Nitrogen Fertilizer Type and Genotype as Drivers of P Acquisition and Rhizosphere Microbiota Assembly in Juvenile Maize Plants
Abstract
1. Introduction
2. Results
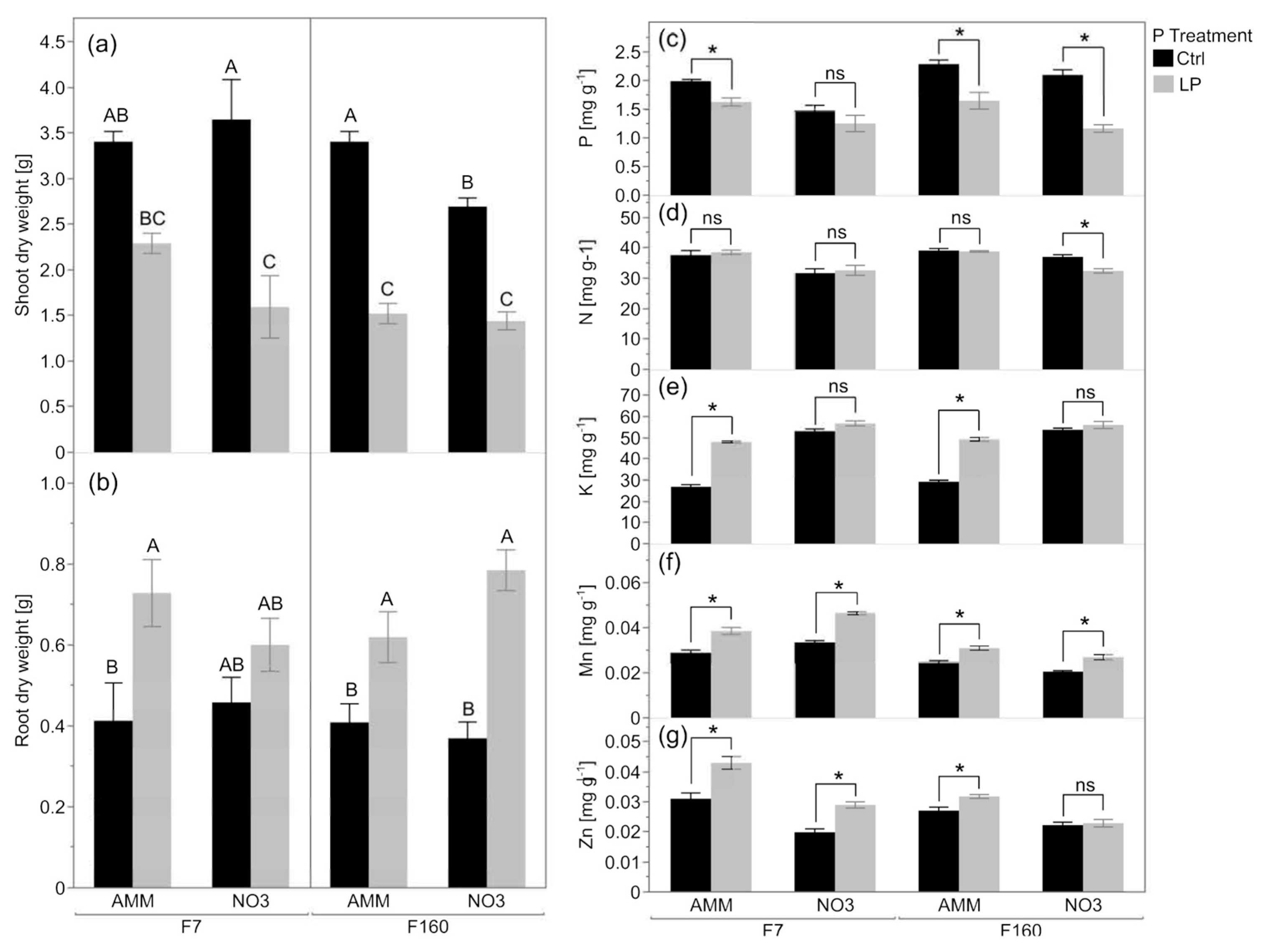
2.1. Low Pi Availability Reduces Maize Biomass and Affects the General Nutritional Status
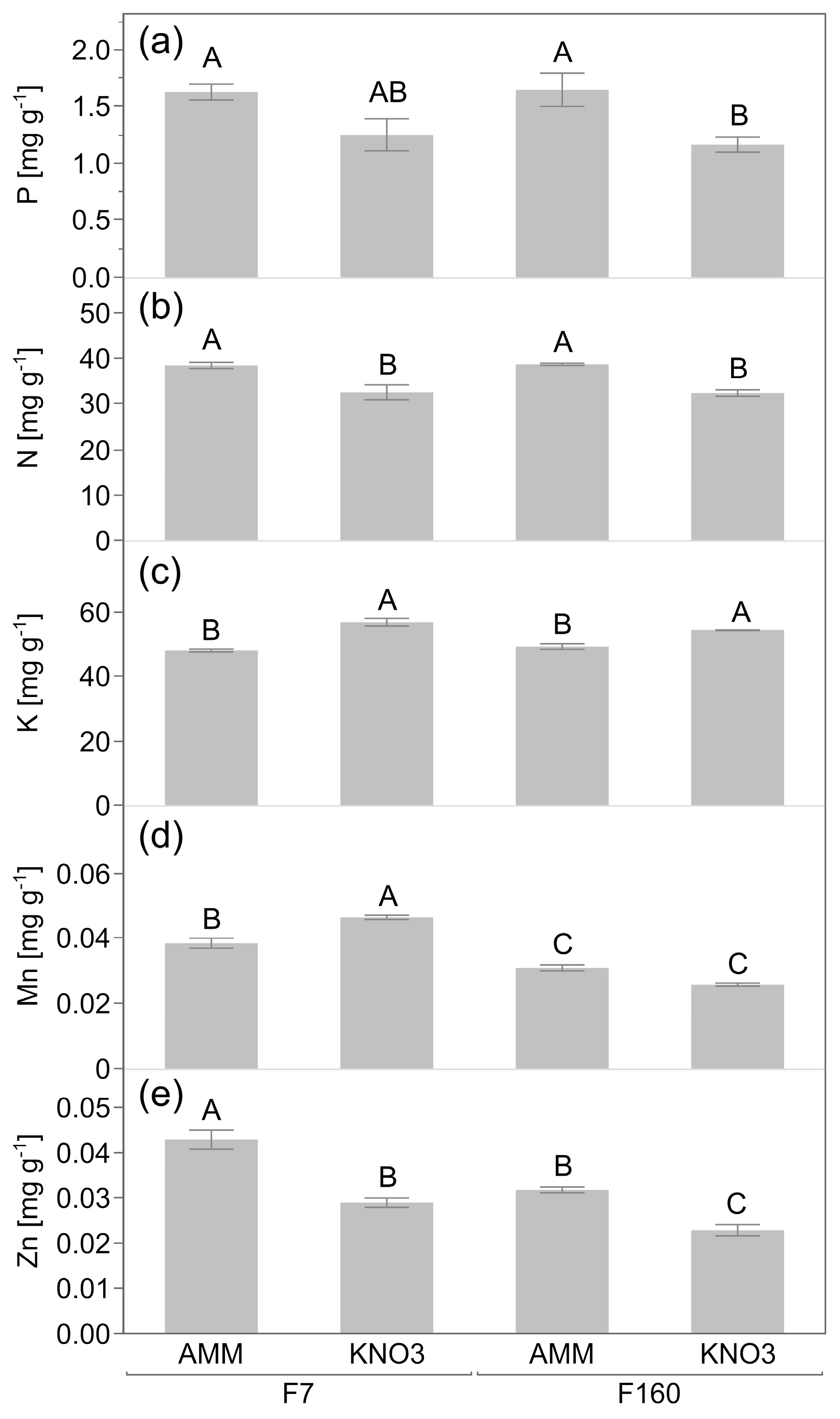
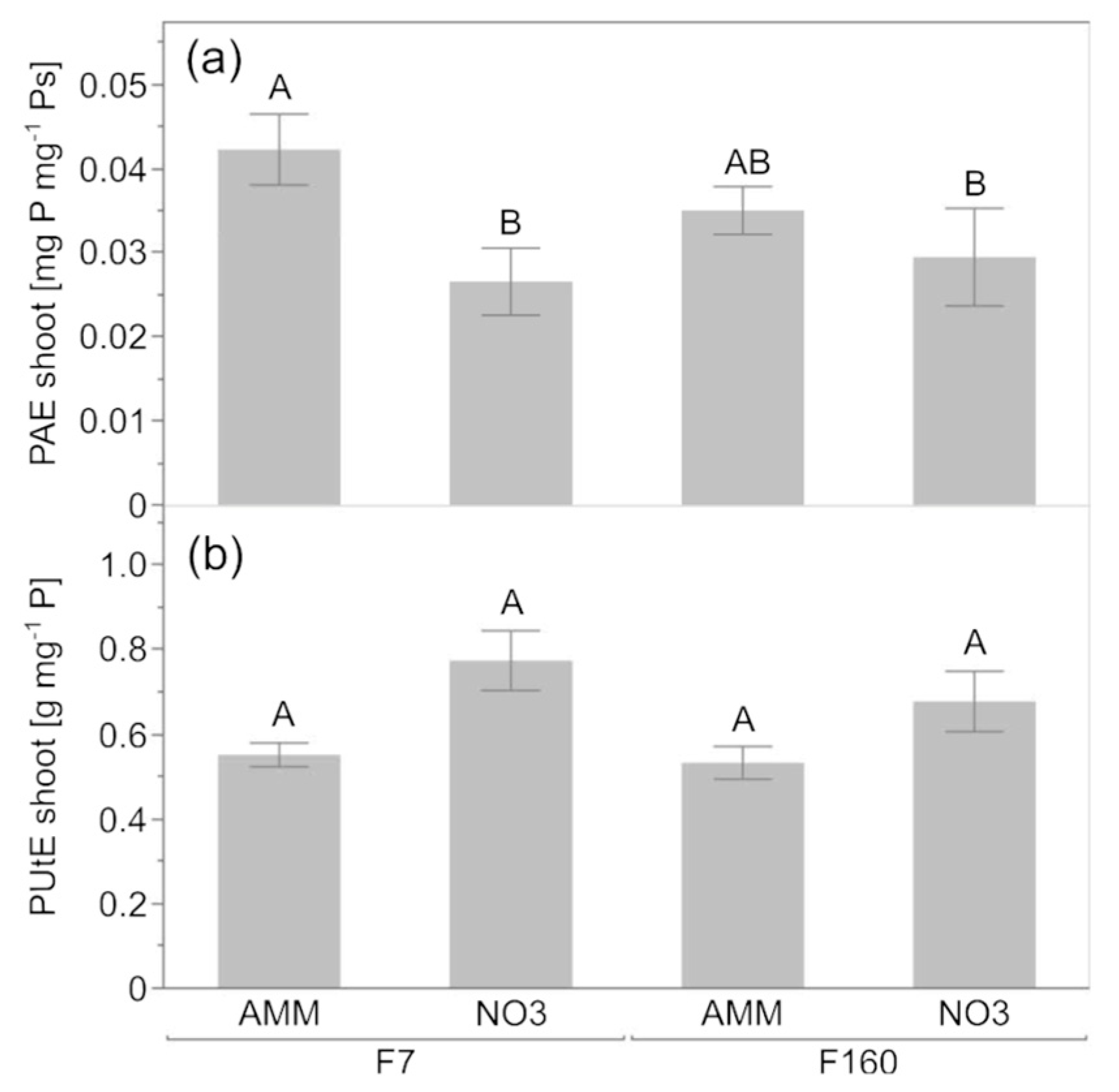
2.2. Effect of Genotype and N Nutrition on Plant Attributes in P-Limited Soil
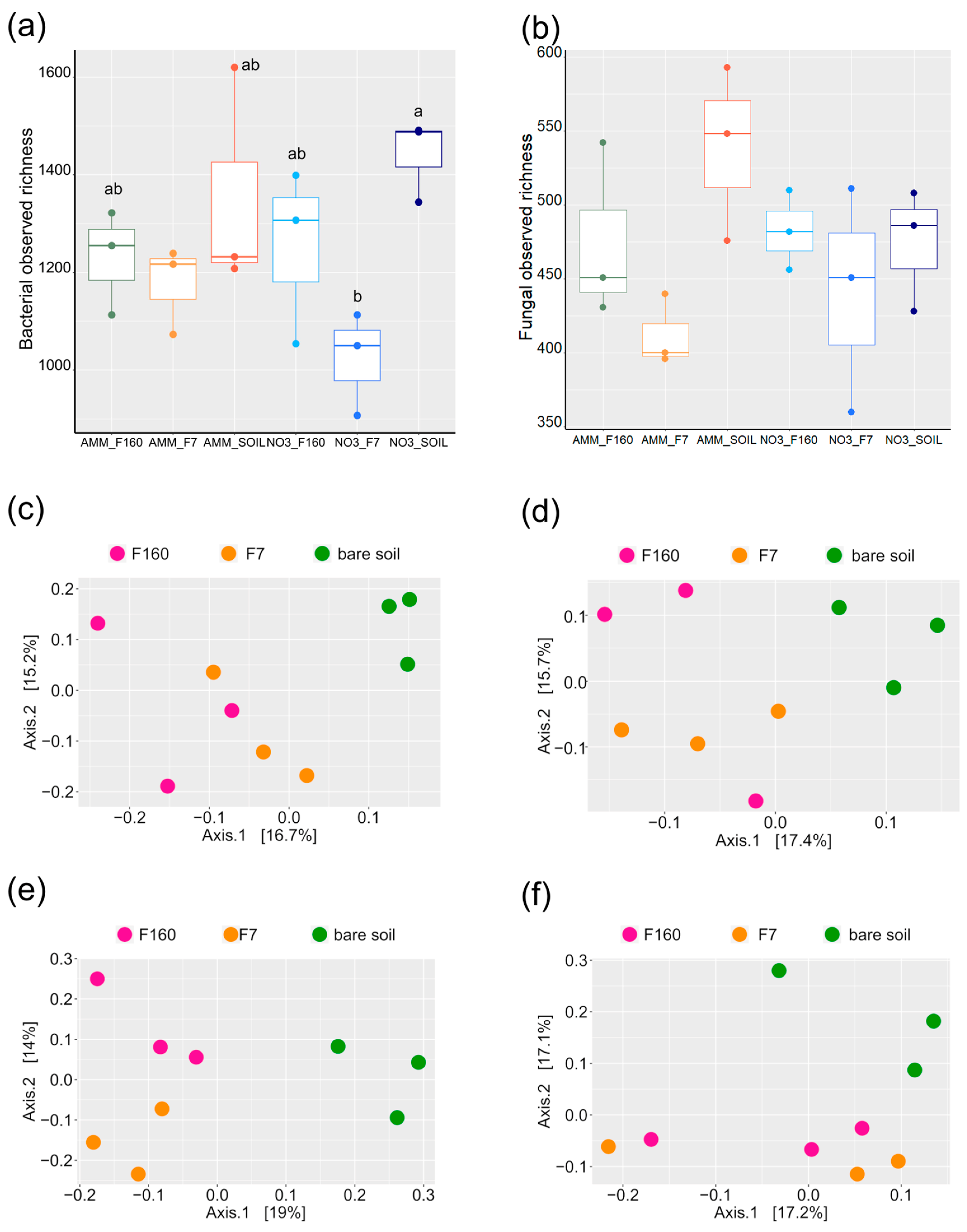
2.3. Microbial Composition of the Anlayzed Samples
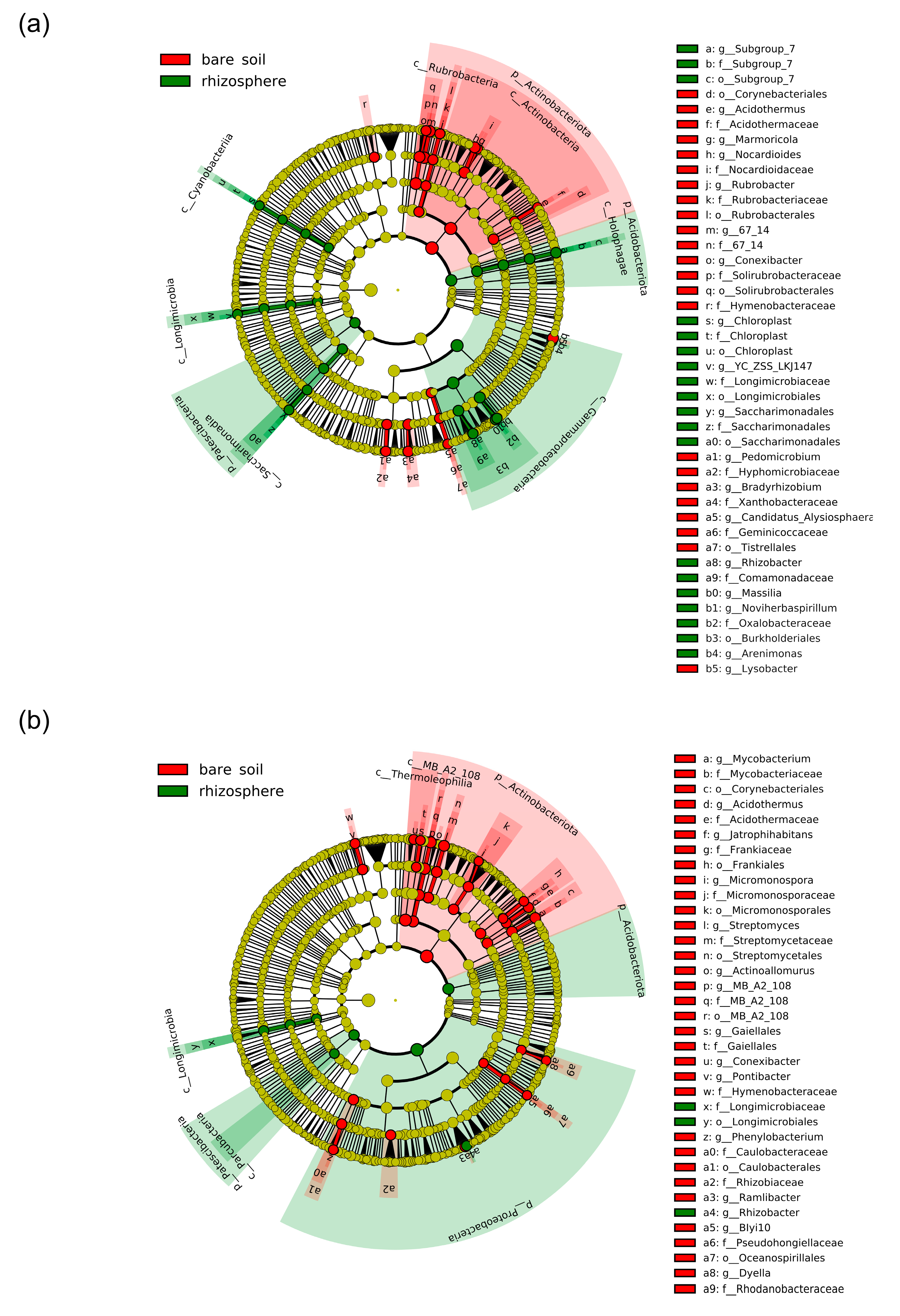
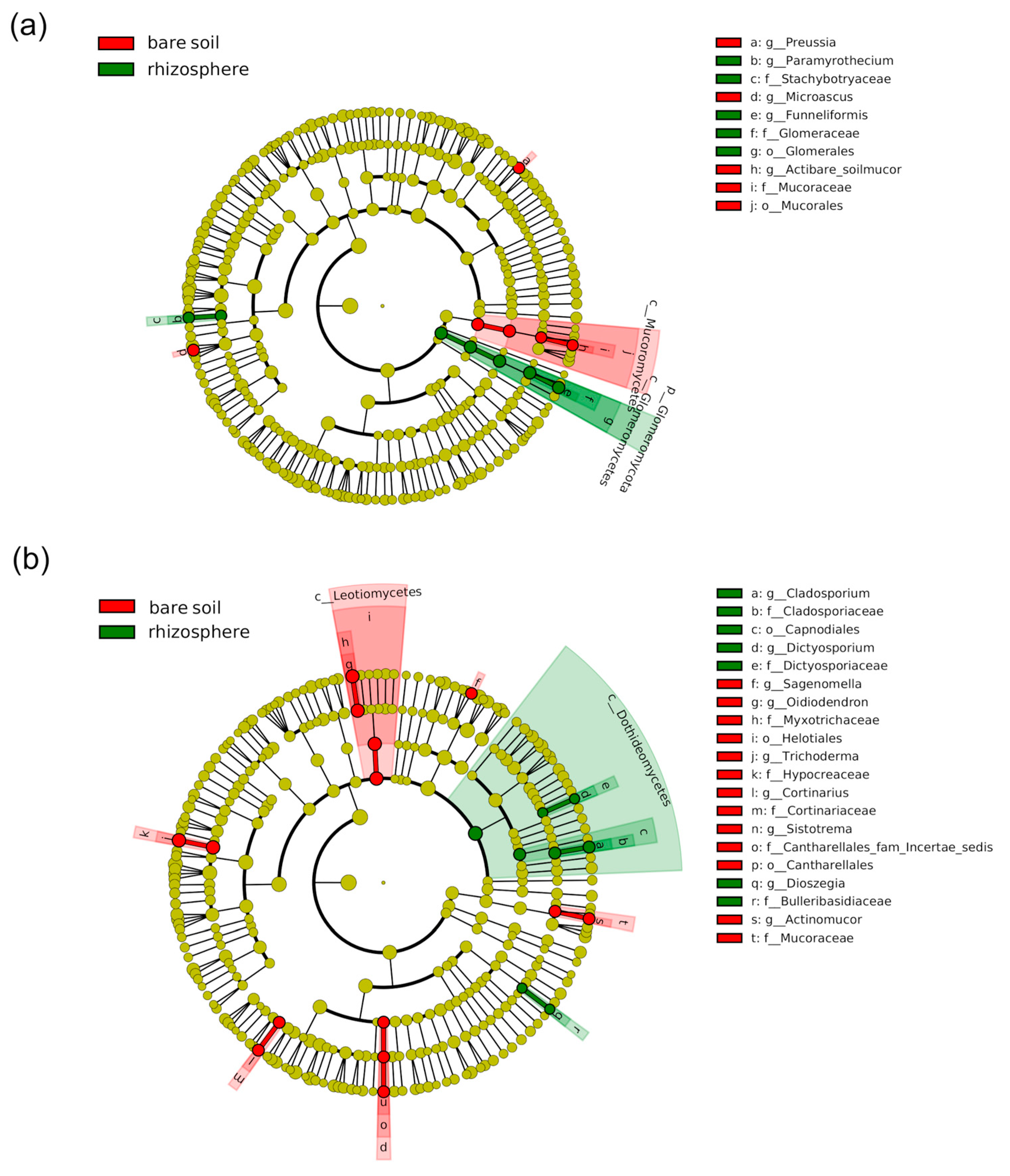
2.4. Effect of Plant Presence and Nitrogen Fertilization on Bacterial and Fungal Communities
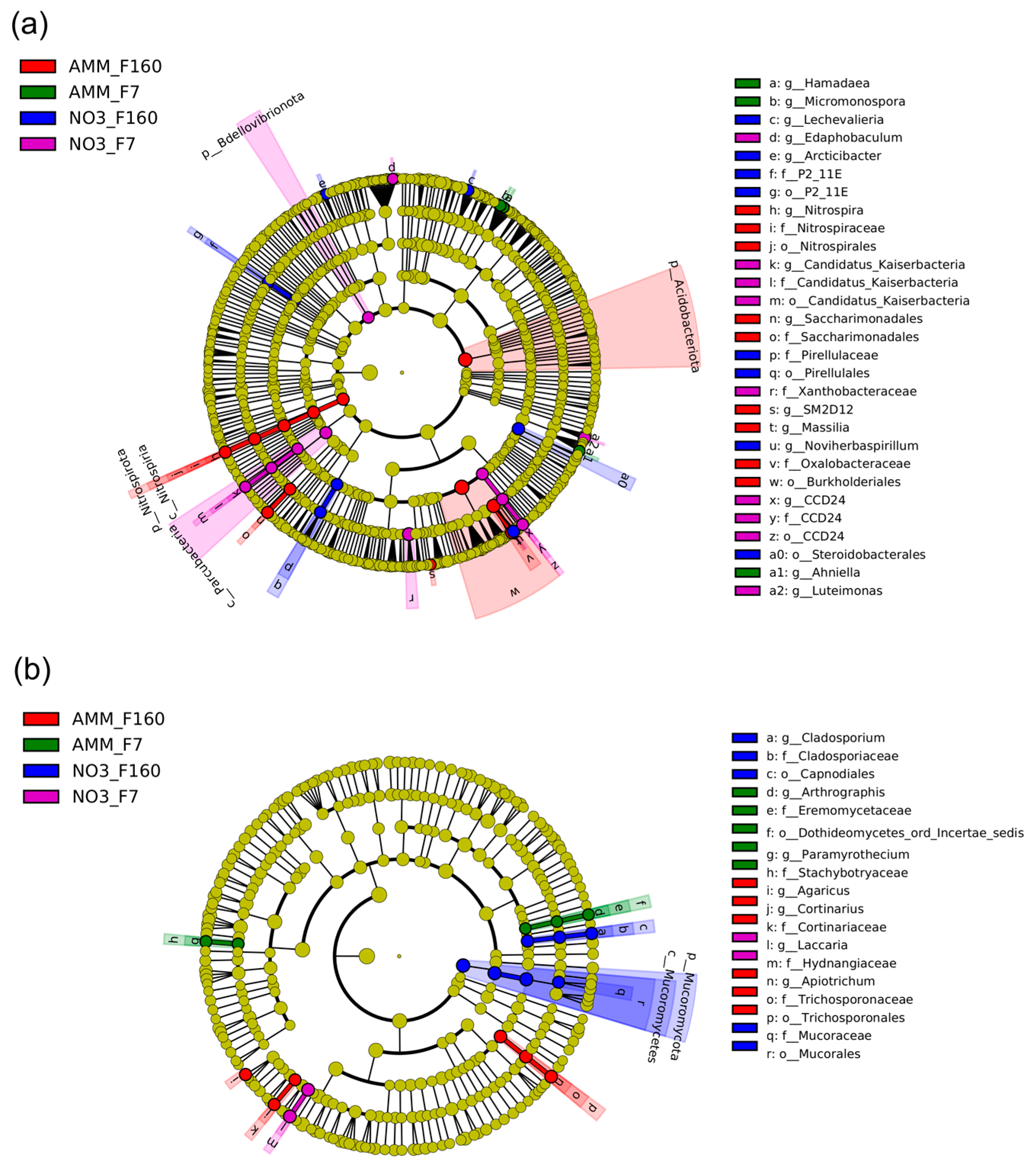
2.5. Differential Effect of N Nutrition and Maize Genotype on Bacterial and Fungal Rhizosphere Communities
3. Discussion
4. Materials and Methods
4.1. Experimental Setup
4.2. Maize Genotype Used in the Study
4.3. Determination of Plant Fitness Parameters
4.4. Definition and Calculation of Phosphorus Use Efficiency
4.5. Root Mycorrhizal Colonization
4.6. Microbiome Analysis
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bieleski, R.L. Phosphate Pools, Phosphate Transport, and Phosphate Availability. Annu. Rev. Plant Physiol. 1973, 24, 225–252. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.-F.; Wu, W.-H. Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 2021, 63, 34–52. [Google Scholar] [CrossRef] [PubMed]
- Barrow, N.J. The effects of pH on phosphate uptake from the soil. Plant Soil 2017, 410, 401–410. [Google Scholar] [CrossRef]
- Penn, C.; Camberato, J. A Critical Review on Soil Chemical Processes that Control How Soil pH Affects Phosphorus Availability to Plants. Agriculture 2019, 9, 120. [Google Scholar] [CrossRef]
- Lynch, J.P.; Brown, K.M. Root strategies for phosphorus acquisition. In The Ecophysiology of Plant-Phosphorus Interactions; Kok, L.J., de Hawkesford, M.J., Stulen, I., White, P.J., Hammond, J.P., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 83–116. ISBN 978-1-4020-8434-8. [Google Scholar]
- Zhu, J.; Mickelson, S.M.; Kaeppler, S.M.; Lynch, J.P. Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theor. Appl. Genet. 2006, 113, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Strock, C.F.; Schneider, H.M.; Sidhu, J.S.; Ajmera, I.; Galindo-Castañeda, T.; Klein, S.P.; Hanlon, M.T. Root anatomy and soil resource capture. Plant Soil 2021, 466, 21–63. [Google Scholar] [CrossRef]
- Fan, M.; Zhu, J.; Richards, C.; Brown, K.M.; Lynch, J.P. Physiological roles for aerenchyma in phosphorus-stressed roots. Funct. Plant Biol. 2003, 30, 493–506. [Google Scholar] [CrossRef]
- Neumann, G. Root Exudates and Nutrient Cycling. In Nutrient Cycling in Terrestrial Ecosystems; Marschner, P., Rengel, Z., Eds.; Springer: Berlin, Heidelberg, 2007; pp. 123–157. ISBN 978-3-540-68026-0. [Google Scholar]
- McKay Fletcher, D.M.; Ruiz, S.; Dias, T.; Petroselli, C.; Roose, T. Linking root structure to functionality: The impact of root system architecture on citrate-enhanced phosphate uptake. New. Phytol. 2020, 227, 376–391. [Google Scholar] [CrossRef]
- Chai, Y.N.; Schachtman, D.P. Root exudates impact plant performance under abiotic stress. Trends Plant Sci. 2022, 27, 80–91. [Google Scholar] [CrossRef]
- Etesami, H.; Jeong, B.R.; Glick, B.R. Contribution of Arbuscular Mycorrhizal Fungi, Phosphate-Solubilizing Bacteria, and Silicon to P Uptake by Plant. Front. Plant Sci. 2021, 12, 699618. [Google Scholar] [CrossRef]
- Kothari, S.K.; Marschner, H.; Römheld, V. Contribution of the VA mycorrhizal hyphae in acquisition of phosphorus and zinc by maize grown in a calcareous soil. Plant Soil 1991, 131, 177–185. [Google Scholar] [CrossRef]
- Battini, F.; Grønlund, M.; Agnolucci, M.; Giovannetti, M.; Jakobsen, I. Facilitation of phosphorus uptake in maize plants by mycorrhizosphere bacteria. Sci. Rep. 2017, 7, 4686. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, X.; Ludewig, U. Arbuscular mycorrhizal colonization outcompetes root hairs in maize under low phosphorus availability. Ann. Bot. 2021, 127, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Mang, M.; Piepho, H.-P.; Melchinger, A.; Ludewig, U. Decline of seedling phosphorus use efficiency in the heterotic pool of flint maize breeding lines since the onset of hybrid breeding. J. Agron. Crop Sci. 2021, 207, 857–872. [Google Scholar] [CrossRef]
- Kaeppler, S.M.; Parke, J.L.; Mueller, S.M.; Senior, L.; Stuber, C.; Tracy, W.F. Variation among Maize Inbred Lines and Detection of Quantitative Trait Loci for Growth at Low Phosphorus and Responsiveness to Arbuscular Mycorrhizal Fungi. Crop Sci. 2000, 40, 358–364. [Google Scholar] [CrossRef]
- Sawers, R.J.H.; Svane, S.F.; Quan, C.; Grønlund, M.; Wozniak, B.; Gebreselassie, M.-N.; González-Muñoz, E.; Chávez Montes, R.A.; Baxter, I.; Goudet, J.; et al. Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root-external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytol. 2017, 214, 632–643. [Google Scholar] [CrossRef]
- Li, X.; Quan, X.; Mang, M.; Neumann, G.; Melchinger, A.; Ludewig, U. Flint maize root mycorrhization and organic acid exudates under phosphorus deficiency: Trends in breeding lines and doubled haploid lines from landraces. J. Plant Nutr. Soil Sci. 2021, 184, 346–359. [Google Scholar] [CrossRef]
- Grunes, D.L. Effect of Nitrogen on the Availability of Soil and Fertilizer Phosphorus to Plants. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 1995; pp. 369–396. [Google Scholar]
- Krouk, G.; Kiba, T. Nitrogen and Phosphorus interactions in plants: From agronomic to physiological and molecular insights. Curr. Opin. Plant Biol. 2020, 57, 104–109. [Google Scholar] [CrossRef]
- Pasley, H.R.; Cairns, J.E.; Camberato, J.J.; Vyn, T.J. Nitrogen fertilizer rate increases plant uptake and soil availability of essential nutrients in continuous maize production in Kenya and Zimbabwe. Nutr. Cycl. Agroecosyst. 2019, 115, 373–389. [Google Scholar] [CrossRef]
- Pedersen, I.F.; Sørensen, P.; Rasmussen, J.; Withers, P.J.A.; Rubæk, G.H. Fertilizer ammonium: Nitrate ratios determine phosphorus uptake by young maize plants. J. Plant Nutr. Soil Sci. 2019, 182, 541–551. [Google Scholar] [CrossRef]
- Wang, P.; Wang, Z.; Pan, Q.; Sun, X.; Chen, H.; Chen, F.; Yuan, L.; Mi, G. Increased biomass accumulation in maize grown in mixed nitrogen supply is mediated by auxin synthesis. J. Exp. Bot. 2019, 70, 1859–1873. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.E.; Kojima, S.; Takahashi, H.; von Wirén, N. Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 2010, 22, 3621–3633. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A. Nitrate Transport, Sensing, and Responses in Plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Ravazzolo, L.; Trevisan, S.; Forestan, C.; Varotto, S.; Sut, S.; Dall’Acqua, S.; Malagoli, M.; Quaggiotti, S. Nitrate and Ammonium Affect the Overall Maize Response to Nitrogen Availability by Triggering Specific and Common Transcriptional Signatures in Roots. Int. J. Mol. Sci. 2020, 21, 686. [Google Scholar] [CrossRef]
- Wang, B.; Zhu, X.; Guo, X.; Qi, X.; Feng, F.; Zhang, Y.; Zhao, Q.; Han, D.; Sun, H. Nitrate Modulates Lateral Root Formation by Regulating the Auxin Response and Transport in Rice. Genes 2021, 12, 850. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; von Wirén, N. Signaling pathways underlying nitrogen-dependent changes in root system architecture: From model to crop species. J. Exp. Bot. 2020, 71, 4393–4404. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Zaidi, A.; Wani, P.A. Role of Phosphate Solubilizing Microorganisms in Sustainable Agriculture—A Review. In Sustainable Agriculture; Lichtfouse, E., Navarrete, M., Debaeke, P., Véronique, S., Alberola, C., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 551–570. ISBN 978-90-481-2665-1. [Google Scholar]
- Ahuja, A.; Ghosh, S.B.; D’Souza, S.F. Isolation of a starch utilizing, phosphate solubilizing fungus on buffered medium and its characterization. Bioresour. Technol. 2007, 98, 3408–3411. [Google Scholar] [CrossRef]
- Matić, S.; Gilardi, G.; Gullino, M.L.; Garibaldi, A. Emergence of Leaf Spot Disease on Leafy Vegetable and Ornamental Crops Caused by Paramyrothecium and Albifimbria Species. Phytopathology 2019, 109, 1053–1061. [Google Scholar] [CrossRef]
- Huo, J.; Yao, Y.; Ben, H.; Tian, T.; Yang, L.; Wang, Y.; Bai, P.; Hao, Y.; Wang, W. First Report of Paramyrothecium foliicola Causing Stem Canker of Cucumber (Cucumis sativus L.) seedlings in China. Plant Dis. 2021, 105, 1859. [Google Scholar] [CrossRef] [PubMed]
- Alewell, C.; Ringeval, B.; Ballabio, C.; Robinson, D.A.; Panagos, P.; Borrelli, P. Global phosphorus shortage will be aggravated by soil erosion. Nat. Commun. 2020, 11, 4546. [Google Scholar] [CrossRef] [PubMed]
- Barry, D.A.J.; Miller, M.H. Phosphorus Nutritional Requirement of Maize Seedlings for Maximum Yield. Agron. J. 1989, 81, 95–99. [Google Scholar] [CrossRef]
- Zhu, J.; Lynch, J.P. The contribution of lateral rooting to phosphorus acquisition efficiency in maize (Zea mays) seedlings. Funct. Plant Biol. 2004, 31, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Niu, L.; Wei, J.; Chen, X.; Chen, Y. Phosphorus Limitation Improved Salt Tolerance in Maize Through Tissue Mass Density Increase, Osmolytes Accumulation, and Na+ Uptake Inhibition. Front. Plant Sci. 2019, 10, 856. [Google Scholar] [CrossRef] [PubMed]
- Postma, J.A.; Dathe, A.; Lynch, J.P. The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol. 2014, 166, 590–602. [Google Scholar] [CrossRef]
- Jia, X.; Liu, P.; Lynch, J.P. Greater lateral root branching density in maize improves phosphorus acquisition from low phosphorus soil. J. Exp. Bot. 2018, 69, 4961–4970. [Google Scholar] [CrossRef]
- Geisseler, D.; Scow, K.M. Long-term effects of mineral fertilizers on soil microorganisms—A review. Soil Biol. Biochem. 2014, 75, 54–63. [Google Scholar] [CrossRef]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front. Microbiol. 2016, 7, 1446. [Google Scholar] [CrossRef]
- Hinsinger, P. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: A review. Plant Soil 2001, 237, 173–195. [Google Scholar] [CrossRef]
- Chen, J.-H.; Barber, S.A. Soil pH and Phosphorus and Potassium Uptake by Maize Evaluated with an Uptake Model. Soil Sci. Soc. Am. J. 1990, 54, 1032–1036. [Google Scholar] [CrossRef]
- Hoffmann, C.; Ladewig, E.; Claassen, N.; Jungk, A. Phosphorus uptake of maize as affected by ammonium and nitrate nitrogenMeasurements and model calculations. Z. Für Pflanz. Und Bodenkd. 1994, 157, 225–232. [Google Scholar] [CrossRef]
- Yang, J.T.; Schneider, H.M.; Brown, K.M.; Lynch, J.P. Genotypic variation and nitrogen stress effects on root anatomy in maize are node specific. J. Exp. Bot. 2019, 70, 5311–5325. [Google Scholar] [CrossRef]
- Mwafulirwa, L.; Paterson, E.; Cairns, J.E.; Daniell, T.J.; Thierfelder, C.; Baggs, E.M. Genotypic variation in maize (Zea mays) influences rates of soil organic matter mineralization and gross nitrification. New. Phytol. 2021, 231, 2015–2028. [Google Scholar] [CrossRef] [PubMed]
- McCormack, M.L.; Dickie, I.A.; Eissenstat, D.M.; Fahey, T.J.; Fernandez, C.W.; Guo, D.; Helmisaari, H.-S.; Hobbie, E.A.; Iversen, C.M.; Jackson, R.B.; et al. Redefining fine roots improves understanding of below-ground contributions to terrestrial biosphere processes. New. Phytol. 2015, 207, 505–518. [Google Scholar] [CrossRef]
- Jansa, J.; Mozafar, A.; Frossard, E. Phosphorus Acquisition Strategies within Arbuscular Mycorrhizal Fungal Community of a Single Field Site. Plant Soil 2005, 276, 163–176. [Google Scholar] [CrossRef]
- Clark, R.B.; Zeto, S.K. Mineral acquisition by arbuscular mycorrhizal plants. J. Plant Nutr. 2000, 23, 867–902. [Google Scholar] [CrossRef]
- Hodge, A.; Fitter, A.H. Substantial nitrogen acquisition by arbuscular mycorrhizal fungi from organic material has implications for N cycling. Proc. Natl. Acad. Sci. USA 2010, 107, 13754–13759. [Google Scholar] [CrossRef]
- Kumar, A.; Shahbaz, M.; Koirala, M.; Blagodatskaya, E.; Seidel, S.J.; Kuzyakov, Y.; Pausch, J. Root trait plasticity and plant nutrient acquisition in phosphorus limited soil. J. Plant Nutr. Soil Sci. 2019, 182, 945–952. [Google Scholar] [CrossRef]
- Garcia, K.; Zimmermann, S.D. The role of mycorrhizal associations in plant potassium nutrition. Front. Plant Sci. 2014, 5, 337. [Google Scholar] [CrossRef]
- Lehmann, A.; Rillig, M.C. Arbuscular mycorrhizal contribution to copper, manganese and iron nutrient concentrations in crops—A meta-analysis. Soil Biol. Biochem. 2015, 81, 147–158. [Google Scholar] [CrossRef]
- Begum, N.; Qin, C.; Ahanger, M.A.; Raza, S.; Khan, M.I.; Ashraf, M.; Ahmed, N.; Zhang, L. Role of Arbuscular Mycorrhizal Fungi in Plant Growth Regulation: Implications in Abiotic Stress Tolerance. Front. Plant Sci. 2019, 10, 1068. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Yano, K. Nitrogen delivery to maize via mycorrhizal hyphae depends on the form of N. supplied. Plant Cell Environ. 2005, 28, 1247–1254. [Google Scholar] [CrossRef]
- Govindarajulu, M.; Pfeffer, P.E.; Jin, H.; Abubaker, J.; Douds, D.D.; Allen, J.W.; Bücking, H.; Lammers, P.J.; Shachar-Hill, Y. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 2005, 435, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Roos, P.; Borggaard, O.K.; Zhu, Y.-G.; Jakobsen, I. Mycorrhiza and root hairs in barley enhance acquisition of phosphorus and uranium from phosphate rock but mycorrhiza decreases root to shoot uranium transfer. New Phytol. 2005, 165, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.K.; George, T.S.; Barrett, G.E.; Hubbard, S.F.; White, P.J. Interactions between root hair length and arbuscular mycorrhizal colonisation in phosphorus deficient barley (Hordeum vulgare). Plant Soil 2013, 372, 195–205. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D. The symbionts forming arbuscular mycorrhizas. In Mycorrhizal symbiosis; ResearchGate: Berlin, Germany, 2008. [Google Scholar]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Francioli, D.; Schulz, E.; Buscot, F.; Reitz, T. Dynamics of Soil Bacterial Communities Over a Vegetation Season Relate to Both Soil Nutrient Status and Plant Growth Phenology. Microb. Ecol. 2018, 75, 216–227. [Google Scholar] [CrossRef]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Custódio, V.; Gonin, M.; Stabl, G.; Bakhoum, N.; Oliveira, M.M.; Gutjahr, C.; Castrillo, G. Sculpting the soil microbiota. Plant J. Cell Mol. Biol. 2022, 109, 508–522. [Google Scholar] [CrossRef]
- Maywald, N.J.; Mang, M.; Pahls, N.; Neumann, G.; Ludewig, U.; Francioli, D. Ammonium fertilization increases the susceptibility to fungal leaf and root pathogens in winter wheat. Front. Plant Sci. 2022, 13, 946584. [Google Scholar] [CrossRef]
- Herrmann, M.; Wegner, C.-E.; Taubert, M.; Geesink, P.; Lehmann, K.; Yan, L.; Lehmann, R.; Totsche, K.U.; Küsel, K. Predominance of Cand. Patescibacteria in Groundwater Is Caused by Their Preferential Mobilization From Soils and Flourishing Under Oligotrophic Conditions. Front. Microbiol. 2019, 10, 1407. [Google Scholar] [CrossRef]
- Dong, H.; Sun, H.; Fan, S.; Jiang, L.; Ma, D. Rhizobacterial communities, enzyme activity, and soil properties affect rice seedling’s nitrogen use. Agron. J. 2021, 113, 633–644. [Google Scholar] [CrossRef]
- Schmidt, J.E.; Kent, A.D.; Brisson, V.L.; Gaudin, A.C.M. Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 2019, 7, 146. [Google Scholar] [CrossRef] [PubMed]
- Kusstatscher, P.; Wicaksono, W.A.; Thenappan, D.P.; Adam, E.; Müller, H.; Berg, G. Microbiome Management by Biological and Chemical Treatments in Maize Is Linked to Plant Health. Microorganisms 2020, 8, 1506. [Google Scholar] [CrossRef] [PubMed]
- Tiziani, R.; Miras-Moreno, B.; Malacrinò, A.; Vescio, R.; Lucini, L.; Mimmo, T.; Cesco, S.; Sorgonà, A. Drought, heat, and their combination impact the root exudation patterns and rhizosphere microbiome in maize roots. Environ. Exp. Bot. 2022, 203, 105071. [Google Scholar] [CrossRef]
- Wolters, B.; Jacquiod, S.; Sørensen, S.J.; Widyasari-Mehta, A.; Bech, T.B.; Kreuzig, R.; Smalla, K. Bulk soil and maize rhizosphere resistance genes, mobile genetic elements and microbial communities are differently impacted by organic and inorganic fertilization. FEMS Microbiol. Ecol. 2018, 94, fiy027. [Google Scholar] [CrossRef]
- Wolińska, A.; Kruczyńska, A.; Podlewski, J.; Słomczewski, A.; Grządziel, J.; Gałązka, A.; Kuźniar, A. Does the Use of an Intercropping Mixture Really Improve the Biology of Monocultural Soils?—A Search for Bacterial Indicators of Sensitivity and Resistance to Long-Term Maize Monoculture. Agronomy 2022, 12, 613. [Google Scholar] [CrossRef]
- Li, X.; Rui, J.; Mao, Y.; Yannarell, A.; Mackie, R. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 2014, 68, 392–401. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef]
- Francioli, D.; van Ruijven, J.; Bakker, L.; Mommer, L. Drivers of total and pathogenic soil-borne fungal communities in grassland plant species. Fungal Ecol. 2020, 48, 100987. [Google Scholar] [CrossRef]
- Leff, J.W.; Bardgett, R.D.; Wilkinson, A.; Jackson, B.G.; Pritchard, W.J.; de Long, J.R.; Oakley, S.; Mason, K.E.; Ostle, N.J.; Johnson, D.; et al. Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. ISME J. 2018, 12, 1794–1805. [Google Scholar] [CrossRef]
- Mohanram, S.; Kumar, P. Rhizosphere microbiome: Revisiting the synergy of plant-microbe interactions. Ann. Microbiol. 2019, 69, 307–320. [Google Scholar] [CrossRef]
- Francioli, D.; Cid, G.; Hajirezaei, M.-R.; Kolb, S. Leaf bacterial microbiota response to flooding is controlled by plant phenology in wheat (Triticum aestivum L.). Sci. Rep. 2022, 12, 11197. [Google Scholar] [CrossRef] [PubMed]
- Francioli, D.; Cid, G.; Kanukollu, S.; Ulrich, A.; Hajirezaei, M.-R.; Kolb, S. Flooding Causes Dramatic Compositional Shifts and Depletion of Putative Beneficial Bacteria on the Spring Wheat Microbiota. Front. Microbiol. 2021, 12, 773116. [Google Scholar] [CrossRef] [PubMed]
- Lewin, S.; Francioli, D.; Ulrich, A.; Kolb, S. Crop host signatures reflected by co-association patterns of keystone Bacteria in the rhizosphere microbiota. Environ. Microbiome 2021, 16, 18. [Google Scholar] [CrossRef]
- Kuffner, M.; de Maria, S.; Puschenreiter, M.; Fallmann, K.; Wieshammer, G.; Gorfer, M.; Strauss, J.; Rivelli, A.R.; Sessitsch, A. Culturable bacteria from Zn- and Cd-accumulating Salix caprea with differential effects on plant growth and heavy metal availability. J. Appl. Microbiol. 2010, 108, 1471–1484. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.-X.; Bi, Q.-F.; Hao, X.-L.; Zhou, G.-W.; Yang, X.-R. Massilia phosphatilytica sp. nov., a phosphate solubilizing bacteria isolated from a long-term fertilized soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 2514–2519. [Google Scholar] [CrossRef]
- Diettrich, J.; Kage, H.; Nett, M. Genomics-inspired discovery of massiliachelin, an agrochelin epimer from Massilia sp. NR 4-1. Beilstein, J. Org. Chem. 2019, 15, 1298–1303. [Google Scholar] [CrossRef]
- Ofek, M.; Hadar, Y.; Minz, D. Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS ONE 2012, 7, e40117. [Google Scholar] [CrossRef]
- Li, C.; Cao, P.; Du, C.; Zhang, X.; Bing, H.; Li, L.; Sun, P.; Xiang, W.; Zhao, J.; Wang, X. Massilia rhizosphaerae sp. nov., a rice-associated rhizobacterium with antibacterial activity against Ralstonia solanacearum. Int. J. Syst. Evol. Microbiol. 2021, 71, 005009. [Google Scholar] [CrossRef]
- Krishnamoorthy, R.; Kim, K.; Subramanian, P.; Senthilkumar, M.; Anandham, R.; Sa, T. Arbuscular mycorrhizal fungi and associated bacteria isolated from salt-affected soil enhances the tolerance of maize to salinity in coastal reclamation soil. Agric. Ecosyst. Environ. 2016, 231, 233–239. [Google Scholar] [CrossRef]
- Chao, L.; Ma, X.; Tsetsegmaa, M.; Zheng, Y.; Qu, H.; Dai, Y.; Li, J.; Bao, Y. Response of Soil Microbial Community Composition and Diversity at Different Gradients of Grassland Degradation in Central Mongolia. Agriculture 2022, 12, 1430. [Google Scholar] [CrossRef]
- Wang, G.; Jin, Z.; Wang, X.; George, T.S.; Feng, G.; Zhang, L. Simulated root exudates stimulate the abundance of Saccharimonadales to improve the alkaline phosphatase activity in maize rhizosphere. Appl. Soil Ecol. 2022, 170, 104274. [Google Scholar] [CrossRef]
- El-Tarabily, K.A.; Nassar, A.H.; Sivasithamparam, K. Promotion of growth of bean (Phaseolus vulgaris L.) in a calcareous soil by a phosphate-solubilizing, rhizosphere-competent isolate of Micromonospora endolithica. Appl. Soil Ecol. 2008, 39, 161–171. [Google Scholar] [CrossRef]
- Hamdali, H.; Bouizgarne, B.; Hafidi, M.; Lebrihi, A.; Virolle, M.J.; Ouhdouch, Y. Screening for rock phosphate solubilizing Actinomycetes from Moroccan phosphate mines. Appl. Soil Ecol. 2008, 38, 12–19. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef]
- Conn, V.M.; Franco, C.M.M. Analysis of the endophytic actinobacterial population in the roots of wheat (Triticum aestivum L.) by terminal restriction fragment length polymorphism and sequencing of 16S rRNA clones. Appl. Environ. Microbiol. 2004, 70, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Hamedi, J.; Mohammadipanah, F. Biotechnological application and taxonomical distribution of plant growth promoting actinobacteria. J. Ind. Microbiol. Biotechnol. 2015, 42, 157–171. [Google Scholar] [CrossRef]
- Ekesi, S.; Maniania, N.K.; Ampong-Nyarko, K.; Onu, I. Effect of Intercropping Cowpea with Maize on the Performance of Metarhizium anisopliae Against Megalurothrips sjostedti (Thysanoptera: Thripidae) and Predators. Environ. Entomol. 1999, 28, 1154–1161. [Google Scholar] [CrossRef]
- Babendreier, D.; Jeanneret, P.; Pilz, C.; Toepfer, S. Non-target effects of insecticides, entomopathogenic fungi and nematodes applied against western corn rootworm larvae in maize. J. Appl. Entomol. 2015, 139, 457–467. [Google Scholar] [CrossRef]
- Jiang, W.; Peng, Y.; Ye, J.; Wen, Y.; Liu, G.; Xie, J. Effects of the Entomopathogenic Fungus Metarhizium anisopliae on the Mortality and Immune Response of Locusta migratoria. Insects 2019, 11, 36. [Google Scholar] [CrossRef]
- Wallenstein, M.D.; Peterjohn, W.T.; Schlesinger, W.H. N Fertilization effects on denitrification and N cycling in an aggrading forest. Ecol. Appl. 2006, 16, 2168–2176. [Google Scholar] [CrossRef] [PubMed]
- Hallin, S.; Jones, C.M.; Schloter, M.; Philippot, L. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 2009, 3, 597–605. [Google Scholar] [CrossRef]
- Wang, F.; Chen, S.; Wang, Y.; Zhang, Y.; Hu, C.; Liu, B. Long-Term Nitrogen Fertilization Elevates the Activity and Abundance of Nitrifying and Denitrifying Microbial Communities in an Upland Soil: Implications for Nitrogen Loss From Intensive Agricultural Systems. Front. Microbiol. 2018, 9, 2424. [Google Scholar] [CrossRef] [PubMed]
- Gericke, S.; Kurmies, B. Die kolorimetrische phosphorsäurebestimmung mit ammonium-vanadat-molybdat und ihre Anwendung in der Pflanzenanalyse. Z. Für Pflanz. Düngung Bodenkd. 1952, 59, 235–247. [Google Scholar]
- Moll, R.H.; Kamprath, E.J.; Jackson, W.A. Analysis and Interpretation of Factors Which Contribute to Efficiency of Nitrogen Utilization 1. Agron.J. 1982, 74, 562–564. [Google Scholar] [CrossRef]
- Brundrett, M.; Melville, L.; Peterson, L. Practical Methods in Mycorrhiza Research: Based on a Workshop Organized in Conjunction with the 9th North American Conference on Mycorrhizae, University of Guelph, Guelph, Ontario, Canada; Mycologue Publications: Vancouver Island, BC, Canada, 1994; pp. 1–161. [Google Scholar]
- Vierheilig, H.; Coughlan, A.P.; Wyss, U.R.S.; Piché, Y. Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 1998, 64, 5004–5007. [Google Scholar] [CrossRef]
- Koske, R.E.; Gemma, J.N. A modified procedure for staining roots to detect VA mycorrhizas. Mycol. Res. 1989, 92, 486–488. [Google Scholar] [CrossRef]
- Trouvelot, A.; Kough, J.L.; Gianinazzi-Pearson, V. Mesure du Taux de Mycorhization VA d’un Système Radiculaire. Recherche de Méthodes d’Estimation Ayant Une Signification Fonctionnelle. Physiol. Genet. Asp. Mycorrhizae 1985, 1986, 217–221. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Leser, T.D.; Amenuvor, J.Z.; Jensen, T.K.; Lindecrona, R.H.; Boye, M.; Møller, K. Culture-independent analysis of gut bacteria: The pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 2002, 68, 673–690. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2020. Available online: https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing (accessed on 1 December 2022).
- Legendre, P.; Gallagher, E.D. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
| Source of Variation (Treatment) | PUE | PUtE | PAE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| df | F | p | df | F | p | df | F | p | |
| Genotype (G) | 1 | 7.2 | 0.0114 | 1 | 5.2 | 0.0292 | 1 | 0.6 | 0.4426 |
| P Treatment (P) | 1 | 65.1 | <0.001 | 1 | 45.0 | <0.001 | 1 | 28.6 | 0.0221 |
| N Treatment (N) | 1 | 6.2 | 0.0183 | 1 | 28.6 | <0.001 | 1 | 21.1 | <0.001 |
| G × P | 1 | 0.4 | 0.5531 | 1 | 6.4 | 0.0170 | 1 | 3.1 | 0.0903 |
| G × N | 1 | 0.2 | 0.6972 | 1 | 1.0 | 0.3235 | 1 | 0.7 | 0.4210 |
| P ×N | 1 | 1.4 | 0.2516 | 1 | 1.1 | 0.3145 | 1 | 1.9 | 0.1757 |
| G × N | 1 | 5.7 | 0.0232 | 1 | 2.9 | 0.0988 | 1 | 0.7 | 0.3964 |
| Genotype | F7 | F160 | ||
|---|---|---|---|---|
| N Treatment | NO3 | AMM | NO3 | AMM |
| Root Traits | ||||
| Root/Shoot | 0.47 (0.12) A | 0.33 (0.05) A | 0.47 (0.07) A | 0.43 (0.07) A |
| Rhizosphere pH | 7.15 (0.07) AB | 7.02 (0.04) B | 7.27 (0.04) A | 7.04 (0.03) B |
| M | 0.09 (0.02) B | 0.15 (0.01) B | 0.15 (0.04) AB | 0.17 (0.04) A |
| TRL [m] | 34.1 (8.3) A | 29.5 (1.7) A | 35.4 (2.6) A | 28.4 (2.4) A |
| SRL [m g−1] | 64.4 (23) A | 43 (5.7) A | 45.7 (4.0) A | 49.5 (9.4) A |
| RD [mm] | 0.35 (0.01) B | 0.38 (0.003) A | 0.31 (0.008) C | 0.33 (0.003) BC |
| FRL [m] | 27.2 (6.8) A | 22.4 (1.4) A | 29.5 (2.3) A | 23.2 (1.9) A |
| RHL [mm] | 0.8 (0.03) AB | 0.82 (0.02) AB | 0.9 (0.03) A | 0.74 (0.03) B |
| Bacteria | Fungi | |||||
|---|---|---|---|---|---|---|
| Parameter | F | R2 | p | F | R2 | p |
| Plant (PL) | 1.949 | 0.102 | 0.001 | 1.598 | 0.089 | 0.001 |
| N treatment (N) | 1.964 | 0.103 | 0.001 | 1.402 | 0.078 | 0.004 |
| PL × N | 1.179 | 0.061 | 0.133 | 0.969 | 0.054 | 0.541 |
| Bacteria | Fungi | |||||
|---|---|---|---|---|---|---|
| Parameter | F | R2 | p | F | R2 | p |
| Genotype (G) | 1.244 | 0.106 | 0.028 | 0.973 | 0.087 | 0.631 |
| N treatment (N) | 1.526 | 0.131 | 0.001 | 1.236 | 0.111 | 0.007 |
| G × N | 1.017 | 0.086 | 0.385 | 0.968 | 0.088 | 0.674 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mang, M.; Maywald, N.J.; Li, X.; Ludewig, U.; Francioli, D. Nitrogen Fertilizer Type and Genotype as Drivers of P Acquisition and Rhizosphere Microbiota Assembly in Juvenile Maize Plants. Plants 2023, 12, 544. https://doi.org/10.3390/plants12030544
Mang M, Maywald NJ, Li X, Ludewig U, Francioli D. Nitrogen Fertilizer Type and Genotype as Drivers of P Acquisition and Rhizosphere Microbiota Assembly in Juvenile Maize Plants. Plants. 2023; 12(3):544. https://doi.org/10.3390/plants12030544
Chicago/Turabian StyleMang, Melissa, Niels Julian Maywald, Xuelian Li, Uwe Ludewig, and Davide Francioli. 2023. "Nitrogen Fertilizer Type and Genotype as Drivers of P Acquisition and Rhizosphere Microbiota Assembly in Juvenile Maize Plants" Plants 12, no. 3: 544. https://doi.org/10.3390/plants12030544
APA StyleMang, M., Maywald, N. J., Li, X., Ludewig, U., & Francioli, D. (2023). Nitrogen Fertilizer Type and Genotype as Drivers of P Acquisition and Rhizosphere Microbiota Assembly in Juvenile Maize Plants. Plants, 12(3), 544. https://doi.org/10.3390/plants12030544








