Effects of Low Nighttime Temperature on Fatty Acid Content in Developing Seeds from Brassica napus L. Based on RNA-Seq and Metabolome
Abstract
1. Introduction
2. Results
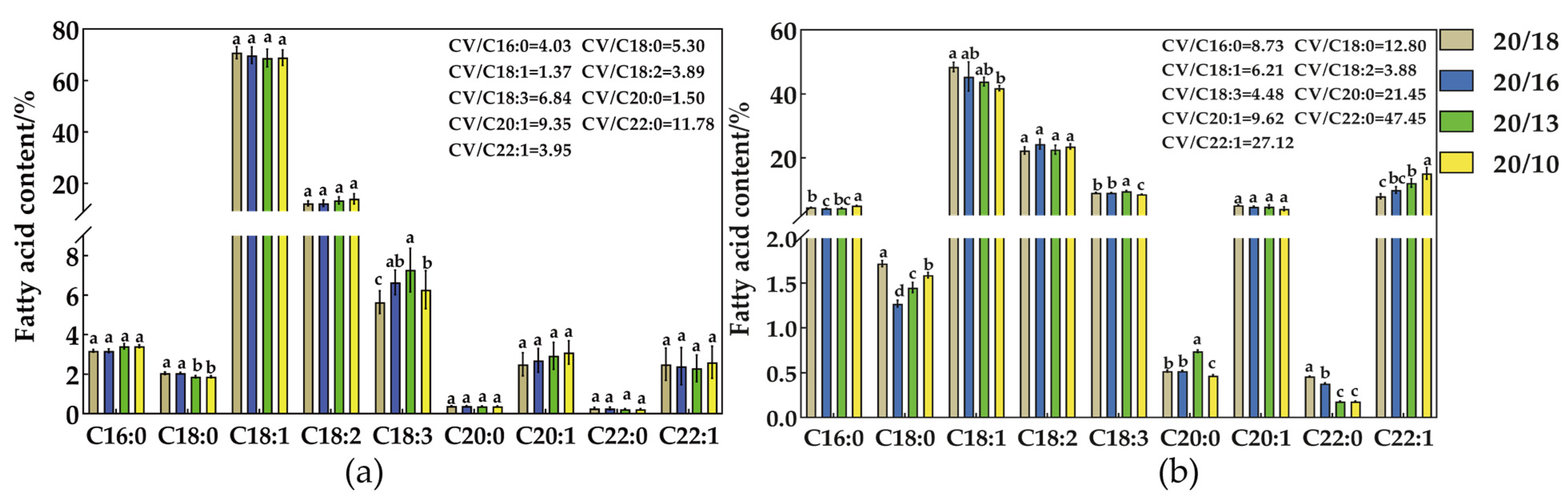
2.1. Effect of Different Nighttime Temperatures on the Fatty Acid Content of Seeds
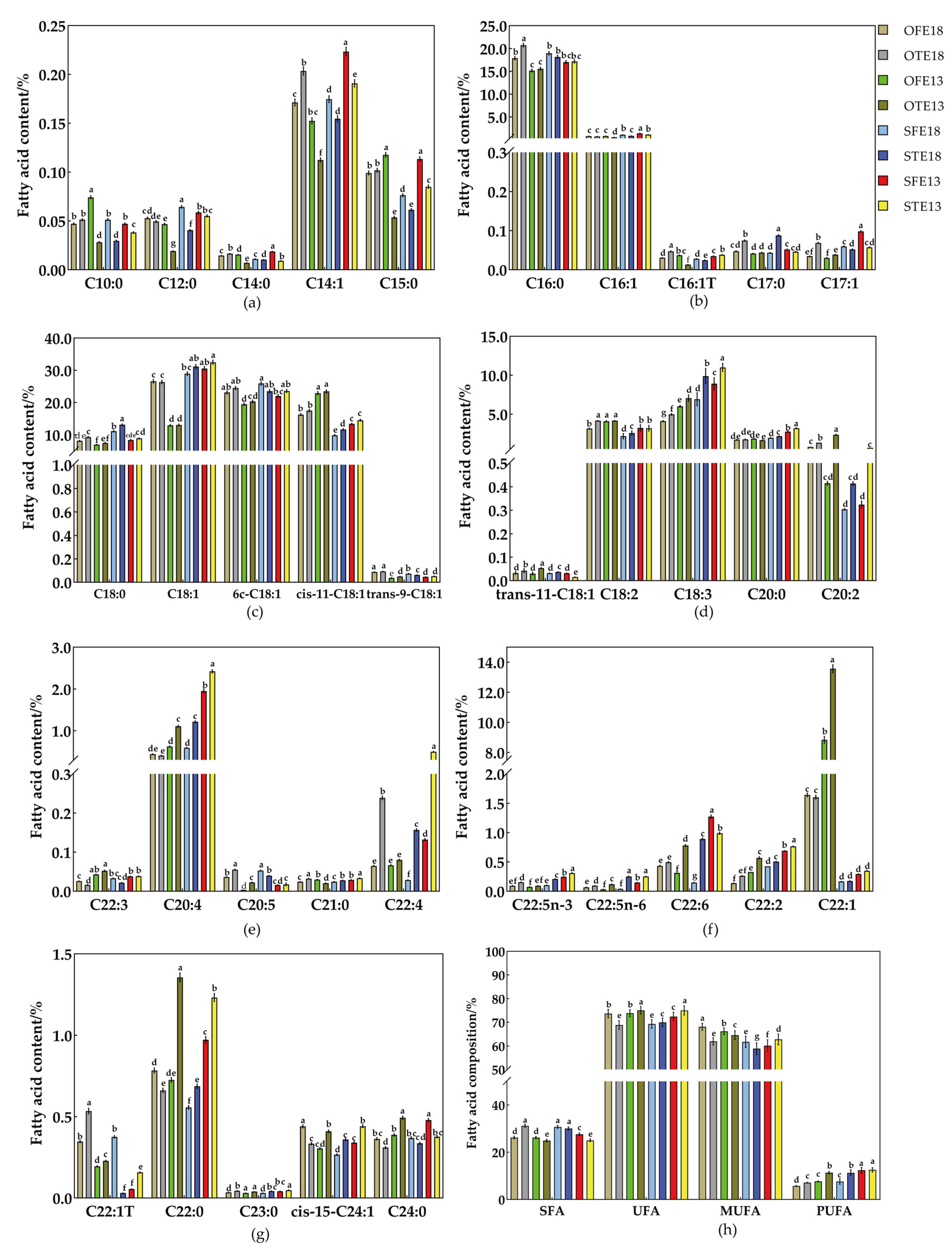
2.2. Qualitative and Quantitative Analyses of Medium- and Long-Chain Fatty Acid Metabolites in Seeds at Different Developmental Stages at Different Nighttime Temperatures
2.3. Transcriptome Analysis of Seeds of at Different Developmental Stages at Different Nighttime Temperatures
2.4. Gene Ontology (GO) Functional Annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis for DEGs at Different Nighttime Temperatures
2.5. DEGs in Pathways Involved in and/or Related to Fatty Acid Metabolism Affected by Different Nighttime Temperatures
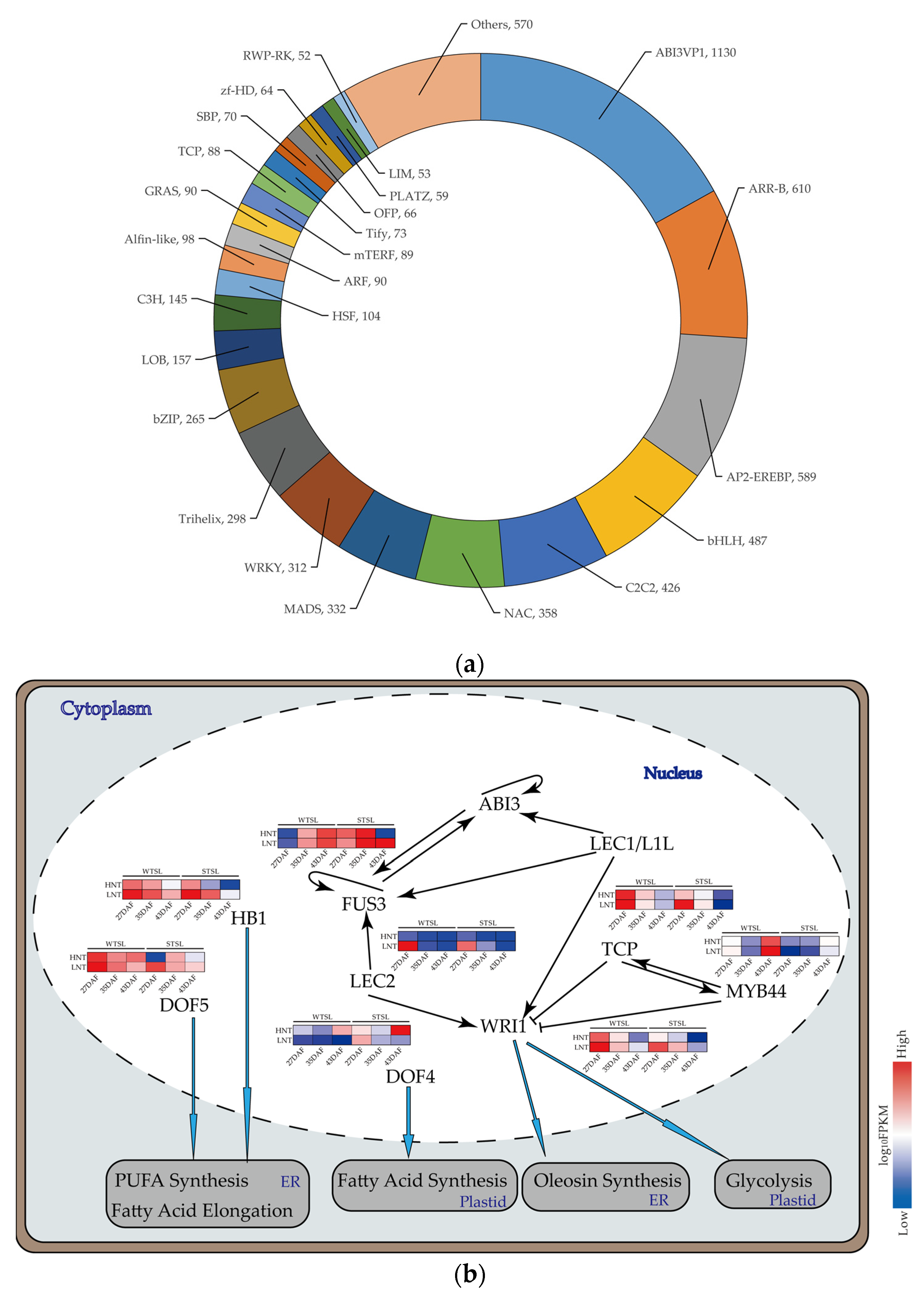
2.6. TFs Related to Metabolism DEGs at Different Nighttime Temperatures
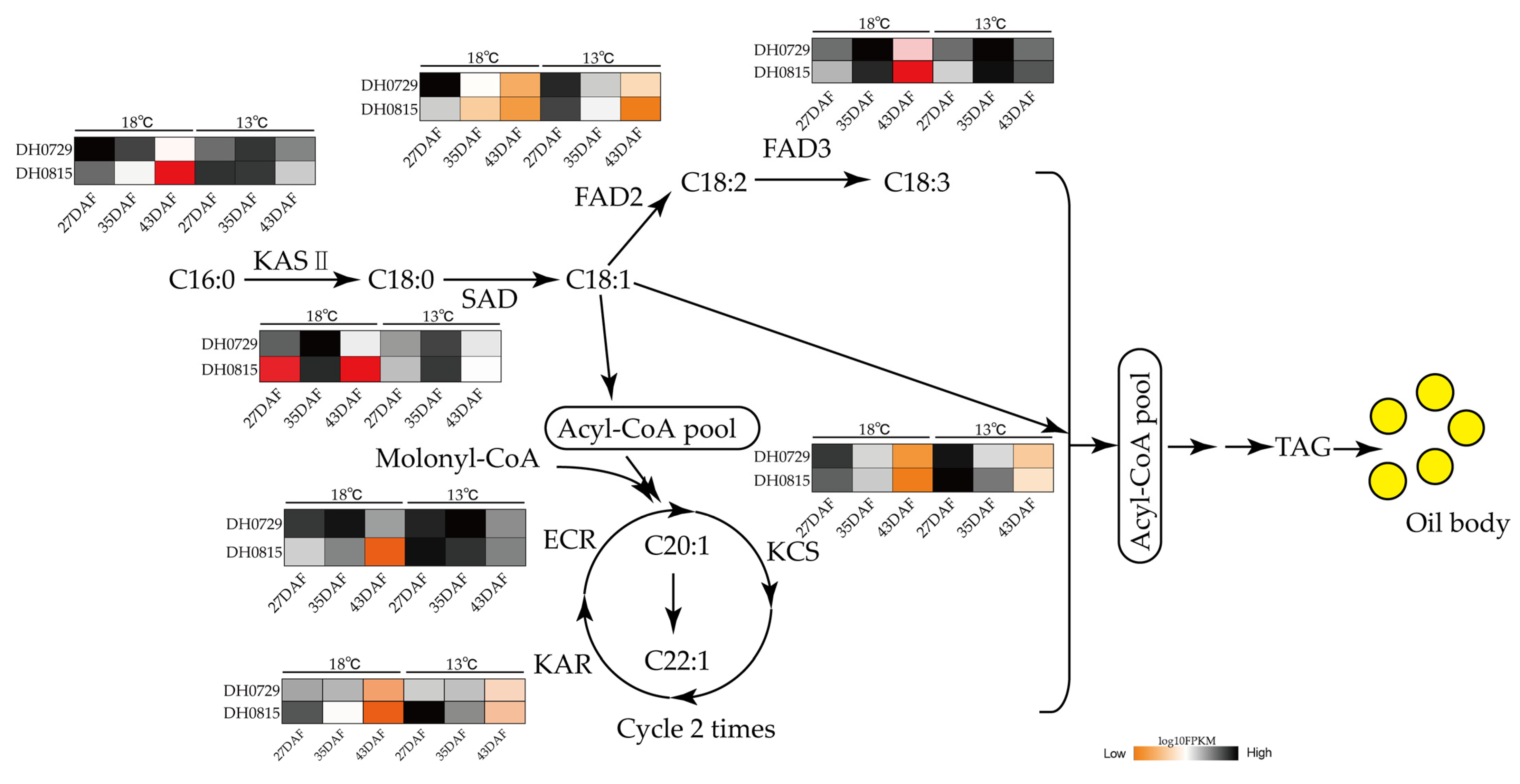
2.7. RNA-Seq Analyses of Fatty Acid Metabolism at Different Nighttime Temperatures
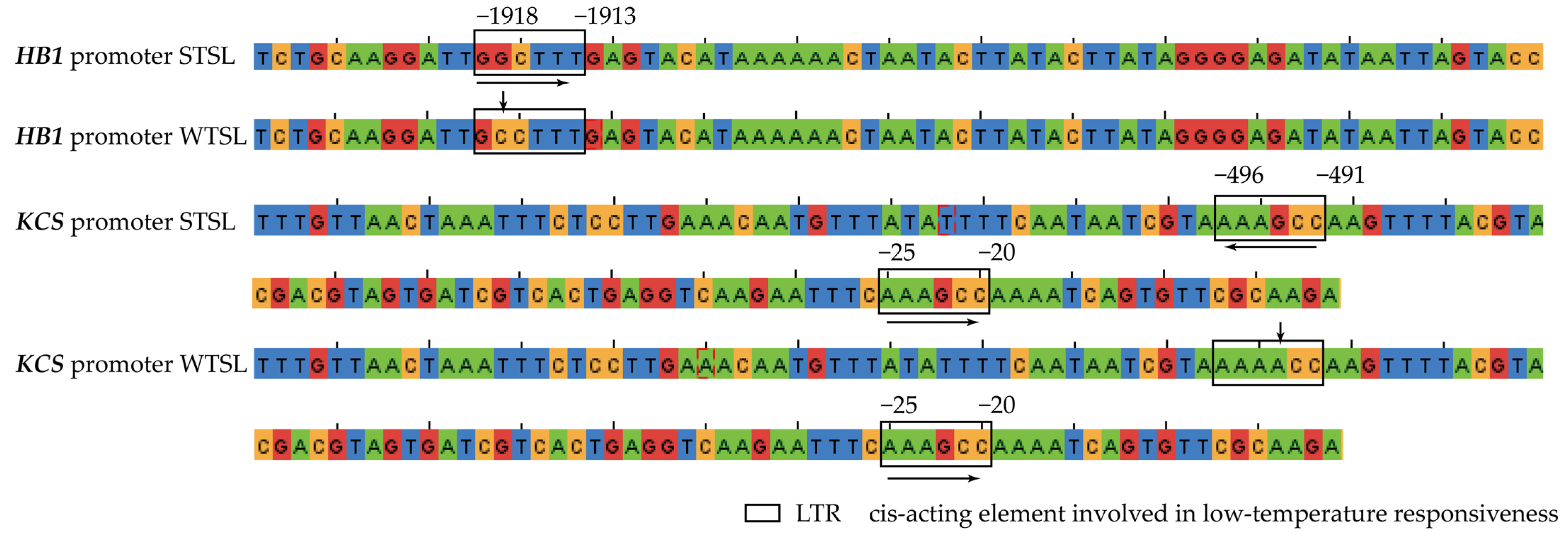
2.8. Prediction of Cis-Acting Elements in DEG Promoters Using plantCARE
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Seed Collection
4.2. Fatty Acid Composition Analysis
4.3. Metabolite Profiling by GC–MS
4.4. RNA Extraction, Illumina Sequencings, and Data Analysis
4.5. Quantitative Real-Time PCR (qRT-PCR)
4.6. Cloning of Cis-Acting Elements in HB1 and KCS Promoters and Cis-Acting Elements of Promoters Using plantCARE
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, S.; Fan, C.; Li, J.; Cai, G.; Yang, Q.; Wu, J.; Yi, X.; Zhang, C.; Zhou, Y. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor. Appl. Genet. 2016, 129, 1203–1215. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Cai, G.; Zu, F.; Zhao, Q.; Qasim, M.U.; Hong, Y.; Fan, C.; Zhou, Y. Comparative transcriptome analysis of developing seeds and silique wall reveals dynamic transcription networks for effective oil production in Brassica napus L. Int. J. Mol. Sci. 2019, 20, 1982. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O’Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a cultivated planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.L.; Zhang, M.L.; Ma, G.J.; Wu, H.; Wu, X.M.; Ren, F.; Li, X.B. Transcriptome profiling analysis reveals the role of silique in controlling seed oil content in Brassica napus. PLoS ONE 2017, 12, e0179027. [Google Scholar] [CrossRef]
- Zhou, L.; Yan, T.; Chen, X.; Li, Z.; Wu, D.; Hua, S.; Jiang, L. Effect of high night temperature on storage lipids and transcriptome changes in developing seeds of oilseed rape. J. Exp. Bot. 2018, 69, 1721–1733. [Google Scholar] [CrossRef]
- Zhu, Y.; Cao, Z.; Xu, F.; Huang, Y.; Chen, M.; Guo, W.; Zhou, W.; Zhu, J.; Meng, J.; Zou, J.; et al. Analysis of gene expression profiles of two near-isogenic lines differing at a QTL region affecting oil content at high temperatures during seed maturation in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 2012, 124, 515–531. [Google Scholar] [CrossRef] [PubMed]
- David, T.C. The effect of temperature on the oil content and fatty acid composition of the oils from several oil seed crops. Can. J. Bot. 1965, 43, 63–69. [Google Scholar]
- Fu, S.X.; Cheng, H.; Qi, C. Microarray analysis of gene expression in seeds of Brassica napus planted in Nanjing (altitude: 8.9 m), Xining (altitude: 2261.2 m) and Lhasa (altitude: 3658 m) with different oil content. Mol. Biol. Rep. 2009, 36, 2375–2386. [Google Scholar] [CrossRef]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef]
- Ye, Y.; Nikovics, K.; To, A.; Lepiniec, L.; Fedosejevs, E.T.; Van Doren, S.R.; Baud, S.; Thelen, J.J. Docking of acetyl-CoA carboxylase to the plastid envelope membrane attenuates fatty acid production in plants. Nat. Commun. 2020, 11, 6191. [Google Scholar] [CrossRef]
- Liu, H.; Gu, J.; Lu, Q.; Li, H.; Hong, Y.; Chen, X.; Ren, L.; Deng, L.; Liang, X. Transcriptomic analysis reveals the high-oleic acid feedback regulating the homologous gene expression of stearoyl-ACP desaturase 2 (SAD2) in Peanuts. Int. J. Mol. Sci. 2019, 20, 3091. [Google Scholar] [CrossRef] [PubMed]
- Bryant, F.M.; Munoz-Azcarate, O.; Kelly, A.A.; Beaudoin, F.; Kurup, S.; Eastmond, P.J. ACYL-ACYL CARRIER PROTEIN DESATURASE 2 and 3 are responsible for making omega-7 fatty acids in the Arabidopsis aleurone. Plant Physiol. 2016, 172, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Kamil, D.; Simon, J.; Ida, L.; Agnieszka, M.; Katarzyna, J.G.; Małgorzata, W.; Sten, S.; Antoni, B. Isoforms of acyl-CoA:diacylglycerol acyltransferase 2 differ substantially in their specificities toward erucic acid. Plant Physiol. 2019, 181, 1468–1479. [Google Scholar]
- Harwood, J.L. Working with randy: The diacylglycerol acyltransferase story. Lipids 2020, 55, 419–423. [Google Scholar] [CrossRef]
- Xue, Y.; Jiang, J.; Yang, X.; Jiang, H.; Du, Y.; Liu, X.; Xie, R.; Chai, Y. Genome-wide mining and comparative analysis of fatty acid elongase gene family in Brassica napus and its progenitors. Gene 2020, 747, 144674. [Google Scholar] [CrossRef]
- Lu, S.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J. 2018, 94, 915–932. [Google Scholar] [CrossRef]
- Tan, H.; Yang, X.; Zhang, F.; Zheng, X.; Qu, C.; Mu, J.; Fu, F.; Li, J.; Guan, R.; Zhang, H.; et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON 1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011, 156, 1577–1588. [Google Scholar] [CrossRef]
- Mu, J.; Tan, H.; Zheng, Q.; Fu, F.; Liang, Y.; Zhang, J.; Yang, X.; Wang, T.; Chong, K.; Wang, X.J.; et al. LEAFY COTYLEDON 1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054. [Google Scholar] [CrossRef]
- Kagaya, Y.; Toyoshima, R.; Okuda, R.; Usui, H.; Yamamoto, A.; Hattori, T. LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE 3. Plant Cell Physiol. 2005, 46, 399–406. [Google Scholar] [CrossRef]
- Rajavel, A.; Klees, S.; Schlüter, J.S.; Bertram, H.; Lu, K.; Schmitt, A.O.; Gültas, M. Unravelling the complex interplay of transcription factors orchestrating seed oil content in Brassica napus L. Int. J. Mol. Sci. 2021, 22, 1033. [Google Scholar] [CrossRef]
- Fernández-Moya, V.; Martínez-Force, E.; Garcés, R. Temperature effect on a high stearic acid sunflower mutant. Phytochemistry 2002, 59, 33–37. [Google Scholar] [CrossRef]
- Chen, M.; Du, X.; Zhu, Y.; Wang, Z.; Hua, S.; Li, Z.; Guo, W.; Zhang, G.; Peng, J.; Jiang, L. Seed fatty acid reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ. 2012, 35, 2155–2169. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, A.S.; Taylor, D.C.; Giblin, M.; Ferrie, A.M.; Ambrose, S.J.; Ross, A.R.; Nelson, K.M.; Irina Zaharia, L.; Sharma, N.; Anderson, M.; et al. Hormonal regulation of oil accumulation in Brassica seeds: Metabolism and biological activity of ABA, 7′-, 8′- and 9′-hydroxy ABA in microspore derived embryos of B. napus. Phytochemistry 2008, 69, 2678–2688. [Google Scholar] [CrossRef]
- Jo, L.; Pelletier, J.M.; Harada, J.J. Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J Integr Plant Biol. 2019, 61, 564–580. [Google Scholar] [CrossRef]
- Kuczynski, C.; McCorkle, S.; Keereetaweep, J.; Shanklin, J.; Schwender, J. An expanded role for the transcription factor WRINKLED1 in the biosynthesis of triacylglycerols during seed development. Front. Plant Sci. 2022, 13, 955589. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, Y.; Zhang, J.; Ji, F.; Jin, F.; Fan, W.; Pei, D. Transcriptome analysis of Walnut (Juglans regia L.) embryos reveals key developmental stages and genes involved in lipid biosynthesis and polyunsaturated fatty acid metabolism. J. Agric. Food Chem. 2021, 69, 377–396. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Zhang, B.; Hao, Y.J.; Huang, J.; Tian, A.G.; Liao, Y.; Zhang, J.S.; Chen, S.Y. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 2007, 52, 716–729. [Google Scholar] [CrossRef] [PubMed]
- Elahi, N.; Duncan, R.W.; Stasolla, C. Modification of oil and glucosinolate content in canola seeds with altered expression of Brassica napus LEAFY COTYLEDON 1. Plant Physiol. Biochem. 2016, 100, 52–63. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, P.; Li, W.; Feng, T.; Shockey, J.; Chen, L.; Zhang, L.; Lü, S. Triacylglycerol biosynthesis in shaded seeds of tung tree (Vernicia fordii) is regulated in part by Homeodomain Leucine Zipper 21. Plant J. 2021, 108, 1735–1753. [Google Scholar] [CrossRef] [PubMed]
- Magrin, G.; Travasso, M.; Rodrlguez, G. Changes in climate and crop production during the 20th century in Argentina. Clim. Change 2005, 72, 229–249. [Google Scholar] [CrossRef]
- Lobell, D.B.; Field, C.B. Global scale climate-crop yield relationships and the impacts of recent warming. Environ. Res. Lett. 2007, 2, 14002. [Google Scholar] [CrossRef]
- García, G.A.; Dreccer, M.F.; Miralles, D.J.; Serrago, R.A. High night temperatures during grain number determination reduce wheat and barley grain yield: A field study. Glob. Change Biol. 2015, 21, 4153–4164. [Google Scholar] [CrossRef] [PubMed]
- Ismaili, A.; Salavati, A.; Mohammadi, P.P. A comparative proteomic analysis of responses to high temperature stress in hypocotyl of Canola (Brassica napus L.). Protein Pept. Lett. 2014, 22, 285–299. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, Q.; Shen, W.; Cram, D.; Fowler, D.B.; Wei, Y.; Zou, J. Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 2015, 27, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Menard, G.N.; Moreno, J.M.; Bryant, F.M.; Munoz-Azcarate, O.; Kelly, A.A.; Hassani-Pak, K.; Kurup, S.; Eastmond, P.J. Genome wide analysis of fatty acid desaturation and its response to temperature. Plant Physiol. 2017, 173, 1594–1605. [Google Scholar] [CrossRef]
- Nesi, N.; Delourme, R.; Brégeon, M.; Falentin, C.; Renard, M. Genetic and molecular approaches to improve nutritional value of Brassica napus L. seed. Comptes Rendus Biol. 2008, 331, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Lang, C.; Wang, F.; Wu, X.; Liu, R.; Zheng, T.; Zhang, D.; Chen, J.; Wu, G. Depressed expression of FAE1 and FAD2 genes modifies fatty acid profiles and storage compounds accumulation in Brassica napus seeds. Plant Sci. 2017, 263, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Singer, S.D.; Weselake, R.J.; Rahman, H. Development and characterization of low α-linolenic acid Brassica oleracea lines bearing a novel mutation in a ‘class a’ FATTY ACID DESATURASE 3 gene. BMC Genet. 2014, 15, 94. [Google Scholar] [CrossRef]
- Wu, G.; Wu, Y.; Xiao, L.; Li, X.; Lu, C. Zero erucic acid trait of rapeseed (Brassica napus L.) results from a deletion of four base pairs in the fatty acid elongase 1 gene. Theor. Appl. Genet. 2008, 116, 491–499. [Google Scholar] [CrossRef]
- Shi, J.H.; Lang, C.X.; Wu, X.L.; Liu, R.H.; Zheng, T.; Zhang, D.Q.; Chen, J.Q.; Wu, G.T. RNAi knockdown of fatty acid elongase1 alters fatty acid composition in Brassica napus. Biochem. Biophys. Res. Commun. 2015, 466, 518–522. [Google Scholar] [CrossRef]
- Wang, P.; Xiong, X.; Zhang, X.; Wu, G.; Liu, F. A Review of erucic acid production in Brassicaceae oilseeds: Progress and prospects for the genetic engineering of high and low-erucic acid rapeseeds (Brassica napus). Front. Plant Sci. 2022, 13, 899076. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Ma, W. WRINKLED1 transcription factor: How much do we know about its regulatory mechanism? Plant Sci. 2018, 272, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, G.; Liu, C.; Liu, S. LEAFY COTYLEDON 2: A regulatory factor of plant growth and seed development. Genes 2021, 12, 1896. [Google Scholar] [CrossRef]
- Miller, C.; Wells, R.; McKenzie, N.; Trick, M.; Ball, J.; Fatihi, A.; Dubreucq, B.; Chardot, T.; Lepiniec, L.; Bevan, M.W. Variation in expression of the HECT E3 ligase UPL3 modulates LEC2 levels, seed size, and crop yields in Brassica napus. Plant Cell 2019, 31, 2370–2385. [Google Scholar] [CrossRef] [PubMed]
- Verdier, J.; Thompson, R.D. Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant Cell Physiol. 2008, 49, 1263–1271. [Google Scholar] [CrossRef]
- Fu, R.; Liu, W.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Comprehensive analysis of the homeodomain-leucine zipper IV transcription factor family in Cucumis sativus. Genome 2013, 56, 395–405. [Google Scholar] [CrossRef]
- Ibáñez-Salazar, A.; Rosales-Mendoza, S.; Rocha-Uribe, A.; Ramírez-Alonso, J.I.; Lara-Hernández, I.; Hernández-Torres, A.; Paz-Maldonado, L.M.; Silva-Ramírez, A.S.; Bañuelos-Hernández, B.; Martínez-Salgado, J.L.; et al. Over-expression of Dof-type transcription factor increases lipid production in Chlamydomonas reinhardtii. J. Biotechnol. 2014, 184, 27–38. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, J.; Li, S.; Liu, L.; Qanmber, G.; Chen, G.; Duan, Z.; Zhao, N.; Wang, G. Systematic characterization of TCP gene family in four cotton species revealed that GhTCP62 regulates branching in Arabidopsis. Biology 2021, 10, 1104. [Google Scholar] [CrossRef]
- Li, D.; Jin, C.; Duan, S.; Zhu, Y.; Qi, S.; Liu, K.; Gao, C.; Ma, H.; Zhang, M.; Liao, Y.; et al. MYB89 transcription factor represses seed oil accumulation. Plant Physiol. 2017, 173, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Salie, M.J.; Thelen, J.J. Regulation and structure of the heteromeric acetyl-CoA carboxylase. Biochim. Biophys. Acta 2016, 1861 Pt B, 1207–1213. [Google Scholar] [CrossRef]
- Sun, F.; Fan, G.; Hu, Q.; Zhou, Y.; Guan, M.; Tong, C.; Li, J.; Du, D.; Qi, C.; Jiang, L.; et al. The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 2017, 92, 452–468. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Lu, Z.C.; Jin, K.M.; Qiu, W.M.; Qiao, G.R.; Han, X.J.; Zhuo, R.Y. SPDE: A multi-functional software for sequence processing and data extraction. Bioinformatics 2021, 20, 3686–3687. [Google Scholar] [CrossRef] [PubMed]


| Daytime/Nighttime Temperature/°C | Sowing to Emergence | Emergence to Bolting | Bolting to Beginning of Flowering | Beginning of Flowering to Full Flowering Stage | Full Flowering to the Final Flowering | Final Flowering to Maturing | Whole Growth Period | |
|---|---|---|---|---|---|---|---|---|
| WTSL | 20/18 | 5.00 ± 1.00 a | 82.00 ± 1.00 a | 5.33 ± 0.58 c | 8.00 ± 1.00 c | 14.00 ± 1.00 c | 35.00 ± 0.58 c | 147.33 ± 4.73 d |
| 20/16 | 4.67 ± 0.58 a | 82.33 ± 1.15 a | 7.00 ± 1.00 c | 9.00 ± 1.00 c | 15.00 ± 1.00 c | 36.67 ± 1.53 c | 154.67 ± 3.21 c | |
| 20/13 | 4.67 ± 0.58 a | 82.00 ± 1.00 a | 10.00 ± 1.00 b | 11.00 ± 1.00 b | 20.00 ± 1.00 b | 45.00 ± 1.00 b | 172.67 ± 2.08 b | |
| 20/10 | 4.67 ± 0.58 a | 82.00 ± 1.00 a | 12.00 ± 1.00 a | 14.00 ± 1.00 a | 24.00 ± 1.00 a | 51.33 ± 0.58 a | 188.00 ± 3.00 a | |
| Mean | 4.75 | 82.08 | 8.58 | 10.50 | 18.25 | 42.00 | 165.67 | |
| SD | 0.17 | 0.17 | 2.99 | 2.65 | 4.65 | 7.61 | 18.30 | |
| CV/% | 3.51 | 0.20 | 34.79 | 25.20 | 25.46 | 18.11 | 11.05 | |
| STSL | 20/18 | 4.67 ± 0.58 a | 81.00 ± 1.00 a | 7.00 ± 1.00 c | 7.00 ± 1.00 d | 15.00 ± 1.00 d | 32.33 ± 0.58 d | 149.00 ± 3.46 d |
| 20/16 | 4.67 ± 0.58 a | 80.00 ± 1.00 a | 8.00 ± 1.00 c | 9.33 ± 0.58 c | 18.00 ± 1.00 c | 36.67 ± 1.15 c | 156.33 ± 0.58 c | |
| 20/13 | 5.00 ± 1.00 a | 81.00 ± 1.00 a | 11.00 ± 1.00 b | 12.33 ± 0.58 b | 24.33 ± 0.58 b | 42.00 ± 1.00 b | 175.00 ± 1.73 b | |
| 20/10 | 4.67 ± 0.58 a | 82.00 ± 1.00 a | 14.33 ± 0.58 a | 14.00 ± 1.00 a | 40.00 ± 1.00 a | 46.33 ± 1.53 a | 203.00 ± 1.73 a | |
| Mean | 4.75 | 81.00 | 10.00 | 10.50 | 24.25 | 39.33 | 170.83 | |
| SD | 0.17 | 0.82 | 3.16 | 3.11 | 11.15 | 6.12 | 24.08 | |
| CV/% | 3.51 | 1.01 | 31.62 | 29.61 | 45.97 | 15.55 | 14.09 |
| Group Comparisons | Total No. of Significantly DEGs | Total No. of Significantly Upregulated DEGs | Total No. of Significantly Downregulated DEGs |
|---|---|---|---|
| SSE18 vs. SFE18 | 7136 | 1604 | 5532 |
| SFE18 vs. STE18 | 10,407 | 4296 | 6111 |
| SSE13 vs. SFE13 | 14,695 | 3928 | 10,767 |
| SFE13 vs. STE13 | 2790 | 795 | 1995 |
| SSE18 vs. SSE13 | 6862 | 4213 | 2649 |
| SFE18 vs. SFE13 | 270 | 199 | 71 |
| STE18 vs. STE13 | 4036 | 1905 | 2131 |
| OSE18 vs. OFE18 | 2033 | 1230 | 803 |
| OFE18 vs. OTE18 | 3675 | 1166 | 2509 |
| OSE13 vs. OFE13 | 7964 | 2823 | 5141 |
| OFE13 vs. OTE13 | 3454 | 1460 | 1994 |
| OSE18 vs. OSE13 | 5804 | 3042 | 2762 |
| OFE18 vs. OFE13 | 1939 | 1120 | 819 |
| OTE18 vs. OTE13 | 1534 | 1000 | 534 |
| Pathway | SSE18 vs. SFE18 | SFE18 vs. STE18 | SSE13 vs. SFE13 | SFE13 vs. STE13 | SSE18 vs. SSE13 | SFE18 vs. SFE13 | STE18 vs. STE13 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | |
| ABC transporters | 2 | 6 | 4 | 10 | 4 | 16 | 2 | 2 | 3 | 0 | 0 | 0 | 0 | 2 |
| Alpha-linolenic acid metabolism | 6 | 8 | 10 | 29 | 3 | 15 | 5 | 4 | 14 | 6 | 3 | 1 | 7 | 12 |
| Arachidonic acid metabolism | 0 | 6 | 4 | 8 | 0 | 12 | 0 | 0 | 6 | 1 | 1 | 0 | 5 | 1 |
| Biosynthesis of unsaturated fatty acids | 1 | 4 | 8 | 17 | 3 | 4 | 1 | 0 | 1 | 7 | 0 | 0 | 1 | 6 |
| Carbon metabolism | 11 | 134 | 29 | 180 | 38 | 159 | 6 | 63 | 33 | 25 | 0 | 1 | 68 | 37 |
| Circadian rhythm—plant | 1 | 4 | 2 | 8 | 3 | 6 | 0 | 4 | 7 | 4 | 0 | 0 | 1 | 2 |
| Citrate cycle (TCA cycle) | 2 | 13 | 9 | 26 | 10 | 16 | 1 | 1 | 4 | 7 | 0 | 0 | 7 | 6 |
| Cutin, suberine, and wax biosynthesis | 20 | 1 | 20 | 14 | 33 | 8 | 8 | 0 | 4 | 15 | 2 | 0 | 0 | 11 |
| Fatty acid biosynthesis | 2 | 28 | 10 | 38 | 2 | 19 | 0 | 1 | 4 | 7 | 0 | 0 | 5 | 9 |
| Fatty acid degradation | 9 | 10 | 13 | 29 | 14 | 15 | 5 | 1 | 6 | 2 | 1 | 0 | 3 | 16 |
| Fatty acid elongation | 2 | 14 | 5 | 29 | 5 | 30 | 1 | 4 | 10 | 5 | 0 | 0 | 4 | 5 |
| Fatty acid metabolism | 3 | 30 | 29 | 34 | 7 | 27 | 1 | 1 | 7 | 9 | 0 | 0 | 5 | 14 |
| Fructose and mannose metabolism | 1 | 44 | 20 | 30 | 5 | 46 | 1 | 15 | 13 | 5 | 2 | 1 | 12 | 6 |
| Glycerolipid metabolism | 11 | 18 | 20 | 31 | 21 | 41 | 3 | 6 | 14 | 11 | 3 | 0 | 6 | 9 |
| Glycerophospholipid metabolism | 6 | 12 | 22 | 29 | 20 | 44 | 4 | 4 | 11 | 11 | 2 | 0 | 9 | 11 |
| Glycolysis/Gluconeogenesis | 8 | 63 | 17 | 75 | 15 | 73 | 4 | 17 | 15 | 19 | 1 | 2 | 22 | 16 |
| Glyoxylate and dicarboxylate metabolism | 5 | 39 | 27 | 65 | 18 | 50 | 1 | 22 | 14 | 7 | 1 | 0 | 26 | 22 |
| Indole alkaloid biosynthesis | 2 | 3 | 3 | 7 | 3 | 6 | 1 | 4 | 4 | 1 | 1 | 0 | 1 | 0 |
| Linoleic acid metabolism | 0 | 1 | 2 | 8 | 1 | 3 | 1 | 0 | 6 | 1 | 1 | 0 | 4 | 1 |
| Oxidative phosphorylation | 5 | 30 | 5 | 26 | 9 | 49 | 3 | 11 | 10 | 4 | 1 | 1 | 6 | 5 |
| Phosphatidylinositol signaling system | 5 | 9 | 3 | 20 | 7 | 35 | 4 | 5 | 11 | 4 | 0 | 0 | 6 | 2 |
| Photosynthesis | 1 | 109 | 25 | 100 | 5 | 71 | 1 | 89 | 1 | 7 | 0 | 1 | 68 | 2 |
| Plant hormone signal transduction | 18 | 52 | 25 | 150 | 55 | 119 | 10 | 17 | 65 | 43 | 3 | 1 | 12 | 55 |
| Propanoate metabolism | 0 | 10 | 2 | 20 | 2 | 11 | 0 | 1 | 3 | 0 | 0 | 0 | 4 | 1 |
| Protein processing in endoplasmic reticulum | 6 | 29 | 20 | 62 | 10 | 100 | 8 | 8 | 37 | 5 | 1 | 1 | 5 | 40 |
| Pyruvate metabolism | 5 | 21 | 24 | 32 | 21 | 31 | 2 | 4 | 9 | 5 | 1 | 0 | 9 | 12 |
| Starch and sucrose metabolism | 19 | 74 | 39 | 95 | 26 | 121 | 10 | 34 | 65 | 25 | 7 | 0 | 28 | 20 |
| Tryptophan metabolism | 14 | 13 | 32 | 18 | 28 | 26 | 2 | 6 | 12 | 25 | 2 | 0 | 15 | 27 |
| Total | 165 | 785 | 429 | 1190 | 368 | 1153 | 85 | 324 | 389 | 261 | 33 | 8 | 339 | 350 |
| Pathway | OSE18 vs. OFE18 | OFE18 vs. OTE18 | OSE13 vs. OFE13 | OFE13 vs. OTE13 | OSE18 vs. OSE13 | OFE18 vs. OFE13 | OTE18 vs. OTE13 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | |
| ABC transporters | 1 | 0 | 1 | 1 | 1 | 4 | 2 | 3 | 2 | 1 | 1 | 1 | 0 | 3 |
| Alpha-linolenic acid metabolism | 3 | 1 | 2 | 7 | 6 | 7 | 5 | 4 | 4 | 6 | 1 | 2 | 9 | 3 |
| Arachidonic acid metabolism | 1 | 1 | 2 | 4 | 1 | 6 | 0 | 2 | 2 | 0 | 1 | 1 | 0 | 1 |
| Biosynthesis of unsaturated fatty acids | 5 | 0 | 0 | 10 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 3 | 1 |
| Carbon metabolism | 20 | 9 | 6 | 57 | 15 | 76 | 16 | 50 | 40 | 36 | 20 | 10 | 21 | 11 |
| Circadian rhythm—plant | 2 | 0 | 4 | 4 | 7 | 4 | 0 | 0 | 5 | 5 | 1 | 0 | 1 | 0 |
| Citrate cycle (TCA cycle) | 5 | 1 | 4 | 14 | 7 | 11 | 5 | 0 | 7 | 14 | 0 | 4 | 6 | 3 |
| Cutin, suberine, and wax biosynthesis | 0 | 0 | 0 | 2 | 19 | 1 | 28 | 1 | 3 | 28 | 2 | 3 | 7 | 0 |
| Fatty acid biosynthesis | 5 | 1 | 2 | 18 | 1 | 0 | 5 | 4 | 5 | 4 | 1 | 7 | 6 | 0 |
| Fatty acid degradation | 2 | 0 | 4 | 5 | 4 | 1 | 12 | 2 | 4 | 11 | 1 | 4 | 8 | 2 |
| Fatty acid elongation | 2 | 1 | 1 | 7 | 2 | 13 | 8 | 5 | 12 | 10 | 2 | 3 | 8 | 0 |
| Fatty acid metabolism | 6 | 1 | 3 | 26 | 3 | 3 | 5 | 4 | 4 | 7 | 4 | 1 | 9 | 1 |
| Fructose and mannose metabolism | 2 | 3 | 3 | 20 | 1 | 21 | 2 | 9 | 10 | 4 | 5 | 9 | 5 | 4 |
| Glycerolipid metabolism | 8 | 0 | 4 | 9 | 10 | 19 | 15 | 4 | 16 | 14 | 2 | 8 | 8 | 4 |
| Glycerophospholipid metabolism | 3 | 1 | 8 | 8 | 13 | 17 | 19 | 5 | 11 | 14 | 2 | 7 | 9 | 2 |
| Glycolysis/Gluconeogenesis | 8 | 0 | 8 | 41 | 7 | 29 | 9 | 11 | 14 | 13 | 6 | 6 | 10 | 5 |
| Glyoxylate and dicarboxylate metabolism | 3 | 6 | 11 | 32 | 9 | 28 | 12 | 16 | 13 | 21 | 6 | 6 | 11 | 5 |
| Indole alkaloid biosynthesis | 0 | 1 | 0 | 3 | 1 | 2 | 0 | 2 | 1 | 1 | 1 | 0 | 1 | 0 |
| Linoleic acid metabolism | 1 | 0 | 0 | 2 | 3 | 3 | 1 | 1 | 3 | 3 | 0 | 2 | 3 | 1 |
| Oxidative phosphorylation | 9 | 1 | 2 | 14 | 3 | 36 | 2 | 7 | 25 | 4 | 3 | 3 | 1 | 1 |
| Phosphatidylinositol signaling system | 4 | 4 | 2 | 4 | 9 | 6 | 1 | 3 | 11 | 5 | 9 | 0 | 1 | 0 |
| Photosynthesis | 3 | 16 | 0 | 78 | 1 | 33 | 2 | 66 | 6 | 5 | 18 | 2 | 21 | 0 |
| Plant hormone signal transduction | 6 | 12 | 12 | 23 | 45 | 47 | 25 | 34 | 24 | 43 | 14 | 8 | 8 | 5 |
| Propanoate metabolism | 3 | 1 | 0 | 7 | 0 | 3 | 2 | 1 | 5 | 3 | 2 | 2 | 1 | 0 |
| Protein processing in endoplasmic reticulum | 12 | 2 | 23 | 31 | 18 | 40 | 6 | 5 | 22 | 13 | 3 | 4 | 4 | 14 |
| Pyruvate metabolism | 6 | 2 | 8 | 16 | 6 | 9 | 11 | 1 | 9 | 19 | 2 | 7 | 5 | 4 |
| Starch and sucrose metabolism | 6 | 3 | 12 | 45 | 22 | 65 | 12 | 19 | 44 | 25 | 8 | 11 | 10 | 6 |
| Tryptophan metabolism | 4 | 0 | 12 | 12 | 19 | 16 | 10 | 4 | 10 | 26 | 1 | 6 | 2 | 8 |
| Total | 130 | 67 | 134 | 500 | 234 | 500 | 215 | 265 | 312 | 337 | 116 | 117 | 178 | 84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mi, C.; Sun, C.; Yuan, Y.; Li, F.; Wang, Q.; Zhu, H.; Hua, S.; Lin, L. Effects of Low Nighttime Temperature on Fatty Acid Content in Developing Seeds from Brassica napus L. Based on RNA-Seq and Metabolome. Plants 2023, 12, 325. https://doi.org/10.3390/plants12020325
Mi C, Sun C, Yuan Y, Li F, Wang Q, Zhu H, Hua S, Lin L. Effects of Low Nighttime Temperature on Fatty Acid Content in Developing Seeds from Brassica napus L. Based on RNA-Seq and Metabolome. Plants. 2023; 12(2):325. https://doi.org/10.3390/plants12020325
Chicago/Turabian StyleMi, Chao, Chao Sun, Yuting Yuan, Fei Li, Qian Wang, Haiping Zhu, Shuijin Hua, and Liangbin Lin. 2023. "Effects of Low Nighttime Temperature on Fatty Acid Content in Developing Seeds from Brassica napus L. Based on RNA-Seq and Metabolome" Plants 12, no. 2: 325. https://doi.org/10.3390/plants12020325
APA StyleMi, C., Sun, C., Yuan, Y., Li, F., Wang, Q., Zhu, H., Hua, S., & Lin, L. (2023). Effects of Low Nighttime Temperature on Fatty Acid Content in Developing Seeds from Brassica napus L. Based on RNA-Seq and Metabolome. Plants, 12(2), 325. https://doi.org/10.3390/plants12020325







