The roles of Arabidopsis C1-2i subclass of C2H2-type zinc-finger transcription factors. Genes 2019, 10, 653. [Google Scholar] [CrossRef]
- Maruyama, K.; Sakuma, Y.; Kasuga, M.; Ito, Y.; Seki, M.; Goda, H.; Shimada, Y.; Yoshida, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004, 38, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Friesen, T.L.; Faris, J.D. Molecular characterization and genomic mapping of the pathogenesis-related protein 1 (PR-1) gene family in hexaploid wheat (Triticum aestivum L.). Mol. Genet. Genom. 2011, 285, 485–503. [Google Scholar] [CrossRef]
- Naz, R.; Bano, A.; Wilson, N.L.; Guest, D.; Roberts, T.H. Pathogenesis-related protein expression in the apoplast of wheat leaves protected against leaf rust following application of plant extracts. Phytopathology 2014, 104, 933–944. [Google Scholar] [CrossRef]
- Kemp, G.; Botha, A.-M.; Kloppers, F.; Pretorius, Z. Disease development and β-1, 3-glucanase expression following leaf rust infection in resistant and susceptible near-isogenic wheat seedlings. Physiol. Mol. Plant Pathol. 1999, 55, 45–52. [Google Scholar] [CrossRef]
- Liu, B.; Xue, X.; Cui, S.; Zhang, X.; Han, Q.; Zhu, L.; Liang, X.; Wang, X.; Huang, L.; Chen, X. Cloning and characterization of a wheat β-1, 3-glucanase gene induced by the stripe rust pathogen Puccinia striiformis f. sp. tritici. Mol. Biol. Rep. 2010, 37, 1045. [Google Scholar] [CrossRef]
- Kasprzewska, A. Plant chitinases-regulation and function. Cell. Mol. Biol. Lett. 2003, 8, 809–824. [Google Scholar] [PubMed]
- Bertini, L.; Caporale, C.; Testa, M.; Proietti, S.; Caruso, C. Structural basis of the antifungal activity of wheat PR4 proteins. FEBS Lett. 2009, 583, 2865–2871. [Google Scholar] [CrossRef] [PubMed]
- Caporale, C.; Di Berardino, I.; Leonardi, L.; Bertini, L.; Cascone, A.; Buonocore, V.; Caruso, C. Wheat pathogenesis-related proteins of class 4 have ribonuclease activity. FEBS Lett. 2004, 575, 71–76. [Google Scholar] [CrossRef]
- De Jesús-Pires, C.; Ferreira-Neto, J.R.; Pacifico Bezerra-Neto, J.; Kido, E.A.; de Oliveira Silva, R.L.; Pandolfi, V.; Wanderley-Nogueira, A.C.; Binneck, E.; da Costa, A.F.; Pio-Ribeiro, G. Plant thaumatin-like proteins: Function, evolution and biotechnological applications. Curr. Protein Pept. Sci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Owens, D.K.; Crosby, K.C.; Runac, J.; Howard, B.A.; Winkel, B.S. Biochemical and genetic characterization of Arabidopsis flavanone 3β-hydroxylase. Plant Physiol. Biochem. 2008, 46, 833–843. [Google Scholar] [CrossRef]
- Han, Y.; Huang, K.; Liu, Y.; Jiao, T.; Ma, G.; Qian, Y.; Wang, P.; Dai, X.; Gao, L.; Xia, T. Functional analysis of two flavanone-3-hydroxylase genes from Camellia sinensis: A critical role in flavonoid accumulation. Genes 2017, 8, 300. [Google Scholar] [CrossRef]
- Rakwal, R.; Agrawal, G.K.; Jwa, N.-S. Characterization of a rice (Oryza sativa L.) Bowman–Birk proteinase inhibitor: Tightly light regulated induction in response to cut, jasmonic acid, ethylene and protein phosphatase 2A inhibitors. Gene 2001, 263, 189–198. [Google Scholar] [CrossRef]
- Wu, S.-W.; Wang, H.-W.; Yang, Z.-D.; Kong, L.-R. Expression comparisons of pathogenesis-related (PR) genes in wheat in response to infection/infestation by Fusarium, Yellow dwarf virus (YDV) aphid-transmitted and hessian fly. J. Integr. Agric. 2014, 13, 926–936. [Google Scholar] [CrossRef]
- Pasquer, F. Effects of Plant Protection Compounds on Wheat Gene Expression. Ph.D. Thesis, University of Zurich, Zürich, Switzerland, 2005. [Google Scholar]
- Carbonero, P.; Díaz, I.; Vicente-Carbajosa, J.; Alfonso-Rubi, J.; Gaddour, K.; Lara, P. Cereal α-amylase/trypsin inhibitors and transgenic insect resistance. In Genetics and Breeding for Crop Quality and Resistance; Springer: Ordrecht, The Netherlands, 1999; pp. 147–158. [Google Scholar] [CrossRef]
- Erdayani, E.; Nagarajan, R.; Grant, N.P.; Gill, K.S. Genome-wide analysis of the HSP101/CLPB gene family for heat tolerance in hexaploid wheat. Sci. Rep. 2020, 10, 3948. [Google Scholar] [CrossRef] [PubMed]
- Biselli, C.; Bagnaresi, P.; Faccioli, P.; Hu, X.; Balcerzak, M.; Mattera, M.G.; Yan, Z.; Ouellet, T.; Cattivelli, L.; Valè, G. Comparative transcriptome profiles of near-isogenic hexaploid wheat lines differing for effective alleles at the 2DL FHB resistance QTL. Front. Plant Sci. 2018, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Douchkov, D.; Johrde, A.; Nowara, D.; Himmelbach, A.; Lueck, S.; Niks, R.; Schweizer, P. Convergent evidence for a role of WIR1 proteins during the interaction of barley with the powdery mildew fungus Blumeria graminis. J. Plant Physiol. 2011, 168, 20–29. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, H.; Li, X. Ubiquitin-specific protease16 interacts with a heavy metal associated isoprenylated plant protein27 and modulates cadmium tolerance. Plant Signal. Behav. 2013, 8, e25680. [Google Scholar] [CrossRef]
- De Abreu-Neto, J.B.; Turchetto-Zolet, A.C.; de Oliveira, L.F.V.; Bodanese Zanettini, M.H.; Margis-Pinheiro, M. Heavy metal-associated isoprenylated plant protein (HIPP): Characterization of a family of proteins exclusive to plants. FEBS J. 2013, 280, 1604–1616. [Google Scholar] [CrossRef]
- Dooner, H.K.; Robbins, T.P.; Jorgensen, R.A. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 1991, 25, 173–199. [Google Scholar] [CrossRef]
- Tan, D.-X.; Hardeland, R.; Manchester, L.C.; Korkmaz, A.; Ma, S.; Rosales-Corral, S.; Reiter, R.J. Functional roles of melatonin in plants, and perspectives in nutritional and agricultural science. J. Exp. Bot. 2012, 63, 577–597. [Google Scholar] [CrossRef]
- Cui, G.; Zhao, X.; Liu, S.; Sun, F.; Zhang, C.; Xi, Y. Beneficial effects of melatonin in overcoming drought stress in wheat seedlings. Plant Physiol. Biochem. 2017, 118, 138–149. [Google Scholar] [CrossRef]
- Jain, D.; Khurana, J.P. Role of pathogenesis-related (PR) proteins in plant defense mechanism. In Molecular Aspects of Plant-Pathogen Interaction; Springer: Singapore, 2018; pp. 265–281. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef]
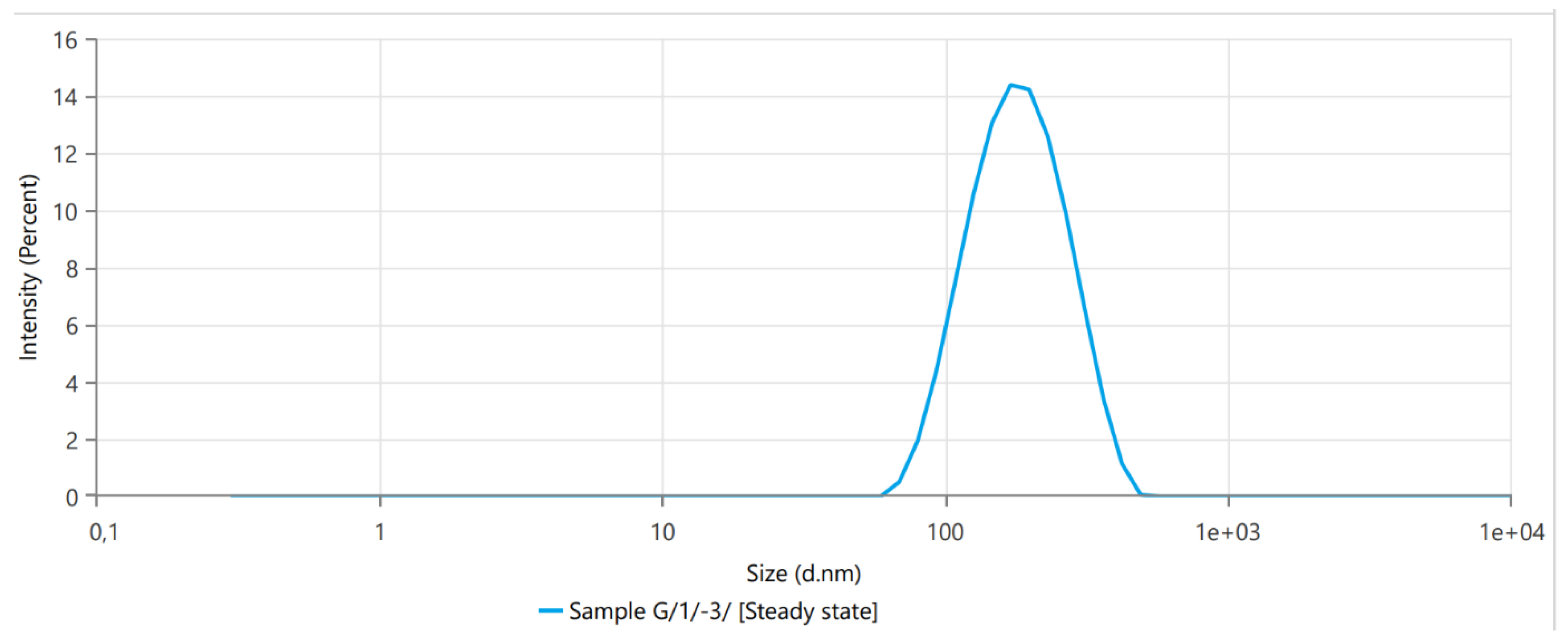
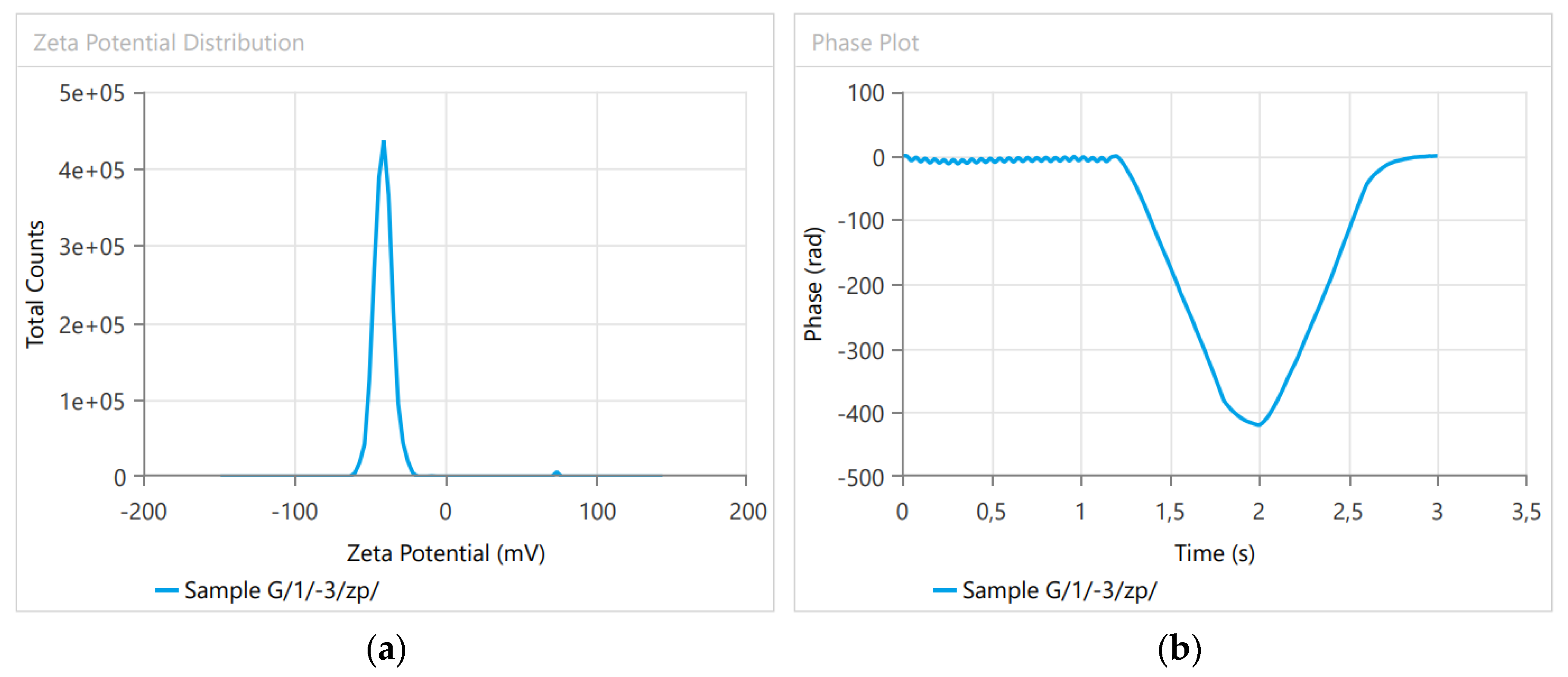

| Name | Mean | Minimum | Maximum |
|---|---|---|---|
| Z-Average (nm) | 149.2 | 134.2 | 164.2 |
| Polydispersity Index (PI) | 0.13035 | 0.1303 | 0.1304 |
| Intercept | 0.951 | 0.949 | 0.953 |
| Peak One Mean by Intensity (nm) | 160.6 | 129.6 | 191.6 |
| Peak One Area by Intensity (%) | 100 | 100 | 100 |
| Derived Mean Count Rate (kcps) | 171,300 | 1.71 × 105 | 1.71 × 105 |
| Name | Mean | Minimum | Maximum |
|---|---|---|---|
| Derived Mean Count Rate (kcps) | 171,300 | 1.71 × 105 | 1.71 × 105 |
| Zeta Potential (mV) | −40.595 | −40.66 | −40.53 |
| Zeta Peak One Mean | −41.18 | −41.43 | −40.93 |
| Zeta Peak Two Mean | −9.2 | −9.25 | −9.15 |
| Conductivity (mS/cm) | 0.007439 | 0.007399 | 0.007479 |
| Wall Zeta Potential (mV) | −66.28 | −66.33 | −66.23 |
| Zeta Deviation (mV) | 8.632 | 8.607 | 8.657 |
| Derived Mean Count Rate (kcps) | 85,150 | 8.52 × 104 | 8.52 × 104 |
| Reference Beam Count Rate (kcps) | 3671.5 | 3645 | 3698 |
| Quality Factor | 4.509 | 4.508 | 4.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kutasy, B.; Kiniczky, M.; Decsi, K.; Kálmán, N.; Hegedűs, G.; Alföldi, Z.P.; Virág, E. ‘Garlic-lipo’4Plants: Liposome-Encapsulated Garlic Extract Stimulates ABA Pathway and PR Genes in Wheat (Triticum aestivum). Plants 2023, 12, 743. https://doi.org/10.3390/plants12040743
Kutasy B, Kiniczky M, Decsi K, Kálmán N, Hegedűs G, Alföldi ZP, Virág E. ‘Garlic-lipo’4Plants: Liposome-Encapsulated Garlic Extract Stimulates ABA Pathway and PR Genes in Wheat (Triticum aestivum). Plants. 2023; 12(4):743. https://doi.org/10.3390/plants12040743
Chicago/Turabian StyleKutasy, Barbara, Márta Kiniczky, Kincső Decsi, Nikoletta Kálmán, Géza Hegedűs, Zoltán Péter Alföldi, and Eszter Virág. 2023. "‘Garlic-lipo’4Plants: Liposome-Encapsulated Garlic Extract Stimulates ABA Pathway and PR Genes in Wheat (Triticum aestivum)" Plants 12, no. 4: 743. https://doi.org/10.3390/plants12040743
APA StyleKutasy, B., Kiniczky, M., Decsi, K., Kálmán, N., Hegedűs, G., Alföldi, Z. P., & Virág, E. (2023). ‘Garlic-lipo’4Plants: Liposome-Encapsulated Garlic Extract Stimulates ABA Pathway and PR Genes in Wheat (Triticum aestivum). Plants, 12(4), 743. https://doi.org/10.3390/plants12040743