Abstract
Salt is harmful to crop production. Therefore, it is important to understand the mechanism of salt tolerance in rice. CIPK genes have various functions, including regulating salt tolerance and other types of stress and nitrogen use efficiency. In rice, OsCIPK24 is known to regulate salt tolerance, but other OsCIPKs could also function in salt tolerance. In this study, we identified another OsCIPK—OsCIPK9—that can regulate salt tolerance. Knockout of OsCIPK9 in rice could improve salt tolerance. Through expression analyses, OsCIPK9 was found to be mainly expressed in the roots and less expressed in mature leaves. Meanwhile, OsCIPK9 had the highest expression 6 h after salt treatment. In addition, we proved the interaction between OsCIPK9 and OsSOS3. The RNA-seq data showed that OsCIPK9 strongly responded to salt treatment, and the transporters related to salt tolerance may be downstream genes of OsCIPK9. Finally, haplotype analyses revealed that Hap6 and Hap8 mainly exist in indica, potentially providing a higher salt tolerance. Overall, a negative regulator of salt tolerance, OsCIPK9, which interacted with OsSOS3 similarly to OsCIPK24 and influenced salt-related transporters, was identified, and editing OsCIPK9 potentially could be helpful for breeding salt-tolerant rice.
1. Introduction
An increase in the output of rice, as a staple food feeding almost half the world, will be required over the next 50 years. Therefore, either a higher yield per ha or more cultivated land is needed for a larger harvest. Making full use of the soil would make higher yields achievable. However, salt in saline–alkali soil damages crop production. Salinity includes osmotic stress, ionic toxicity, and nutritional deficiencies, which inhibit rice development [1].
As a major abiotic stressor, researchers have focused on the mechanism of salt tolerance using quantitative trait loci (QTL) mapping [2] and genome-wide association studies (GWASs) [3,4]. With progress in rice genome sequencing, related genes that could be used for rice breeding, such as SKC1, have been cloned [5]. SKC1 is a high-affinity K+ transporter which was cloned from Pokkali with extreme salt resistance. OsWRKY53 is a key regulator cloned from GWAS which can regulate the expression of OsMKK10.2 in promoting ion homeostasis and trans-represses SKC1 [4]. The superior alleles identified could be useful for breeding rice with salt tolerance. Meanwhile, greater yields would be obtained in saline–alkali soils with the use of salt-tolerant genes [6].
In addition to genes like SKC1, CIPK family genes have an important role in salt tolerance. CIPK family genes are plant-specific proteins which interact with CBL and serve as major downstream signaling components [7,8]. The CBL-CIPK network plays a vital role in salinity stress, disease defense, drought tolerance, and other stresses [9]. In Arabidopsis, the SOS (Salt Overly Sensitive) pathway was the first well-studied pathway, and this acts as an example for other networks. The AtCBL4–AtCIPK24 complex activates downstream AtSOS1, which affects Na+ extrusion and long-distance Na+ transport [10]. Also, AtCIPK24 can form an AtCIPK24–GI complex to delay the flowering time under saline conditions [11]. After that, another network, AtCBL10-AtCIPK8, was identified, which also regulates AtSOS1 [12]. In addition to salinity stress, the ATCBL4-AtCIPK6 pathway regulates K+ allocation [13]. The AtCBL9-AtCIPK3 network can affect seed dormancy through activating ABR1 in the nucleus [14]. In short, CIPK family genes have multiple functions in Arabidopsis. In rice, CIPKs can phosphorylate many transporters that have multiple functions in various processes, such as nitrogen uptake [15], K+ uptake [16], and microbe-associated molecular pattern-induced defense [17]. OsCIPK9, 14, 15 regulates microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression in cultured cells [18]. Twelve OsCIPK genes, including OsCIPK9, have been demonstrated to be induced by salinity stress [19]. In addition, CIPKs are upregulated during panicle development and abiotic stress [18]. In salt tolerance, OsSOS2 (OsCIPK24) and OsSOS3 (OsCBL4) play vital roles [20]. The calcium-binding protein OsSOS3/CBL4 can sense the cytosolic calcium signal elicited by salt stress, then interact with and activate OsSOS2 (OsCIPK24). Then, activated OsSOS2/OsCIPK24 phosphorylates and activates OsSOS1 to regulate Na+ homeostasis and improve rice tolerance to salt stress [21]. In rice, the CIPK family has 33 members, with OsCIPK24 regulating salt tolerance [18], but other OsCIPKs which may also have a role in salt tolerance are still unknown. A previous study showed that the mutant Oscipk9 showed a mild salt tolerance [16], but the possible mechanisms of salt tolerance and the haplotypes of OsCIPK9 are still unclear.
Here, we proved that OsCIPK9 is a negative regulator of salt tolerance in rice using CRISPR-Cas9. OsCIPK9 knockouts of rice showed an increased salt tolerance, and overexpression lines were more sensitive. The Na+ concentration changed significantly after salt treatment. OsSOS3 showed a higher expression in the OsCIPK9-cas line, and OsSOS3 interacted with OsCIPK9. As determined through RNA-seq data, OsCIPK9 strongly responded to salt treatment. We found obvious changes in the expression of transporters like OsKAT1, especially under saline conditions. Finally, we analyzed the haplotypes of OsCIPK9 and showed that the haplotype of OsCIPK9 had an obvious subpopulation classification.
2. Results
2.1. OsCIPK9 Negatively Regulated Salt Tolerance
To study the specific role of OsCIPK9 in rice, we constructed knockout lines using CRISPR-Cas9. One knockout line (OsCIPK9-cas) with a base insertion was identified (Figure 1a). The insertion caused a frameshift, and translation stopped after 148 aa (Figure 1a). The protein sequence of the knockout line showed that the domain lacked 140 normal amino acids (143–283) and contained six mutated amino acids (from 143 to 148) compared to that of the wild type (Figure 1b). We also created overexpression lines for further function validation. The overexpression lines OsCIPK9-OE2 and OsCIPK9-OE3 had approximately 9- and 5-fold higher expressions than the wild type (Nipponbare), respectively (Figure 1c).
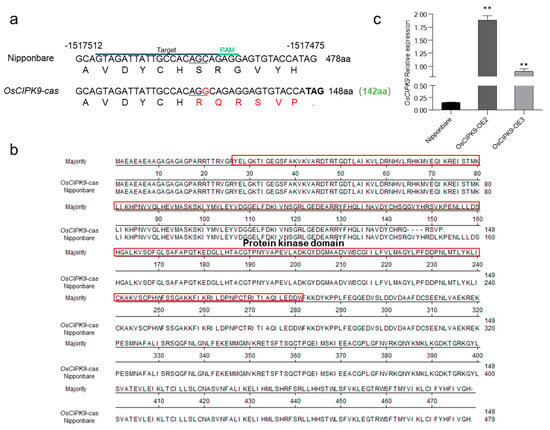
Figure 1.
Transgenic line construction. (a) Targeted mutagenesis of OsCIPK9 with CRISPR/Cas9. The mutant alleles have 1 nucleotide insertion in OsCIPK9-cas. The base shown in red represents the inserted nucleotide. The amino acids in red represent mutated amino acids. The line below in nucleotides represents the positions which translated the mutated amino acids. (b) The result of protein alignment between Nipponbare and OsCIPK9-cas. The domain was predicted in Pfam (pfam.xfam.org, accessed on 30 August 2022) presented in red square. (c) The expression of Nipponbare and overexpression lines (OsCIPK9-OE2, OsCIPK9-OE3). **: p < 0.01. Statistical significance (versus Nipponbare) was calculated using a Student’s t-test.
To verify whether OsCIPK9 could function in salt tolerance, we treated the transgenic lines and wild type with 0 and 120 mM NaCl. As a result, the knockout line (OsCIPK9-cas) showed a higher tolerance to salt treatment, while the overexpression lines were more sensitive (Figure 2a–c). The OsCIPK9-OE2 and OsCIPK9-OE3 lines had a lower relative fresh weight compared with the WT, while that of the knockout line was higher (Figure 2d,e). We also measured the Na+ and K+ concentrations of whole seedlings. In the shoots, the four lines showed no differences in Na+ concentrations and Na+/K+ content ratios under no-salt conditions (Figure S1), but a discrepancy was observed under salt treatment, in which these values were higher in the OE lines and lower in the knockout lines (Figure 2f,h). In the roots, the Na+ concentration of the OE lines was lower than the wild type and the knockout line under no-salt conditions, but displayed almost no difference under saline conditions (Figure S1). The Na+/K+ content ratio was higher in the OE lines than in the knockout lines (Figure 2g,i). Thus, OsCIPK9 conferred salt tolerance in rice, and knocking out this gene improved salt tolerance through regulating the Na concentration.
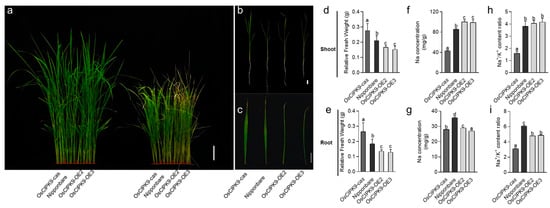
Figure 2.
The phenotype of Nipponbare and transgenic lines under 0 and 120 mM NaCl conditions. (a–c) The phenotype of the wild type (Nipponbare), knockout line (OsCIPK9-cas) and overexpression lines (OsCIPK9-OE2, OsCIPK9-OE3) under 0 mM and 120 mM NaCl conditions, bar = 10, 2, 2 cm. (d,f,h) Comparison of Nipponbare, knockout line (OsCIPK9-cas) and overexpression lines (OsCIPK9-OE2, OsCIPK9-OE3) in terms of relative fresh weights, Na concentrations per plant and Na+/K+ content ratios in shoots under 120 mM NaCl conditions. n = 3. (e,g,i) Comparison of Nipponbare, the knockout line (OsCIPK9-cas) and overexpression lines (OsCIPK9-OE2, OsCIPK9-OE3) in terms of relative fresh weights, Na concentrations per plant and Na+/K+ content ratios in roots under 120 mM NaCl conditions. n = 3. The data are presented as means ± SDs. Statistical significance (versus Nipponbare) was calculated using a Student’s t-test. Different letters indicate significant difference, p < 0.05.
2.2. Expression Pattern of OsCIPK9
To determine the expression pattern of OsCIPK9, various tissues were collected and the mRNA abundance of OsCIPK9 was examined using RT-PCR analysis. The results showed that OsCIPK9 was most expressed in the roots and least expressed in mature leaves (Figure 3a). Also, we measured the expression at different time points after salt treatment. The results show that OsCIPK9 had the highest expression 6 h after salt treatment (Figure 3b). The knock-out line showed lower expression from 1 h to 7 days, with the largest difference observed after 6 h. These results indicate that OsCIPK9 responded to salt treatment, and knocking out the gene reduced the expression under salt treatment.
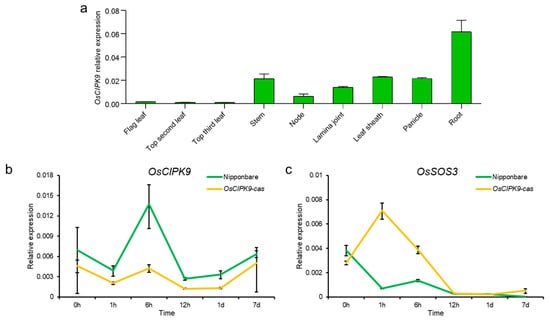
Figure 3.
The expression of OsCIPK9 and OsSOS3. (a) The expression of OsCIPK9 in various tissues. (b,c) Comparison of the expression of OsCIPK9 and OsSOS3 between Nipponbare and the knockout line (OsCIPK9-cas) under 0 and 120 mM NaCl conditions. n = 3. The data are presented as means ± SDs.
2.3. OsCIPK9 Interacted with OsSOS3
According to the change in Na+ concentrations in different transgenic lines, including the knock-out and overexpression lines, we examined whether OsSOS3 in the SOS pathway could interact with OsCIPK9 similar to OsSOS2/OsCIPK24. Firstly, OsSOS3 showed a lower expression in Nipponbare compared with OsCIPK9-cas, while OsCIPK9 showed a higher expression (Figure 3b,c). Thus, we supposed that OsSOS3 may interact with OsCIPK9, playing an opposing role in salt tolerance. To test this hypothesis, we analyzed the interaction between OsSOS3 and OsCIPK9 using a yeast two-hybrid (Y2H) assay. OsSOS3 strongly interacted with OsCIPK9 (Figure 4a). Also, we verified the interaction between OsSOS3 and OsCIPK9 using an in vivo firefly luciferase complementation imaging (LCI) assay in Nicotiana benthamiana leaf epidermal cells (Figure 4b). Taken together, these results suggest that OsSOS3 could interact with OsCIPK9 in rice.
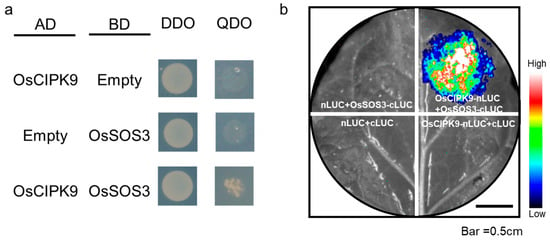
Figure 4.
OsCIPK9 physically interacts with OsSOS3. (a) Y2H assay showing that OsCIPK9 can interact with OsSOS3 (A). AD, activation domain; BD, binding domain; DDO, SD/–Trp/–Leu; QDO, SD/–Trp/–Leu/–His/–Ade. (b) LCI assay showing that OsCIPK9 interacts with OsSOS3 in leaf cells of N. benthamiana. Colored scale bar indicates the luminescence intensity in counts per second (cps). CLUC, C terminus of LUC; NLUC, N terminus of LUC.
2.4. Transcriptome Analysis of OsCIPK9
To identify possible downstream genes influenced by OsCIPK9, we performed a transcriptome analysis using knockout lines with different treatments (0 and 120 mM salt). Many upregulated and downregulated DEGs (differential expression genes) were identified, but the number of up- or downregulated genes under NaCl treatment was much greater than under control conditions (Figure 5a). The DEGs included 2285 regulated genes without salt treatment and 9190 genes under salt treatment. Only 1338 genes were found for both treatments (Figure 5b). A GO analysis showed that genes in the plasma membrane of cellular components were identified, but the number of genes under salt treatment (1190) was more than three times that under no salt treatment (358) (Figure 5c and Figure S2). In biological processes, more genes were clustered in cell wall organization under no salt treatment, while the ratio increased under salt treatment. Additionally, more genes were grouped into translation and carbohydrate metabolic processes under salt treatment (Figure S3). The GO molecular function analysis showed that the DNA-binding transcription faction activity process had a large number of genes with a lower Q value under no salt treatment. Under salt treatment, the process with the most regulated genes was structural constituents of the ribosome (Figure S4). Additionally, a KEGG analysis revealed almost no difference between Nipponbare and OsCIPK9-cas without NaCl treatment (Figure 5d). A large difference was identified under 120 mM NaCl treatment, as expected (Figure 5d). In the KEGG analysis, almost all processes differed, including transport and catabolism, signal transduction, and membrane transport. The processes consisted of the functions of CIPKs, which have been reported previously [22]. The GO and KEGG results indicated that OsCIPK9 strongly reacted to salt treatment and played a vital role in salt tolerance, especially in membranes.
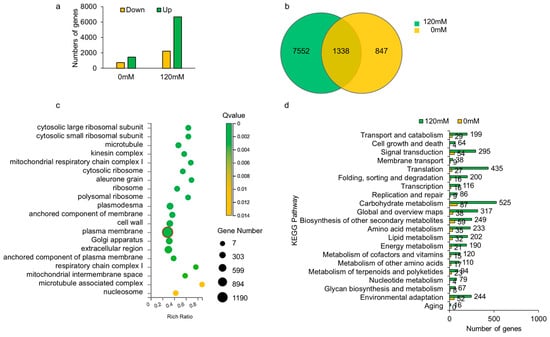
Figure 5.
The analysis of RNA-seq data. (a) The number of DEGs after 0 mM and 120 mM NaCl treatment. (b) The overlap of genes identified after 0 mM and 120 mM NaCl treatment. (c) GO analysis of genes identified by RNA-seq. The red circle highlighted the gene number of the plasma membrane identified by GO analysis. (d) KEGG analysis of genes identified by RNA-seq.
The RNA-seq analysis showed that OsCIPK9 affected membrane transport. To identify possible influencing genes in the membrane, we investigated the expression of previously reported transporters, such as SKC1 and OsKAT1, in the RNA-seq data. Luckily, we found that SKC1, OsKAT1, and OsNHX1 expression was higher in OsCIPK9-cas than in Nipponbare under salt treatment (Figure 6a–c). In particular, the expression of OsKAT1 rose almost 15-fold under salt treatment, but only 2-fold under no salt treatment (Figure 6b). To verify the expression results, we used real-time PCR to measure the expression of the three transporters and obtained results similar to the RNA-seq data (Figure 6d,e). Therefore, we concluded that OsCIPK9 conferred salt tolerance by affecting the expression of salt-related transporters expressed in the membrane, like OsKAT1.
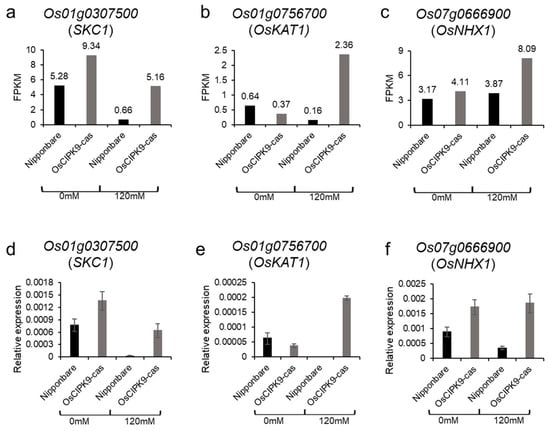
Figure 6.
The expression of related transporters. (a–c) The FPKM of SKC1, OsKAT1, and OsNHX1. (d–f) The relative expressions of SKC1, OsKAT1, and OsNHX1.
2.5. The Haplotypes of OsCIPK9
OsCIPK9 plays a role in salt tolerance, and the haplotype of OsCIPK9 may be helpful in breeding or for germplasm improvement in rice. To determine the haplotypes of OsCIPK9, we analyzed them using SNP data from the Rice3K database. OsCIPK9 contains 21 SNPs with 4 missense variants and 12 SNPs in the promoter (Figure 7a). All nine haplotypes were identified based on SNP variants. Hap1 predominantly emerged in japonica, mostly in temperate Japonica (55.5%), while Hap6 and Hap8 (89.2%) were identified in indica. Hap2 and Hap9 were identified only in the Aus subpopulation. Hap3 and Hap7 were identified in the Basmati subpopulation. Hap5 was identified in various subpopulations but was rarely found in Indica (Figure 7b). A previous study demonstrated that the tolerance level of INDICA was higher than that of japonica at the seedling stage [23]. Therefore, varieties with Hap6 and Hap8 may have a higher salt tolerance than those with Hap1. Subsequently, we analyzed the haplotype network. The results showed that an unknown haplotype connected the haplotypes, mostly in Japonica and Indica (Figure 7c). Hap7, which was found in almost all Aus varieties, was the key haplotype connecting the present haplotypes in Japonica and Indica (Figure 7c). These results indicate that OsCIPK9 has a subpopulation classification, and Hap7 is the key haplotype that connects the haplotypes existing in Japonica and Indica.
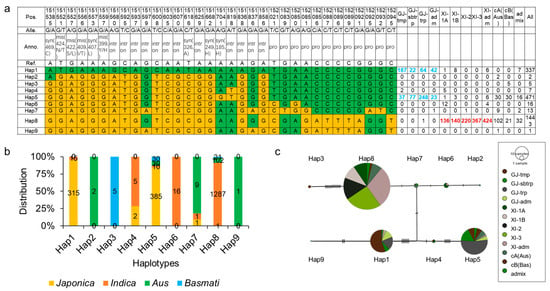
Figure 7.
Analysis of haplotypes of OsCIPK9. (a) Haplotypes of OsCIPK9 based on the Rice3K database. The green represented the same nucleotides with the reference. The yellow represented the variations compared with the reference. (b) The distribution of subpopulations in each haplotype. (c) The gene network of different haplotypes. Pos: position; Alle: allele; Anno: annotation; GJ: japonica, tmp: temperate, sbtrp: subtropical, trp: tropical, adm: admix. XI: indica, Bas: basmati, Pro: promoter variation, Syn: synonymous variation, Mis: missense variation, Intron: intron variation.
3. Discussion
In a previous study, OsCIPK9 was found to regulate ammonium-dependent root growth [15]. Also, AtCIPK23 was found to function in salt tolerance and nitrogen use, including nitrate and ammonium in Arabidopsis. Here, knocking out OsCIPK9 increased the tolerance to salt stress at the seedling stage. Thus, this gene has multiple functions in nitrogen use and salt tolerance. In our results, OsCIPK9-cas also showed higher expressions of OsAMT1.1 and OsAMT2.1 than Nipponbare under salt stress, especially OsAMT1.1, with a two-fold higher expression than the wild type (Figure S5), indicating that OsCIPK9 plays a role not only in salt tolerance but also in ammonium transport. Overall, OsCIPK9 could regulate nitrogen use and salt tolerance like AtCPIK23.
In this study, OsSOS3 was found to interact with OsCIPK9. In a previous study, OsSOS2 was found to be activated by OsSOS3 [24]. Secondly, OsKAT1 and some other transporter genes were differentially expressed. In a previous study, researchers found that CIPK genes could phosphorylate transporters such as AtCIPK23 and CHL1 in Arabidopsis [25]. The phosphorylation of CHL1 has different functions in nitrate transport and sensing. In addition, AtCIPK23 can phosphorylate AtAMT1.1 and influence ammonium uptake [26]. OsSOS2 can phosphorylate OsSOS1, which is a Na+/H+ antiporter 1, and regulates salt tolerance in rice [27]. Further studies should be performed to reveal whether OsSOS3 can activate OsCIPK9 and whether OsCIPK9 can phosphorylate the transporters detected in our study, which would provide more powerful evidence for salt tolerance.
AtSOS3 (AtCBL4) could function in salt tolerance by interacting with AtSOS2 (AtCIPK24) [10]. In our study, we proved that OsSOS3 could also interact with OsCIPK9. This result indicates that the CBL-CIPK network plays a vitally important role in plant salt tolerance. Also, AtSOS3 (AtCBL4) regulates K+ homeostasis through the CBL4-CIPK6-AKT2 pathway [13]. Moreover, AtSOS3 (AtCBL4) was found to be involved in auxin transport [28]. In future studies, more phenotypes may be studied in the OsSOS3-OsCIPK9 pathway.
Other OsCIPKs like OsCIPK04 also responded to salt treatment according to a previous study [19]. With the development of knockout technology, transgenic lines can be produced more easily than before. Our study showed that OsCIPK9 could improve salt tolerance using the knockout line. Future studies could focus on other OsCIPKs which may regulate salt tolerance, and rapidly validate the function using CRISP technology. With more OsCIPKs being identified in the future, we may identify a more comprehensive OsCIPK pathway in salt tolerance.
A haplotype analysis showed that lots of variations were present between japonica and indica. Japonica had a different evolutionary process compared to indica [29]. Thus, these variations may cause a change in OsCIPK9’s function in salt tolerance. Complementation experiments and functional studies of various haplotypes should be conducted in the future. More evidence about the function or causal SNPs of different haplotypes would help in molecular breeding, especially MAS (Marker Assistant Selection).
With progress in gene editing, more editing strategies, such as one-base editing and primer editing, have been used for crop improvements [30]. OsCIPK9 is a negative regulator of salt treatment; therefore, it is better to edit the genomic or promoter region of OsCIPK9 to produce new alleles for breeding. A haplotype analysis showed that the promoter had more polymorphisms than the gene in our study. Therefore, editing the promoter and creating more lines with differential expressions [31] would be useful for breeding based on the evaluation of other agronomic traits.
4. Conclusions
In summary, we identified a negative regulator of salt tolerance in rice, OsCIPK9. In addition, we proposed the possible mechanism in which OsCIPK9 is involved through interaction with OsSOS3. The most affected cellular component was that of the plasma membrane, and the downstream genes of OsCIPK9 may be the transporters located in the plasma membrane, like OsKAT1, as observed through RNA-seq analyses. Finally, we suggest the possible pathway of salt tolerance in rice in which OsCIPK9 is involved (Figure 8). Our study shows that other OsCIPKs could also function in salt tolerance, like OsCIPK24. In addition, OsSOS3, which can interact not only with OsCIPK24 but also with OsCIPK9, regulates salt tolerance in rice. More importantly, our study investigated more OsCIPKs which may regulate salt tolerance in rice.
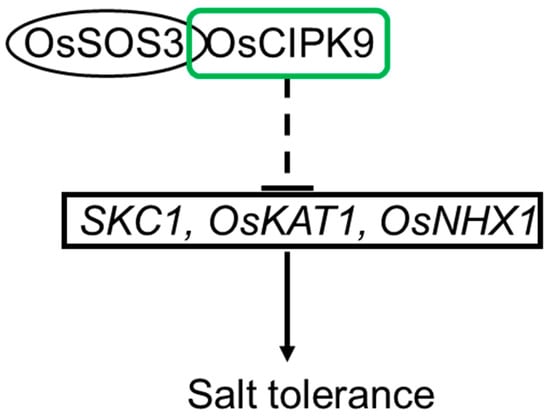
Figure 8.
The pathway of OsCIPK9 in the salt tolerance of rice.
5. Materials and Methods
5.1. Plant Materials and Transgenic Lines Construction
The Line japonica Nipponbare was used as the wild type, and CRISPR/Cas9 technology was used to produce the knockout line (OsCIPK9-cas). The pam sequence is shown in Figure 1a. The full CDS of OsCIPK9 was cloned from the cDNA of Nipponbare, and the pCUbi1390 recombination vector was constructed. Overexpression lines were developed using Agrobacterium-mediated genetic transformation. Two independent overexpression lines (OsCIPK9-OE2 and OsCIPK9-OE3) were isolated from transgenic lines. The expression of overexpression lines was determined using real-time PCR.
5.2. Salt Treatment and Phenotypic Identification
The seeds of all lines were geminated and grown in Yoshida nutrition solution for 14 days. NaCl concentrations of 120 mM and 0 mM (CK) were used for the treatment and control, respectively. The traits, including fresh weight and Na+ and K+ concentrations, were measured 7 days after NaCl treatment. Relative fresh weight = fresh weight under no salt conditions − fresh weight under salt treatment. Na+ and K+ concentrations were determined according to the method described by Wang et al. [32].
5.3. Y2H Assays
A Y2H assay was performed with a MatchMaker GAL4 Two-Hybrid System (Clontech, Mountain View, CA, USA, https://www.takarabio.com/, accessed on 14 August 2023). The full-length coding sequences of genes (OsCIPK9 and OsSOS3) of interest were cloned from the cDNA of Nipponbare into pGADT7 and pGBKT7 (Clontech, Mountain View, CA, USA, https://www.takarabio.com/, accessed on 14 August 2023), and different combinations of constructs were transformed together into the yeast (Saccharomyces cerevisiae) AH109 strain. Positive transformants were selected on synthetic dropout (SD/−Leu/−Trp, DDO) nutrient media, while the interactions were screened in SD medium (SD/−Leu/−Trp/−His/−Ade, QDO).
5.4. Firefly LCI Assays
The coding sequences of OsCIPK9 and OsSOS3 were cloned into the pCAMBIA-nLUC or pCAMBIA-cLUC vectors. These constructed vectors were introduced into the Agrobacterium tumefaciens strain EHA105. Various combinations of EHA105 strains were used to infiltrate N. benthamiana leaves. The relative LUC activity was measured by using a Nightshade LB 985 system (Berthold Technologies, Baden, Germany, 10 August 2023), as described previously [33].
5.5. RNA-Seq
Additionally, the roots of different lines under 0 and 120 mM NaCl treatments were collected for RNA-seq analysis. The total RNA of three independent plants was isolated using a plant RNA purification reagent (Invitrogen, Shanghai, China, 10 November 2022). RNA-seq was performed using the BGI T7 platform, and analyses of DEGs (Differentially Expressed Genes), KEGG (Kyoto Encyclopedia of Genes and Genomes), and GO (Gene Ontology) were performed using Dr. Tom website produced by BGI, China (https://biosys.bgi.com, accessed on 20 January 2023). The expression of all genes produced via RNA-seq is listed in Table S1.
5.6. Real-Time PCR
The total RNA of different tissues and roots at different time points was extracted using an RNA prep Pure Plant Kit (Tiangen Biotech, Beijing, China). Then, ~1 μg of total RNA was reverse-transcribed into cDNA using a PrimeSciptTM Reverse Transcriptase kit (Takara, Shiga, Japan, www.takarabio.com, accessed on 5 March 2023). Quantitative RT-PCR assays were performed using an SYBR Premix Ex Taq™ kit (Takara, Shiga, Japan, www.takarabio.com, accessed on 5 March 2023). Real-time PCR was performed in a real-time PCR machine (I-Cycle, Bio-Rad, Hercules, CA, USA ). The primers are listed in Table S2. The rice UBQ (Os03g0234350) gene was used as an internal control.
5.7. Haplotype Analysis
SNP data of Rice3K were downloaded from the Rice SNP-Seek Database (https://snp-seek.irri.org/, accessed on 5 June 2022). The haplotypes were separated based on all SNP variants of the OsCIPK9 gene and promoter (~2K upstream sequence of OsCIPK9). The subpopulations of rice were divided into japonica (temperate, subtropical, tropical, and admix), indica (1A,1B,2,3, and admix), Aus, Basmati, and admix. The haplotype network was constructed using PopART software (https://popart.maths.otago.ac.nz/, accessed on 5 July 2022) [34].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12213723/s1, Figure S1: Comparison of Nipponbare (Nip), knock-out line (OsCIPK9-cas) and overexpression lines (OsCIPK9-OE2, OsCIPK9-OE3) Na concentrations per plant and Na+/K+ content ratios in shoot and root under 0 mM NaCl conditions; Figure S2: The result of GO cellular component analysis between OsCIPK9-cas and Nipponbare under 0 mM NaCl treatment; Figure S3: The result of GO biological process analysis between OsCIPK9-cas and Nipponbare under 0 mM and 120 mM NaCl treatment; Figure S4: The result of GO molecular function analysis between OsCIPK9-cas and Nipponbare under 0 mM and 120 mM NaCl treatment; Figure S5: The FPKM of OsAMT1.1 and OsAMT2.1 in Nipponbare and knock-out line (OsCIPK9-cas) under 0 mM and 120 mM NaCl conditions; Table S1: The expression of all genes produced by RNA-seq; Table S2: The list of primers.
Author Contributions
Y.Z., B.W. and Z.Z. directed the project. Z.Z. and W.T. performed the experiments. Z.S., J.L., B.Y., Y.L., B.W., D.X. and J.Y. participated in the experiments. Z.Z. and Y.Z. wrote the manuscript. Y.Z. and B.W. revised and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Lianyungang “521 Project” scientific research project, The Financial Grant Support Program of Lianyungang City (QNJJ1803; QNJJ2201), Special Project of Science and Technology in Northern Jiangsu Province (LYG-SZ201930), The Natural Science Foundation of Jiangsu Province (BK20221276) supported this study. The funding agencies had no role in the study design, data collection and analysis, decision to publish or manuscript preparation.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Qin, H.; Li, Y.X.; Huang, R.F. Advances and Challenges in the Breeding of Salt-Tolerant Rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Kota, S.; Flowers, T.J. Salt tolerance in rice: Seedling and reproductive stage QTL mapping come of age. Theor. Appl. Genet. 2021, 134, 3495–3533. [Google Scholar] [CrossRef] [PubMed]
- Nayyeripasand, L.; Garoosi, G.A.; Ahmadikhah, A. Genome-Wide Association Study (GWAS) to Identify Salt-Tolerance QTLs Carrying Novel Candidate Genes in Rice During Early Vegetative Stage. Rice 2021, 14, 9. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhu, C.; Xuan, W.; An, H.; Tian, Y.; Wang, B.; Chi, W.; Chen, G.; Ge, Y.; Li, J.; et al. Genome-wide association studies identify OsWRKY53 as a key regulator of salt tolerance in rice. Nat. Commun. 2023, 14, 3550. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Zhu, G.; Zhou, G.; Younas, M.U.; Suliman, M.S.E.; Liu, J.; Zhu, Y.M.; Salih, E.G.I. Integrated approaches for increasing plant yield under salt stress. Front. Plant Sci. 2023, 14, 1215343. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Liu, B.; Sun, X.; Sun, X.; Zheng, C. New functions of CIPK gene family are continue to emerging. Trends Plant Sci. 2022, 49, 6647–6658. [Google Scholar] [CrossRef]
- Shi, J.; Kim, K.N.; Ritz, O.; Albrecht, V.; Gupta, R.; Harter, K.; Luan, S.; Kudla, J. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 1999, 11, 2393–2405. [Google Scholar]
- Mao, J.; Mo, Z.; Yuan, G.; Xiang, H.; Visser, R.G.F.; Bai, Y.; Liu, H.; Wang, Q.; van der Linden, C.G. The CBL-CIPK network is involved in the physiological crosstalk between plant growth and stress adaptation. Plant Cell Environ. 2022, 46, 3012–3022. [Google Scholar] [CrossRef]
- Qiu, Q.S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef]
- Kim, W.Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Xia, Y.; Xie, Q.; Cao, Y.; Wang, Z.; Hao, G.; Song, J.; Zhou, Y.; Jiang, X. The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 2020, 71, 1801–1814. [Google Scholar] [CrossRef] [PubMed]
- Held, K.; Pascaud, F.; Eckert, C.; Gajdanowicz, P.; Hashimoto, K.; Corratgé-Faillie, C.; Offenborn, J.N.; Lacombe, B.; Dreyer, I.; Thibaud, J.B.; et al. Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 2011, 21, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, S.K.; Kanwar, P.; Yadav, A.K.; Sharma, C.; Kumar, A.; Pandey, G.K. Arabidopsis CBL interacting protein kinase 3 interacts with ABR1, an APETALA2 domain transcription factor, to regulate ABA responses. Plant Sci. 2017, 254, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Y.H.; Kumar, V.; Han, X.; Kim, S.H.; Jeong, J.H.; Kim, C.M.; Gao, Y.; Han, C.D. CBL-INTERACTING PROTEIN KINASE 9 regulates ammonium-dependent root growth downstream of IDD10 in rice (Oryza sativa). Ann. Bot. 2019, 124, 947–960. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Alnayef, M.; Bose, J.; Chen, Z.H.; Venkataraman, G.; Zhou, M.X.; Shabala, L.; Yu, M. Revealing the Role of the Calcineurin B-Like Protein-Interacting Protein Kinase 9 (CIPK9) in Rice Adaptive Responses to Salinity, Osmotic Stress, and K+ Deficiency. Plants 2021, 10, 1513. [Google Scholar] [CrossRef] [PubMed]
- Kurusu, T.; Hamada, J.; Nokajima, H.; Kitagawa, Y.; Kiyoduka, M.; Takahashi, A.; Hanamata, S.; Ohno, R.; Hayashi, T.; Okada, K.; et al. Regulation of Microbe-Associated Molecular Pattern-Induced Hypersensitive Cell Death, Phytoalexin Production, and Defense Gene Expression by Calcineurin B-Like Protein-Interacting Protein Kinases, OsCIPK14/15, in Rice Cultured Cells. Plant Physiol. 2010, 153, 678–692. [Google Scholar] [CrossRef]
- Kanwar, P.; Sanyal, S.K.; Tokas, I.; Yadav, A.K.; Pandey, A.; Kapoor, S.; Pandey, G.K. Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium. 2014, 56, 81–95. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, Y.M.; Xiong, L.Z. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef]
- Kolukisaoglu, U.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef]
- Liu, C.; Mao, B.; Yuan, D.; Chu, C.; Duan, M. Salt tolerance in rice: Physiological responses and molecular mechanisms. Crop J. 2022, 10, 13–25. [Google Scholar] [CrossRef]
- Li, J.; Long, Y.; Qi, G.N.; Li, J.; Xu, Z.J.; Wu, W.H.; Wang, Y. The Os-AKT1 Channel Is Critical for K+ Uptake in Rice Roots and Is Modulated by the Rice CBL1-CIPK23 Complex. Plant Cell 2014, 26, 3387–3402. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Choi, W.Y.; Ko, J.C.; Kim, T.S.; Gregorio, G.B. Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta 2003, 216, 1043–1046. [Google Scholar] [CrossRef] [PubMed]
- El Mahi, H.; Pérez-Hormaeche, J.; De Luca, A.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.J.P.P. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.H.; Lin, S.H.; Hu, H.C.; Tsay, Y.F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Straub, T.; Ludewig, U.; Neuhauser, B. The Kinase CIPK23 Inhibits Ammonium Transport in Arabidopsis thaliana. Plant Cell 2017, 29, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef]
- Kumar Meena, M.; Kumar Vishwakarma, N.; Tripathi, V.; Chattopadhyay, D. CBL-interacting protein kinase 25 contributes to root meristem development. J. Exp. Bot. 2019, 70, 133–147. [Google Scholar] [CrossRef]
- Gutaker, R.M.; Groen, S.C.; Bellis, E.S.; Choi, J.Y.; Pires, I.S.; Bocinsky, R.K.; Slayton, E.R.; Wilkins, O.; Castillo, C.C.; Negrao, S.; et al. Genomic history and ecology of the geographic spread of rice. Nat. Plants 2020, 6, 492–502. [Google Scholar] [CrossRef]
- Mishra, R.; Joshi, R.K.; Zhao, K.J. Base editing in crops: Current advances, limitations and future implications. Plant Biotechnol. J. 2020, 18, 20–31. [Google Scholar] [CrossRef]
- Zeng, D.C.; Liu, T.L.; Ma, X.L.; Wang, B.; Zheng, Z.Y.; Zhang, Y.L.; Xie, X.R.; Yang, B.W.; Zhao, Z.; Zhu, Q.L.; et al. Quantitative regulation of Waxy expression by CRISPR/Cas9-based promoter and 5’UTR-intron editing improves grain quality in rice. Plant Biotechnol. J. 2020, 18, 2385–2387. [Google Scholar] [CrossRef]
- Wang, R.; Jing, W.; Xiao, L.Y.; Jin, Y.K.; Shen, L.; Zhang, W.H. The Rice High-Affinity Potassium Transporter1;1 Is Involved in Salt Tolerance and Regulated by an MYB-Type Transcription Factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
