The Beneficial Effects of Inoculation with Selected Nodule-Associated PGPR on White Lupin Are Comparable to Those of Inoculation with Symbiotic Rhizobia
Abstract
:1. Introduction
2. Results
2.1. Isolation and Molecular Characterization of Nodule-Associated Bacteria
2.2. PGP Properties and Enzymatic Activities of the Isolates
2.3. pH and Temperature Growth Range and Tolerance to Salt and Heavy Metals
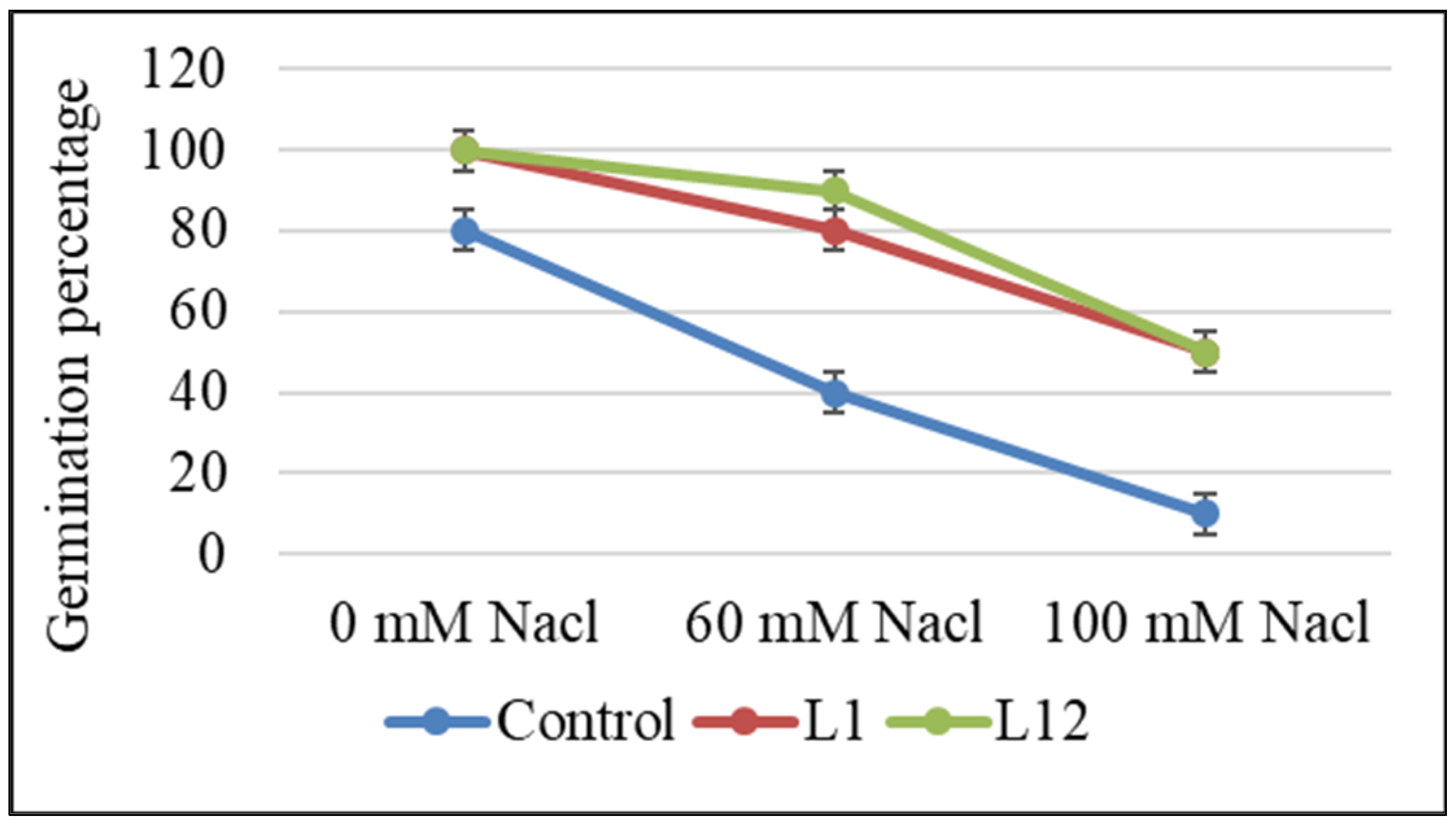
2.4. Effects of Selected PGPR Strains on Germination, Growth, and Nodulation of L. albus Plants
3. Discussion
4. Materials and Methods
4.1. Soil Samples Collection and Characterization
4.2. Isolation and Bacterial Growth
4.3. Analysis of Diversity and Identification of Isolates
4.3.1. DNA Isolation and PCR Amplification
4.3.2. Phylogenetic Analysis
4.4. Plant Growth-Promoting Traits of Selected Nodule-Associated Bacteria
4.4.1. Nitrogen Fixation
4.4.2. Phosphate Solubilization
4.4.3. Indole Acetic Acid (IAA) Production
4.4.4. 1-Amino-Cyclopropane-1-Carboxylate (ACC) Deaminase Activity
4.4.5. Siderophore Production
4.4.6. Zinc Solubilization Assay
4.4.7. Lipase Production
4.4.8. Cellulase Production
4.4.9. Pectinase Activity
4.5. Tolerance of Isolates to Acid and Alkaline pH, Temperature, Sodium Chloride and Heavy Metals
4.6. Lupin Seed Germination Assay
4.7. Seed Inoculation and Plant Growth
4.8. Statistical Analyses
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lucas, M.M.; Stoddard, F.L.; Annicchiarico, P.; Frías, J.; Martínez-Villaluenga, C.; Sussmann, D.; Duranti, M.; Seger, A.; Zander, P.M.; Pueyo, J.J. The future of lupin as a protein crop in Europe. Front. Plant Sci. 2015, 6, 705. [Google Scholar] [CrossRef]
- Leporatti, M.L.; Ghedira, K. Comparative analysis of medicinal plants used in traditional medicine in Italy and Tunisia. J. Ethnobiol. Ethnomed. 2009, 5, 31. [Google Scholar] [CrossRef]
- Ishaq, A.R.; El-Nashar, H.A.S.; Younis, T.; Mangat, M.A.; Shahzadi, M.; Ul Haq, A.S.; El-Shazly, M. Genus Lupinus (Fabaceae): A review of ethnobotanical, phytochemical and biological studies. J. Pharm. Pharmacol. 2022, 74, 1700–1717. [Google Scholar] [CrossRef] [PubMed]
- Siger, A.; Czubinski, J.; Kachlicki, P.; Dwiecki, K.; Lampart-Szczapa, E.; Nogala-Kalucka, M. Antioxidant activity and phenolic content in three lupin species. J. Food Compos. Anal. 2012, 25, 190–197. [Google Scholar] [CrossRef]
- Quiñones, M.A.; Fajardo, S.; Fernández-Pascual, M.; Lucas, M.M.; Pueyo, J.J. Nodulated white lupin plants growing in contaminated soils accumulate unusually high mercury concentrations in their nodules, roots and especially cluster roots. Horticulturae 2021, 7, 302. [Google Scholar] [CrossRef]
- Quiñones, M.A.; Ruiz-Díez, B.; Fajardo, S.; López-Berdonces, M.A.; Higueras, P.L.; Fernández-Pascual, M. Lupinus albus plants acquire mercury tolerance when inoculated with an Hg-resistant Bradyrhizobium strain. Plant Physiol. Biochem. 2013, 73, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Pueyo, J.J.; Quiñones, M.A.; Coba de la Peña, T.; Fedorova, E.E.; Lucas, M.M. Nitrogen and phosphorus interplay in lupin root nodules and cluster roots. Front. Plant Sci. 2021, 12, 644218. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, M.A.; Lucas, M.M.; Pueyo, J.J. Adaptive mechanisms make lupin a choice crop for acidic soils affected by aluminum toxicity. Front. Plant Sci. 2022, 12, 810692. [Google Scholar] [CrossRef] [PubMed]
- Msaddak, A.; Mars, M.; Quiñones, M.A.; Lucas, M.M.; Pueyo, J.J. Lupin, a unique legume that is nodulated by multiple microsymbionts: The role of horizontal gene transfer. Int. J. Mol. Sci. 2023, 24, 6496. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Rajendran, G.; Patel, M.H.; Joshi, S.J. Isolation and characterization of nodule-associated Exiguobacterium sp. from the root nodules of Fenugreek (Trigonella foenum-graecum) and their possible role in plant growth promotion. Int. J. Microbiol. 2012, 2012, 693982. [Google Scholar] [CrossRef]
- De Meyer, S.E.; De Beuf, K.; Vekeman, B.; Willems, A. A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol. Biochem. 2015, 83, 1–11. [Google Scholar] [CrossRef]
- Velázquez, E.; Valverde, A.; Rivas, R.; Gomis, V.; Peix, Á.; Gantois, I.; Igual, J.M.; León-Barrios, M.; Willems, A.; Mateos, P.F.; et al. Strains nodulating Lupinus albus on different continents belong to several new chromosomal and symbiotic lineages within Bradyrhizobium. Anton. Leeuw. 2010, 97, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Kloepper, J.W.; Schroth, M.N. Plant growth-promoting rhizobacteria on radishes. In Proceedings of the IVth International Conference on Plant Pathogenic Bacteria Volume 2, Station de Pathologie Vegetale et Phyto-Bacteriologie, Angers, France, 27 August–2 September 1978; pp. 879–882. [Google Scholar]
- Pinter, I.F.; Salomon, M.V.; Berli, F.; Bottini, R.; Piccoli, P. Characterization of the As(III) tolerance conferred by plant growth promoting rhizobacteria to in vitro-grown grapevine. Appl. Soil Ecol. 2017, 109, 60–68. [Google Scholar] [CrossRef]
- Pucciariello, C.; Boscari, A.; Tagliani, A.; Brouquisse, R.; Perata, P. Exploring legume-rhizobia symbiotic models for waterlogging tolerance. Front. Plant Sci. 2019, 10, 578. [Google Scholar] [CrossRef] [PubMed]
- Chamkhi, I.; El Omari, N.; Balahbib, A.; El Menyiy, N.; Benali, T.; Ghoulam, C. Is the rhizosphere a source of applicable multi-beneficial microorganisms for plant enhancement? Saudi J. Biol. Sci. 2022, 29, 1246–1259. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef]
- Etesami, H.; Jeong, B.R.; Glick, B.R. Contribution of arbuscular mycorrhizal fungi, phosphate–solubilizing bacteria, and silicon to P uptake by plant. Front. Plant Sci. 2021, 12, 699618. [Google Scholar] [CrossRef] [PubMed]
- Lurthy, T.; Cantat, C.; Jeudy, C.; Declerck, P.; Gallardo, K.; Barraud, C.; Leroy, F.; Ourry, A.; Lemanceau, P.; Salon, C.; et al. Impact of bacterial siderophores on iron status and ionome in pea. Front. Plant Sci. 2020, 11, 730. [Google Scholar] [CrossRef]
- Kang, S.-M.; Shahzad, R.; Khan, M.A.; Hasnain, Z.; Lee, K.-E.; Park, H.-S.; Kim, L.-R.; Lee, I.-J. Ameliorative effect of indole-3-acetic acid- and siderophore-producing Leclercia adecarboxylata MO1 on cucumber plants under zinc stress. J. Plant Interact. 2021, 16, 30–41. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Torre, S.; Bessadok, K.J.; Flores-Duarte, N.D.; Rodríguez-Llorente, I.; Caviedes, M.A.; Pajuelo, E. Helping legumes under stress situations: Inoculation with beneficial microorganisms. In Legume Crops; IntechOpen: Rijeka, Croatia, 2020; pp. 1–20. [Google Scholar]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Chandwani, S.; Amaresan, N. Role of ACC deaminase producing bacteria for abiotic stress management and sustainable agriculture production. Environ. Sci. Pollut. Res. 2022, 29, 22843–22859. [Google Scholar] [CrossRef] [PubMed]
- Alemneh, A.A.; Zhou, Y.; Ryder, M.H.; Denton, M.D. Mechanisms in plant growth-promoting rhizobacteria that enhance legume–rhizobial symbioses. J. Appl. Microbiol. 2020, 129, 1133–1156. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, A.; Matsushita, N.; Hougetsu, T. Nodulation in black locust by the gammaproteobacteria Pseudomonas sp. and the betaproteobacteria Burkholderia sp. Syst. Appl. Microbiol. 2010, 33, 269–274. [Google Scholar] [CrossRef]
- Martínez-Hidalgo, P.; Hirsch, A.M. The Nodule Microbiome: N2-fixing rhizobia do not live alone. Phytobiomes J. 2017, 1, 70–82. [Google Scholar] [CrossRef]
- Bessadok, K.; Navarro-Torre, S.; Pajuelo, E.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Caviedes, M.Á.; Fterich, A.; Mars, M.; Rodríguez-Llorente, I.D. The ACC-deaminase producing bacterium Variovorax sp. CT7.15 as a tool for improving Calicotome villosa nodulation and growth in arid regions of Tunisia. Microorganisms 2020, 8, 541. [Google Scholar] [CrossRef] [PubMed]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E.; Bedmar, E.J. Bacterial associations with legumes. Crit. Rev. Plant Sci. 2015, 34, 17–42. [Google Scholar] [CrossRef]
- Ferchichi, N.; Toukabri, W.; Boularess, M.; Smaoui, A.; Mhamdi, R.; Trabelsi, D. Isolation, identification and plant growth promotion ability of endophytic bacteria associated with lupine root nodule grown in Tunisian soil. Arch. Microbiol. 2019, 201, 1333–1349. [Google Scholar] [CrossRef]
- Tounsi-Hammami, S.; Le Roux, C.; Dhane-Fitouri, S.; De Lajudie, P.; Duponnois, R.; Ben Jeddi, F. Genetic diversity of rhizobia associated with root nodules of white lupin (Lupinus albus L.) in Tunisian calcareous soils. Syst. Appl. Microbiol. 2019, 42, 448–456. [Google Scholar] [CrossRef]
- Mahanty, T.; Bhattacharjee, S.; Goswami, M.; Bhattacharyya, P.; Das, B.; Ghosh, A.; Tribedi, P. Biofertilizers: A potential approach for sustainable agriculture development. Environ. Sci. Pollut. Res. 2017, 24, 3315–3335. [Google Scholar] [CrossRef]
- Nieto, J.J.; Fernández-Castillo, R.; Márquez, M.C.; Ventosa, A.; Quesada, E.; Ruiz-Berraquero, F. Survey of metal tolerance in moderately halophilic eubacteria. Appl. Environ. Microbiol. 1989, 55, 2385–2390. [Google Scholar] [CrossRef]
- Coutinho, H.L.C.; Oliveira, V.M.; Lovato, A.; Maia, A.H.N.; Manfio, G.P. Evaluation of the diversity of rhizobia in Brazilian agricultural soils cultivated with soybeans. Appl. Soil Ecol. 1999, 13, 159–167. [Google Scholar] [CrossRef]
- Durán, D.; Rey, L.; Sánchez-Cañizares, C.; Navarro, A.; Imperial, J.; Ruiz-Argueso, T. Genetic diversity of indigenous rhizobial symbionts of the Lupinus mariae-josephae endemism from alkaline-limed soils within its area of distribution in Eastern Spain. Syst. Appl. Microbiol. 2013, 36, 128–136. [Google Scholar] [CrossRef] [PubMed]
- McInnes, A. Structure and diversity among rhizobial strains, populations and communities? A review. Soil Biol. Biochem. 2004, 36, 1295–1308. [Google Scholar] [CrossRef]
- Rivas, R.; Peix, A.; Mateos, P.F.; Trujillo, M.E.; Martínez-Molina, E.; Velázquez, E. Biodiversity of populations of phosphate solubilizing rhizobia that nodulate chickpea in different Spanish soils. Plant Soil 2006, 287, 23–33. [Google Scholar] [CrossRef]
- Young, J.M.; Kuykendall, L.D.; Martínez-Romero, E.; Kerr, A.; Sawada, H. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J. Syst. Evol. Microbiol. 2001, 51, 89–103. [Google Scholar] [CrossRef]
- Zahradník, J.; Nunvar, J.; Pařízková, H.; Kolářová, L.; Palyzová, A.; Marešová, H.; Grulich, M.; Kyslíková, E.; Kyslík, P. Agrobacterium bohemicum sp. nov. isolated from poppy seed wastes in Central Bohemia. Syst. Appl. Microbiol. 2018, 41, 184–190. [Google Scholar] [CrossRef]
- Aserse, A.A.; Räsänen, L.A.; Assefa, F.; Hailemariam, A.; Lindström, K. Phylogeny and genetic diversity of native rhizobia nodulating common bean (Phaseolus vulgaris L.) in Ethiopia. Syst. Appl. Microbiol. 2012, 35, 120–131. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Liu, Y.; Wang, E.T.; Ren, C.; Liu, W.; Xu, H.; Wu, H.; Jiang, N.; Li, Y.; et al. Genetic diversity and community structure of rhizobia nodulating Sesbania cannabina in saline–alkaline soils. Syst. Appl. Microbiol. 2016, 39, 195–202. [Google Scholar] [CrossRef]
- Valdez-Nuñez, R.A.; Castro-Tuanama, R.; Castellano-Hinojosa, A.; Bedmar, E.J.; Ríos-Ruiz, W.F. PGPR characterization of non-nodulating bacterial endophytes from root nodules of Vigna unguiculata (L.) Walp. In Microbial Probiotics for Agricultural Systems; Zúñiga-Dávila, D., González-Andrés, F., Ormeño-Orrillo, E., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 111–126. ISBN 978-3-030-17596-2. [Google Scholar]
- Waraczewska, Z.; Niewiadomska, A.; Wolna-Maruwka, A.; Sulewska, H.; Budka, A.; Pilarska, A.A. The effect of in vitro coinoculation on the physiological parameters of white lupine plants (Lupinus albus L.). Appl. Sci. 2022, 12, 12382. [Google Scholar] [CrossRef]
- Zakhia, F.; Jeder, H.; Willems, A.; Gillis, M.; Dreyfus, B.; De Lajudie, P. Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microb. Ecol. 2006, 51, 375–393. [Google Scholar] [CrossRef]
- Deng, Z.S.; Zhao, L.F.; Kong, Z.Y.; Yang, W.Q.; Lindström, K.; Wang, E.T.; Wei, G.H. Diversity of endophytic bacteria within nodules of the Sphaerophysa salsula in different regions of Loess Plateau in China. FEMS Microbiol. Ecol. 2011, 76, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Leite, H.A.C.; Silva, A.B.; Gomes, F.P.; Gramacho, K.P.; Faria, J.C.; de Souza, J.T.; Loguercio, L.L. Bacillus subtilis and Enterobacter cloacae endophytes from healthy Theobroma cacao L. trees can systemically colonize seedlings and promote growth. Appl. Microbiol. Biotechnol. 2013, 97, 2639–2651. [Google Scholar] [CrossRef] [PubMed]
- Kilian, M.; Steiner, U.; Krebs, B.; Junge, H.; Schmiedeknecht, G.; Hain, R. FZB24® Bacillus subtilis—Mode of action of a microbial agent enhancing pant vitality. Pflanzenschutz Nachr. Bayer 2000, 1, 72–93. [Google Scholar]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Klimova, S.Y.; Shestakov, A.I.; Botina, S.G.; Netrusov, A.I. Orchid-associated bacteria produce indole-3-acetic acid, promote seed germination, and increase their microbial yield in response to exogenous auxin. Arch. Microbiol. 2007, 188, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Viveros, O.; Jorquera, M.A.; Crowley, D.E.; Gajardo, G.; Mora, M.L. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nutr. 2010, 10, 293–319. [Google Scholar] [CrossRef]
- Miransari, M.; Smith, D.L. Plant hormones and seed germination. Environ. Exp. Bot. 2014, 99, 110–121. [Google Scholar] [CrossRef]
- Bashan, Y.; de-Bashan, L.E. Inoculant preparation and formulations for Azospirillum spp. In Handbook for Azospirillum; Cassán, F.D., Okon, Y., Creus, C.M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 469–485. ISBN 978-3-319-06541-0. [Google Scholar]
- Ahmad, F.; Ahmad, I.; Khan, M.S. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef]
- Panwar, M.; Tewari, R.; Nayyar, H. Microbial consortium of plant growth-promoting rhizobacteria improves the performance of plants growing in stressed soils: An overview. In Phosphate Solubilizing Microorganisms; Khan, M., Zaidi, A., Musarrat, J., Eds.; Springer: Cham, Switzerland, 2014; pp. 257–285. ISBN 978-3-319-08215-8. [Google Scholar]
- Jha, C.K.; Saraf, M. Plant growth promoting rhizobacteria (PGPR): A review. J. Agric. Res. Dev. 2015, 5, 108–119. [Google Scholar] [CrossRef]
- Babalola, O.O. Beneficial bacteria of agricultural importance. Biotechnol. Lett. 2010, 32, 1559–1570. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, P.; Fang, L.; Zhang, Q.-A.; Yan, C.; Chen, J. Isolation and characterization of phosphate-solubilizing bacteria from mushroom residues and their effect on tomato plant growth promotion. Pol. J. Microbiol. 2017, 66, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Trivedi, G.; Saraf, M. Iron biofortification in mungbean using siderophore producing plant growth promoting bacteria. Environ. Sust. 2018, 1, 357–365. [Google Scholar] [CrossRef]
- Turan, M.; Kitir, N.; Alkaya, Ü.; Günes, A.; Tüfenkçi, S.; Yildirim, E.; Nikerel, E. Making soil more accessible to plants: The case of plant growth promoting rhizobacteria. In Plant Growth; Rigobelo, E.C., Ed.; InTech: Rijeka, Croatia, 2016; pp. 61–69. ISBN 978-953-51-2771-0. [Google Scholar]
- Gupta, G.; Parihar, S.S.; Ahirwar, N.K.; Snehi, N.K. Plant growth promoting rhizobacteria (PGPR): Current and future prospects for development of sustainable agriculture. J. Microb. Biochem. Technol. 2015, 7, 96–102. [Google Scholar] [CrossRef]
- Selim, S.M.; Zayed, M.S. Role of biofertilizers in sustainable agriculture under abiotic stresses. In Microorganisms for Green Revolution; Panpatte, D.G., Jhala, Y.K., Vyas, R.V., Shelat, H.N., Eds.; Springer: Singapore, 2017; pp. 281–301. ISBN 978-981-10-6240-7. [Google Scholar]
- Kang, B.G.; Kim, W.T.; Yun, H.S.; Chang, S.C. Use of plant growth-promoting rhizobacteria to control stress responses of plant roots. Plant Biotechnol. Rep. 2010, 4, 179–183. [Google Scholar] [CrossRef]
- Menéndez, E.; Robledo, M.; Jiménez-Zurdo, J.I.; Velázquez, E.; Rivas, R.; Murray, J.D.; Mateos, P.F. Legumes display common and host-specific responses to the rhizobial cellulase CelC2 during primary symbiotic infection. Sci. Rep. 2019, 9, 13907. [Google Scholar] [CrossRef]
- Mwenda, G.M.; Hill, Y.J.; O’Hara, G.W.; Reeve, W.G.; Howieson, J.G.; Terpolilli, J.J. Competition in the Phaseolus vulgaris-Rhizobium symbiosis and the role of resident soil rhizobia in determining the outcomes of inoculation. Plant Soil 2023, 487, 61–77. [Google Scholar] [CrossRef]
- Msaddak, A.; Rey, L.; Imperial, J.; Palacios, J.M.; Mars, M.; Pueyo, J.J. Phylogenetic analyses of rhizobia isolated from nodules of Lupinus angustifolius in Northern Tunisia reveal Devosia sp. as a new microsymbiont of lupin species. Agronomy 2021, 11, 1510. [Google Scholar] [CrossRef]
- Nonnoi, F.; Chinnaswamy, A.; García de la Torre, V.S.; Coba de la Peña, T.; Lucas, M.M.; Pueyo, J.J. Metal tolerance of rhizobial strains isolated from nodules of herbaceous legumes (Medicago spp. and Trifolium spp.) growing in mercury-contaminated soils. Appl. Soil Ecol. 2012, 61, 49–59. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Chenna, R. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kirchhof, G.; Reis, V.M.; Baldani, J.I.; Eckert, B.; Dobereiner, J.; Hartmann, A. Occurrence, physiological and molecular analysis of endophytic diazotrophic bacteria in gramineous energy plants. Plant Soil 1997, 194, 45–55. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Glickmann, E.; Dessaux, Y. A critical examination of the specificity of the Salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl. Environ. Microbiol. 1995, 61, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Honma, M.; Shimomura, T. Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 1978, 42, 1825–1831. [Google Scholar] [CrossRef]
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- Fasim, F.; Ahmed, N.; Parsons, R.; Gadd, G.M. Solubilization of zinc salts by a bacterium isolated from the air environment of a tannery. FEMS Microbiol. Lett. 2002, 213, 1–6. [Google Scholar] [CrossRef]
- Sharma, S.K. Characterization of zinc-solubilizing Bacillus isolates and their potential to influence zinc assimilation in soybean seeds. J. Microbiol. Biotechnol. 2012, 22, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Elbeltagy, A.; Nishioka, K.; Suzuki, H.; Sato, T.; Sato, Y.-I.; Morisaki, H.; Mitsui, H.; Minamisawa, K. Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci. Plant Nutr. 2000, 46, 617–629. [Google Scholar] [CrossRef]
- Barra, P.J.; Inostroza, N.G.; Acuña, J.J.; Mora, M.L.; Crowley, D.E.; Jorquera, M.A. Formulation of bacterial consortia from avocado (Persea americana Mill.) and their effect on growth, biomass and superoxide dismutase activity of wheat seedlings under salt stress. Appl. Soil Ecol. 2016, 102, 80–91. [Google Scholar] [CrossRef]
- Penrose, D.M.; Moffatt, B.A.; Glick, B.R. Determination of 1-aminocycopropane-1-carboxylic acid (ACC) to assess the effects of ACC deaminase-containing bacteria on roots of canola seedlings. Can. J. Microbiol. 2001, 47, 77–80. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil; University of California: Berkeley, CA, USA, 1938. [Google Scholar]


| Strain | Nitrogen Fixation | Phosphate Solubilization | IAA Production * | ACC Deaminase Activity ** | Siderophore Production | Enzyme Activities | ||
|---|---|---|---|---|---|---|---|---|
| Lipase | Cellulase | Pectinase | ||||||
| L1 | + | ++ | 8.93 | 22.02 ± 0.56 | ++ | + | + | - |
| L4 | - | - | 1.65 | - | - | + | - | - |
| L5 | - | ± | - | - | - | + | - | - |
| L6 | ± | - | - | - | ± | + | - | - |
| L9 | - | - | - | - | - | - | - | - |
| L12 | + | ++ | 8.89 | 22.16 ± 0.29 | ++ | + | + | - |
| L13 | + | + | - | - | + | + | - | - |
| L15 | + | + | 12.32 | - | + | + | - | - |
| L18 | + | - | 8.45 | - | - | - | + | - |
| L19 | ± | - | 8.95 | - | - | - | - | - |
| L21 | + | - | 4.82 | - | - | - | + | - |
| Strain | pH | Temperature (°C) | NaCl MTC (mM) | Heavy Metals MTC (μM) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Zn | Cd | Cr | Pb | Ni | Cu | ||||
| L1 | 4–12 | 25–40 | 850 | 750 | 200 | 150 | 1750 | 2000 | 2000 |
| L4 | 4–12 | 25–40 | 340 | 750 | 50 | 450 | 2000 | 1000 | 2000 |
| L5 | 4–12 | 25–40 | 1200 | 500 | 75 | 450 | 2000 | 1000 | 2000 |
| L6 | 4–12 | 25–40 | 1200 | 500 | 25 | 250 | 2000 | 1500 | 1500 |
| L9 | 7–9 | 25–40 | 170 | 150 | 00 | 100 | 1750 | 1000 | 1500 |
| L12 | 4–12 | 25–40 | 850 | 750 | 200 | 150 | 2000 | 1500 | 2000 |
| L13 | 4–12 | 25–40 | 1200 | 750 | 100 | 450 | 2000 | 2000 | 2000 |
| L15 | 4–12 | 25–40 | 850 | 750 | 500 | 150 | 1750 | 2000 | 2000 |
| L18 | 4–10 | 25–40 | 340 | 750 | 150 | 50 | 2000 | 1000 | 2000 |
| L19 | 4–12 | 25–40 | 340 | 750 | 100 | 100 | 1750 | 1000 | 1000 |
| L21 | 4–10 | 25–40 | 340 | 750 | 150 | 150 | 2000 | 1000 | 2000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Msaddak, A.; Quiñones, M.A.; Mars, M.; Pueyo, J.J. The Beneficial Effects of Inoculation with Selected Nodule-Associated PGPR on White Lupin Are Comparable to Those of Inoculation with Symbiotic Rhizobia. Plants 2023, 12, 4109. https://doi.org/10.3390/plants12244109
Msaddak A, Quiñones MA, Mars M, Pueyo JJ. The Beneficial Effects of Inoculation with Selected Nodule-Associated PGPR on White Lupin Are Comparable to Those of Inoculation with Symbiotic Rhizobia. Plants. 2023; 12(24):4109. https://doi.org/10.3390/plants12244109
Chicago/Turabian StyleMsaddak, Abdelhakim, Miguel A. Quiñones, Mohamed Mars, and José J. Pueyo. 2023. "The Beneficial Effects of Inoculation with Selected Nodule-Associated PGPR on White Lupin Are Comparable to Those of Inoculation with Symbiotic Rhizobia" Plants 12, no. 24: 4109. https://doi.org/10.3390/plants12244109
APA StyleMsaddak, A., Quiñones, M. A., Mars, M., & Pueyo, J. J. (2023). The Beneficial Effects of Inoculation with Selected Nodule-Associated PGPR on White Lupin Are Comparable to Those of Inoculation with Symbiotic Rhizobia. Plants, 12(24), 4109. https://doi.org/10.3390/plants12244109









