Improvement of Resistance to Clubroot Disease in the Ogura CMS Restorer Line R2163 of Brassica napus
Abstract
1. Introduction
2. Results
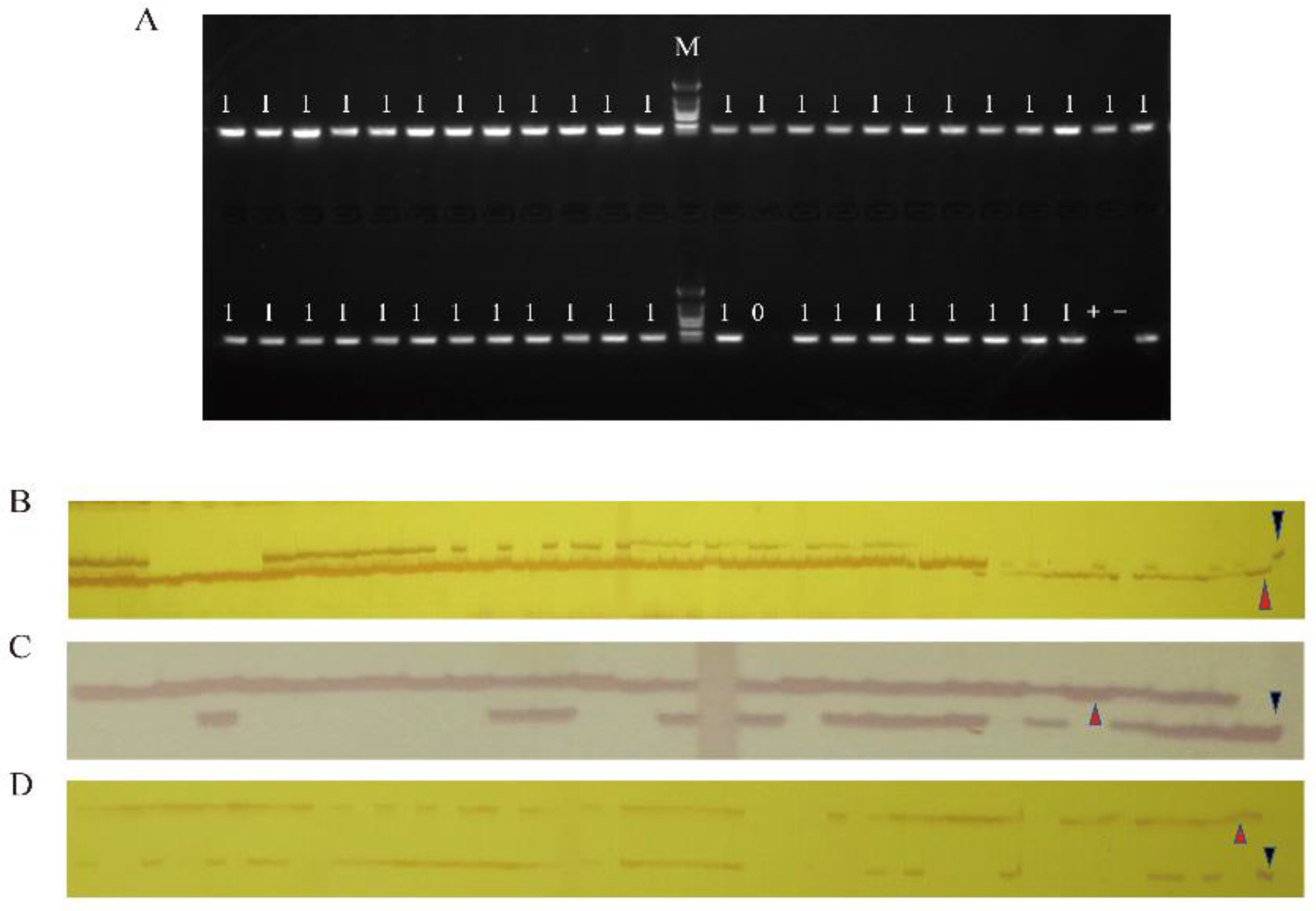
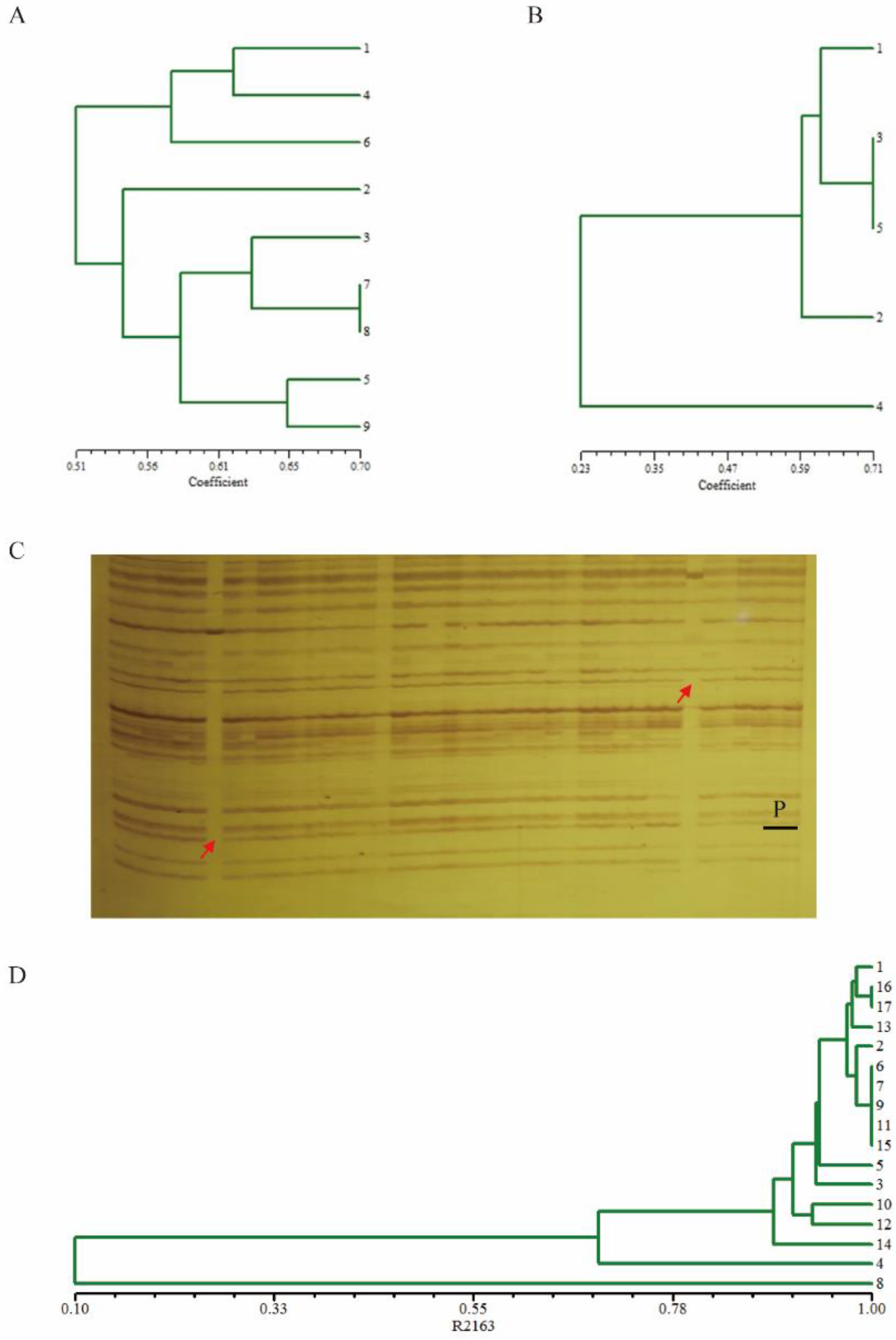
2.1. Marker-Assisted Introgression of PbBa8.1 into R2163
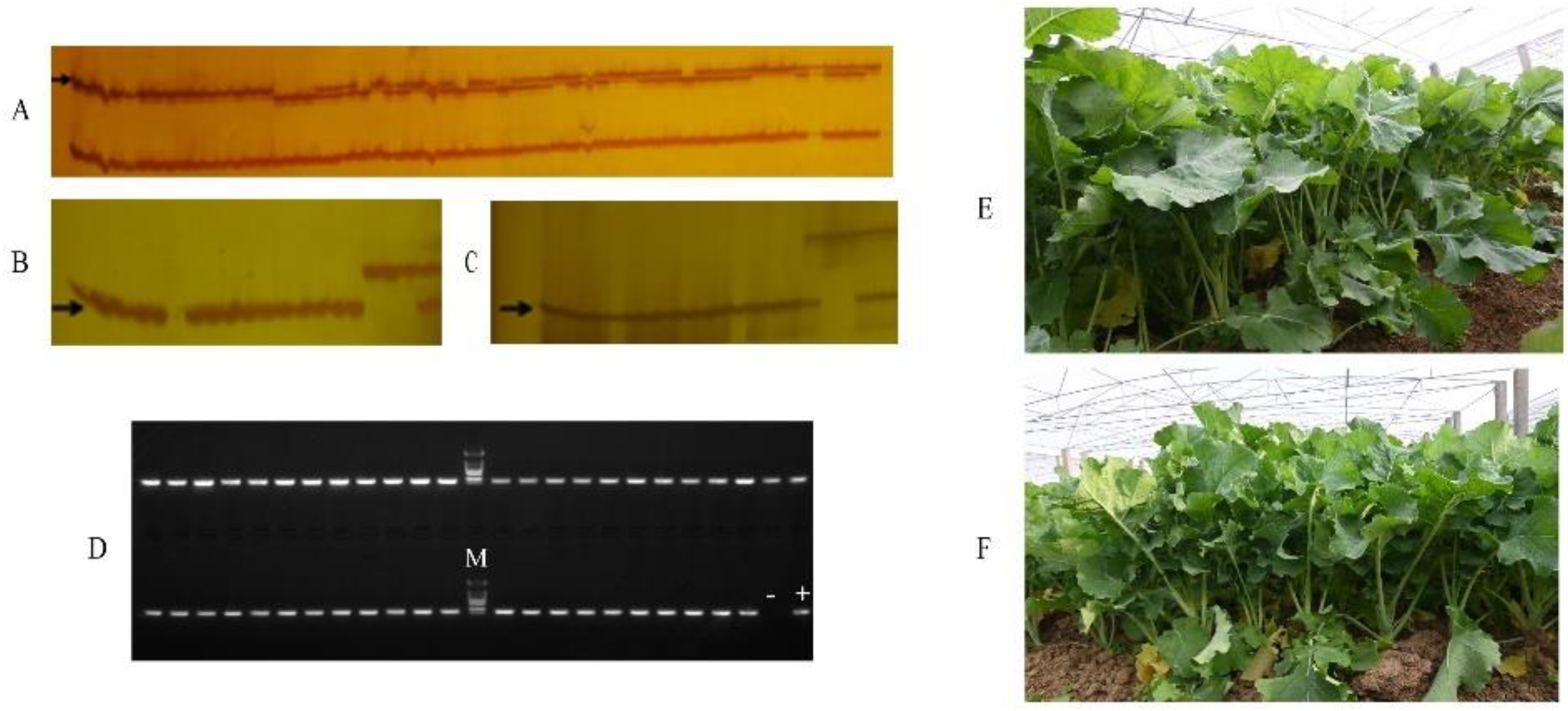
2.2. Preparation and Clubroot Resistance Identification of the Hybrids Kenyouza 741R and Huayouza 706R
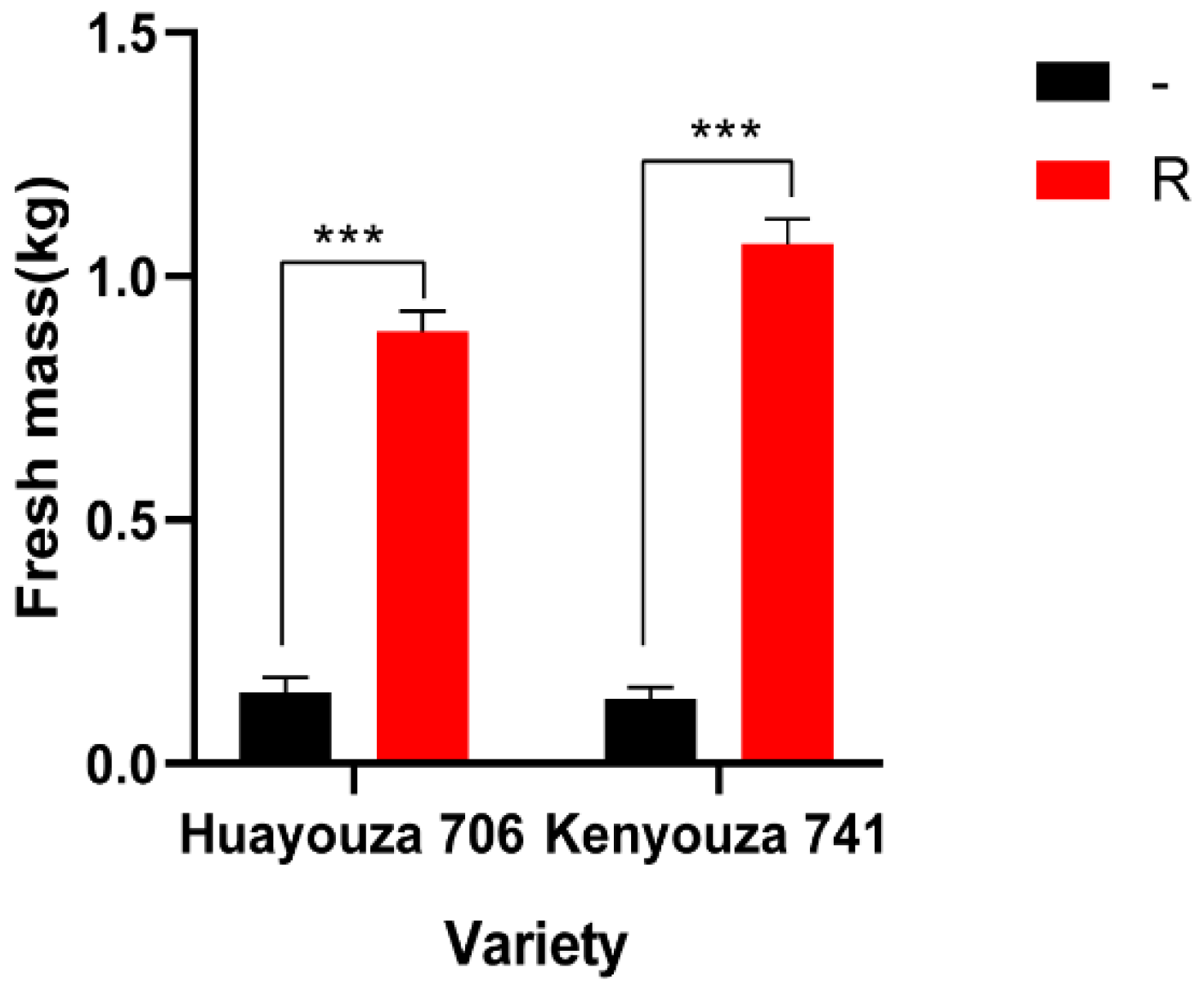
2.3. Clubroot Resistance Evaluation
2.4. Evaluation of Agro-Morphological Characters of the Improved Kenyouza 741R and Huayouza 706R
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Technical Route of R2163R Breeding
4.2. Preparation of Hybrids of Kenyouza 741R and Huayouza 706R
4.3. Evaluation of Clubroot Resistance in Kenyouza 741R and Huayouza 706R
4.4. Foreground and Background Selection
4.5. DNA Extraction and PCR System
4.6. Evaluation of Agro-Morphological Characters of the Improved Kenyouza 741R and Huayouza 706r
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Girke, A.; Schierholt, A.; Becker, H.C. Extending the rapeseed gene pool with resynthesized Brassica napus II: Heterosis. Theor. Appl. Genet. 2012, 124, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Ogura, H. Studies on the new male sterility in Japanese radish, with special references on the utilization of this sterility towards the practical raising of hybrid seeds. Mem. Fac. Agric. Kagoshima Univ. 1968, 6, 40–75. [Google Scholar]
- Bannerot, H.; Boulidard, L.; Chupeau, Y. Unexpected difficulties met with the radish cytoplasm in Brassica oleracea. Eucarpia Crucif. Newslett. 1977, 2, 16. [Google Scholar]
- Bellaoui, M.; Grelon, M.; Pelletier, G.; Budar, F. The restorer Rfo gene acts post-translationally on the stability of the ORF138 Ogura CMS-associated protein in reproductive tissues of rapeseed cybrids. Plant Mol. Biol. 1999, 40, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, G.; Primard, C.; Vedel, F.; Chetrit, P.; Renard, M. Intergeneric cytoplasmic hybridization in Cruciferae by protoplast fusion. MGG Mol. Gen. Genet. 1983, 191, 244–250. [Google Scholar] [CrossRef]
- Giancola, S.; Marhadour, S.; Desloire, S.; Clouet, V.; Falentin-Guyomarc’h, H.; Laloui, W.; Falentin, C.; Pelletier, G.; Renard, M.; Bendahmane, A.; et al. Characterization of a radish introgression carrying the Ogura fertility restorer gene Rfo in rapeseed, using the Arabidopsis genome sequence and radish genetic mapping. Theor. Appl. Genet. 2003, 107, 1442–1451. [Google Scholar] [CrossRef]
- Primard-Brisset, C.; Poupard, J.P.; Horvais, R.; Eber, F.; Pelletier, G.; Renard, M.; Delourme, R. A new recombined double low restorer line for the Ogu-INRA cms in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2005, 111, 736–746. [Google Scholar] [CrossRef]
- Wu, Z. Mechanism and Utilization of Alloplasmic Male-Sterile Lines in Brassica Juncea. Ph.D. Dissertation, Huazhong Agricultural University, Wuhan, China, 2020. [Google Scholar]
- Dixon, G.R. Clubroot (Plasmodiophora brassicae Woronin)–an agricultural and biological challenge worldwide. Can. J. Plant Pathol. 2014, 36 (Suppl. 1), 5–18. [Google Scholar] [CrossRef]
- Wang, J.; Huang, Y.; Hu, X.L.; Niu, Y.Z.; Li, X.L.; Liang, Y. Study on symptom, yield loss of clubroot and modality of Plasmodiophora brassicae in rape. Chin. J. Oil Crop Sci. 2008, 30, 112. [Google Scholar]
- Kato, T.; Hatakeyama, K.; Fukino, N.; Matsumoto, S. Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa. Breed Sci. 2013, 63, 116–124. [Google Scholar] [CrossRef]
- Kageyama, K.; Asano, T. Life Cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 2009, 28, 203–211. [Google Scholar] [CrossRef]
- Dixon, G.R. The Occurrence and Economic Impact of Plasmodiophora brassicae and Clubroot Disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Friberg, H.; Lagerlöf, J.; Rämert, B. Germination of Plasmodiophora brassicae resting spores stimulated by a non-host plant. Eur. J. Plant Pathol. 2005, 113, 275. [Google Scholar] [CrossRef]
- Niwa, R.; Kumei, T.; Nomura, Y.; Yoshida, S.; Osaki, M.; Ezawa, T. Increase in soil pH due to Ca-rich organic matter application causes suppression of the clubroot disease of crucifers. Soil Biol. Biochem. 2007, 39, 778–785. [Google Scholar] [CrossRef]
- Peng, G.; Falk, K.C.; Gugel, R.K.; Franke, C.; Yu, F.Q.; James, B.; Strelkov, S.E.; Hwang, S.F.; Mcgregor, L. Sources of resistance to Plasmodiophora brassicae (clubroot) pathotypes virulent on canola. Can. J. Plant Pathol. 2014, 36, 89–99. [Google Scholar] [CrossRef]
- Hirani, A.H.; Gao, F.; Liu, J.; Fu, G.H.; Wu, C.R.; McVetty, P.B.; Duncan, R.W.; Li, G. Combinations of independent dominant loci conferring clubroot resistance in all four turnip accessions (Brassica rapa) from the European clubroot differential set. Front. Plant Sci. 2018, 9, 1628. [Google Scholar] [CrossRef]
- Rahman, H.; Peng, G.; Yu, F.; Falk, K.C.; Kulkarni, M.; Selvaraj, G. Genetics and breeding for clubroot resistance in Canadian spring canola (Brassica napus L.). Can. J. Plant Pathol. 2014, 36 (Suppl. 1), 122–134. [Google Scholar] [CrossRef]
- Manzanares-Dauleux, M.J.; Delourme, R.; Baron, F.; Thomas, G. Mapping of one major gene and of QTLs involved in resistance to clubroot in Brassica napus. Theor. Appl. Genet. 2000, 101, 885–891. [Google Scholar] [CrossRef]
- Werner, S.; Diederichsen, E.; Frauen, M.; Schondelmaier, J.; Jung, C. Genetic mapping of clubroot resistance genes in oilseed rape. Theor. Appl. Genet. 2008, 116, 363. [Google Scholar] [CrossRef]
- Huang, Z.; Peng, G.; Liu, X.J.; Deora, A.; Falk, K.C.; Gossen, B.D.; Mcdonald, M.R.; Yu, F.Q. Fine Mapping of a Clubroot Resistance Gene in Chinese Cabbage Using SNP Markers Identified from Bulked Segregant RNA Sequencing. Front. Plant Sci. 2017, 8, 1448. [Google Scholar] [CrossRef]
- Huang, Z.; Peng, G.; Gossen, B.D.; Yu, F. Fine mapping of a clubroot resistance gene from turnip using SNP markers identified from bulked segregant RNA-Seq. Mol. Breed. 2019, 39, 131. [Google Scholar] [CrossRef]
- Karim, M.M.; Dakouri, A.; Zhang, Y.; Chen, Q.L.; Peng, G.; Strelkov, S.E.; Gossen, B.D.; Yu, F.Q. Two Clubroot-Resistance Genes, Rcr3 and Rcr9wa, Mapped in Brassica rapa Using Bulk Segregant RNA Sequencing. Int. J. Mol. Sci. 2020, 21, 5033. [Google Scholar] [CrossRef]
- Laila, R.; Park, J.I.; Robin, A.H.K.; Natarajan, S.; Vijayakumar, H.; Shirasawa, K.; Isobe, S.; Kim, H.T.; Nou, I.S. Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage (Brassica rapa L.). BMC Plant Biol. 2019, 19, 13. [Google Scholar] [CrossRef]
- Choi, S.R.; Oh, S.H.; Chhapekar, S.S.; Dhandapani, V.; Lee, C.Y.; Rameneni, J.J.; Ma, Y.; Choi, G.J.; Lee, S.S.; Lim, Y.P. Quantitative trait locus mapping of clubroot resistance and Plasmodiophora brassicae pathotype banglim-specific marker development in Brassica rapa. Int. J. Mol. Sci. 2020, 21, 4157. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Lamara, M.; Wei, Y.; Hu, H.; Parkin, I.A.; Gossen, B.D.; Peng, G.; Yu, F.Q. Clubroot resistance gene Rcr6 in Brassica nigra resides in a genomic region homologous to chromosome A08 in B. rapa. BMC Plant Biol. 2019, 19, 224. [Google Scholar] [CrossRef] [PubMed]
- Kamei, A.; Tsuro, M.; Kubo, N.; Hayashi, T.; Wang, N.; Fujimura, T.; Hirai, M. QTL mapping of clubroot resistance in radish (Raphanus sativus L.). Theor. Appl. Genet. 2010, 120, 1021–1027. [Google Scholar] [CrossRef]
- Yoshikawa, H. Breeding for Clubroot Resistance in Chinese Cabbage; Asian Vegetable Research and Development Center Publication (AVRDC): Shanhua, Taiwan, 1981. [Google Scholar]
- Matsumoto, E.; Yasui, C.; Ohi, M.; Tsukada, M. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis). Euphytica 1998, 104, 79–86. [Google Scholar] [CrossRef]
- Piao, Z.Y.; Deng, Y.Q.; Choi, S.R.; Park, Y.J.; Lim, Y.P. SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor. Appl. Genet. 2004, 108, 1458–1465. [Google Scholar] [CrossRef]
- Hatakeyama, K.; Niwa, T.; Kato, T.; Ohara, T.; Kakizaki, T.; Matsumoto, S. The tandem repeated organization of NB-LRR genes in the clubroot-resistant CRb locus in Brassica rapa L. Mol. Genet. Genom. 2017, 292, 397–405. [Google Scholar] [CrossRef]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Matsumoto, S.; Hirai, M. Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 2003, 107, 997–1002. [Google Scholar] [CrossRef]
- Suwabe, K.; Tsukazaki, H.; Iketani, H.; Hatakeyama, K.; Kondo, M.; Fujimura, M.; Nunome, T.; Fukuoka, H.; Hirai, M.; Matsumoto, S. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: The genetic origin of clubroot resistance. Genetics 2006, 173, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Hirai, M.; Harada, T.; Kubo, N.; Tsukada, M.; Suwabe, K.; Matsumoto, S. A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor. Appl. Genet. 2004, 108, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Pang, W.X.; Fu, P.Y.; Li, X.N.; Zhan, Z.X.; Yu, S.; Piao, Z.Y. Identification and Mapping of the Clubroot Resistance Gene CRd in Chinese Cabbage (Brassica rapa ssp. pekinensis). Front. Plant Sci. 2018, 9, 653. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Saito, A.; Hayashida, N.; Taguchi, G.; Matsumoto, E. Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theor. Appl. Genet. 2008, 117, 759–767. [Google Scholar] [CrossRef]
- Chen, J.J.; Jing, J.; Zhan, Z.X.; Zhang, T.; Zhang, C.Y.; Piao, Z.Y. Identification of Novel QTLs for Isolate-Specific Partial Resistance to Plasmodiophora brassicae in Brassica rapa. PLoS ONE 2013, 8, e85307. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Song, T.; Falk, K.C.; Zhang, X.G.; Liu, X.J.; Chang, A.; Lahlali, R.; McGregor, L.; Gossen, B.D.; Yu, F.Q.; et al. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genom. 2014, 15, 1166. [Google Scholar] [CrossRef]
- Yu, F.Q.; Zhang, X.G.; Peng, G.; Falk, K.C.; Strelkov, S.E.; Gossen, B.D. Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 2017, 7, 4516. [Google Scholar] [CrossRef]
- Zhan, Z.X.; Jiang, Y.F.; Zhu, Z.Y.; Zhang, C.S.; Yang, Q.Y.; Li, Q.; Hou, Z.K.; Gong, J.F.; Cheng, Y.G.; Wu, J.S.; et al. Development of close linked marker to PbBa8.1 conferring canola resistance to Plasmodiophora brassicae. Chin. J. Oil Crop Sci. 2015, 37, 766. [Google Scholar]
- Hu, X.Y.; Sullivan-Gilbert, M.; Kubik, T.; Danielson, J.; Hnatiuk, N.; Marchione, W.; Greene, T.; Thompson, S.A. Mapping of the Ogura fertility restorer gene Rfo and development of Rfo allele-specific markers in canola (Brassica napus L.). Mol. Breed. 2008, 22, 663–674. [Google Scholar] [CrossRef]
- Wen, Y.C.; Zhang, S.F.; Wang, J.P.; Zhu, J.C.; Zhao, L.; Cao, J.H. Screening and Preliminary Studies of Restorers of Radish Cytoplasmic Male Sterile Lines in Brassica napus L. Acta Agric. Boreali-Sin. 2010, 25, 102–106. [Google Scholar]
- Chen, W.J.; Li, M.; Wang, T.H.; Hui, R.K.; Tu, J.X.; Fu, T.D. Development of new restorer materials with Ogu CMS in Brassica napus. Agric. Sci. Technol. 2013, 14, 18. [Google Scholar]
- Wang, T.H.; Chen, W.J.; Li, M.; Tu, J.X.; Guo, Y.M. Research progress on heterosis utilization of radish cytoplasmic male sterility in Brassica napus L. Mol. Plant Breed. 2017, 15, 2777–2783. [Google Scholar]
- Chen, W.J.; Li, M.; Wang, T.H.; Hui, R.K.; Tu, J.X.; Fu, T.D. Creative process of a new restorer material with Ogu CMS in Brassica nupus. Sci. Agric. Sin. 2012, 45, 1465–1474. [Google Scholar]
- Wen, Y.C.; Zhang, S.F.; Wang, J.P.; Zhu, J.C.; He, J.P.; Cai, D.F.; Cao, J.H.; Zhao, L.; Wang, D.G. Studies on Rapid Improvement Techniques of Radish Cytoplasmic Male Sterile Restorers in Brassica napus L. Acta Agric. Boreali-Sin. 2016, 31, 102–109. [Google Scholar]
- Zhao, X.W.; Li, B.; Zhang, K.; Hu, K.N.; Yi, B.; Wen, J.; Ma, C.Z.; Shen, J.X.; Fu, T.D.; Tu, J.X. Breeding signature of combining ability improvement revealed by a genomic variation map from recurrent selection population in Brassica napus. Sci. Rep. 2016, 6, 29553. [Google Scholar] [CrossRef]
- Fei, W.X.; Hwang, S.H.; Wang, S.F.; Wu, X.Y.; Gao, Z.M.; Li, Q.S.; Hou, S.M.; Rong, S.B.; Jiang, Y.F.; Lei, W.X.; et al. Pathotype identification of Plasmodiophora brassicae and resistance of rapeseed variety resources to clubroot disease. Chin. J. Oil Crop Sci. 2016, 38, 626–639. [Google Scholar]
- Gao, L.; Cao, Y.H.; Xia, Z.H.; Jiang, G.H.; Liu, G.Z.; Zhang, W.X.; Zhai, W.X. Do transgenesis and marker-assisted backcross breeding produce substantially equivalent plants?—A comparative study of transgenic and backcross rice carrying bacterial blight resistant gene Xa21. BMC Genom. 2013, 14, 738. [Google Scholar] [CrossRef]
- Sanchez, A.C.; Brar, D.S.; Huang, N.; Li, Z.; Khush, G.S. Sequence tagged site marker-assisted selection for three bacterial blight resistance genes in rice. Crop Sci. 2000, 40, 792–797. [Google Scholar] [CrossRef]
- Whittaker, J.C.; Curnow, R.N.; Haley, C.S.; Thompson, R. Using marker-maps in marker-assisted selection. Genet. Res. 1995, 66, 255–265. [Google Scholar] [CrossRef]
- Hospital, F.; Moreau, L.; Lacoudre, F.; Charcosset, A.; Gallais, A. More on the efficiency of marker-assisted selection. Theor. Appl. Genet. 1997, 95, 1181–1189. [Google Scholar] [CrossRef]
- Moreau, L.; Charcosset, A.; Hospital, F.; Gallais, A. Marker-assisted selection efficiency in populations of finite size. Genetics 1998, 148, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Hospital, F.; Chevalet, C.; Mulsant, P. Using markers in gene introgression breeding programs. Genetics 1992, 132, 1199–1210. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.T.; Doyle, J.L. Isolation of Plant DNA from Fresh Tissue; Focus: San Francisco, CA, USA, 1990; Volume 12, pp. 13–15. [Google Scholar]



| Variety | Number of Diseased Plants | Total Number of Plants Surveyed | Disease Incidence % |
|---|---|---|---|
| Kenyouza 741 | 393 | 393 | 100.00 |
| Kenyouza 741R | 21 | 384 | 5.47 |
| Huayouza 706 | 365 | 365 | 100.00 |
| Huayouza 706R | 3 | 372 | 0.81 |
| Variety | Fresh Mass of Aerial Parts of 20 Plants (kg) | p-Value | |||
|---|---|---|---|---|---|
| Ⅰ | Ⅱ | Ⅲ | Average Value | ||
| Kenyouza 741 | 0.16 | 0.12 | 0.12 | 0.13 | 0.000016 |
| Kenyouza 741R | 1.12 | 1.06 | 1.02 | 1.07 | |
| Huayouza 706 | 0.14 | 0.12 | 0.18 | 0.15 | 0.000008 |
| Huayouza 706R | 0.92 | 0.90 | 0.84 | 0.89 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Li, J.; Ma, M.; Li, B.; Zhou, Y.; Pan, Y.; Fan, Y.; Yi, B.; Tu, J. Improvement of Resistance to Clubroot Disease in the Ogura CMS Restorer Line R2163 of Brassica napus. Plants 2022, 11, 2413. https://doi.org/10.3390/plants11182413
Chen J, Li J, Ma M, Li B, Zhou Y, Pan Y, Fan Y, Yi B, Tu J. Improvement of Resistance to Clubroot Disease in the Ogura CMS Restorer Line R2163 of Brassica napus. Plants. 2022; 11(18):2413. https://doi.org/10.3390/plants11182413
Chicago/Turabian StyleChen, Jiao, Jiahui Li, Mengya Ma, Bao Li, Yuanwei Zhou, Yongzhong Pan, Youjun Fan, Bin Yi, and Jinxing Tu. 2022. "Improvement of Resistance to Clubroot Disease in the Ogura CMS Restorer Line R2163 of Brassica napus" Plants 11, no. 18: 2413. https://doi.org/10.3390/plants11182413
APA StyleChen, J., Li, J., Ma, M., Li, B., Zhou, Y., Pan, Y., Fan, Y., Yi, B., & Tu, J. (2022). Improvement of Resistance to Clubroot Disease in the Ogura CMS Restorer Line R2163 of Brassica napus. Plants, 11(18), 2413. https://doi.org/10.3390/plants11182413







