Identification and Characterisation of Pseudomonas savastanoi pv. savastanoi as the Causal Agent of Olive Knot Disease in Croatian, Slovenian and Portuguese Olive (Olea europaea L.) Orchards
Abstract
1. Introduction
2. Results
2.1. Distribution and Incidence of Olive Knot Disease Symptoms in Surveyed Countries
2.2. Biochemical (LOPAT), Physiological and Molecular Tests for Characterisation of Pss Isolates from Infected Olive Materials
3. Discussion
4. Materials and Methods
4.1. Plant Material Sampling
4.2. Bacterial Isolation and Characterisation
4.3. Molecular Identification of Pss
4.4. Protein Mass Spectra Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gomila, M.; Busquets, A.; Mulet, M.; García-Valdés, E.; Lalucat, J. Clarification of taxonomic status within the Pseudomonas syringae species group based on a phylogenomic analyses. Front. Microbiol. 2017, 8, 2422. [Google Scholar] [CrossRef] [PubMed]
- Berge, O.; Monteil, C.L.; Bartoli, C.; Chandeysson, C.; Guilbaud, C.; Sands, D.C.; Morris, C.E. A user’s guide to a database of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS ONE 2014, 9, e105547. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, Y.; Taguchi, F.; Mukaihara, T. Pathogenicity and virulence factors of Pseudomonas syringae. J. Gen. Plant Pathol. 2013, 79, 285–296. [Google Scholar] [CrossRef]
- Morris, C.E.; Ramirez, N.; Berge, O.; Lacroix, C.; Monteil, C.; Chandeysson, C.; Guilbaud, C.; Blischke, A.; Auđur Sigurbjörnsdóttir, M.; Vilhelmsson, O.P. Pseudomonas syringae on plants in iceland has likely evolved for several milion years outside the reach of processes that mix this bacterial complex across Earth’s temperate zones. Pathogens 2022, 11, 357. [Google Scholar] [CrossRef] [PubMed]
- Caballo-Ponce, E.; Murillo, J.; Martínez-Gil, M.; Moreno-Pérez, A.; Pintado, A.; Ramos, C. Knots Untie: Molecular Determinants Involved in Knot Formation Included by Pseudomonas savastanoi in Woody Hosts. Front. Plant Sci. 2017, 8, 1089. [Google Scholar] [CrossRef]
- FAOSTAT. Food and Agriculture Organization of the United Nations. 2022. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 27 May 2022).
- Lanza, B.; Ninfali, P. Antioxidants in extra virgin olive oil and table olives: Connections between agriculture and processing for health choices. Antioxidants 2020, 9, 41. [Google Scholar] [CrossRef]
- Notario, A.; Sánchez, R.; Luaces, P.; Sanz, C.; Pérez, A.G. The infestation of olive fruits by Bactrocera oleae (Rossi) modifies the expression of key genes in the biosynthesis of volatile and phenolic compounds and alters the composition of virgin oilve oil. Molecules 2022, 27, 1650. [Google Scholar] [CrossRef]
- Quesada, J.M.; Penyalver, R.; Pérez-Panadés, J.; Salcedo, C.I.; Carbonell, E.A.; López, M.M. Dissemination of Pseudomonas savastanoi pv. savastanoi populations and subsequent appearance of olive knot disease. Plant Pathol. 2010, 59, 262–269. [Google Scholar] [CrossRef]
- Lin, N.-C.; Martin, G.B. Pto- and Prf-Mediated Recognition of AvrPto and AvrPtoB Restricts the Ability of Diverse Pseudomonas syringae Pathovars to Infect tomato. Mol. Plant Microbe Interact. 2007, 20, 806–815. [Google Scholar] [CrossRef]
- Moreno-Pérez, A.; Pintado, A.; Murillo, J.; Caballo-Ponce, E.; Tegli, S.; Moretti, C.; Rodríguez-Pelenzuela, P.; Ramos, C. Host range determinants of Pseudomonas savastanoi pathovars of woody hosts revealed by comparative genomics and cross-pathogenicity tests. Front. Plant Sci. 2020, 11, 973. [Google Scholar] [CrossRef]
- Tegli, S.; Gori, A.; Cerboneschi, M.; Cipriani, M.G.; Sisto, A. Type Three Secretion System in Pseudomonas savastanoi Pathovars: Does timing matter? Genes 2011, 2, 957–979. [Google Scholar] [CrossRef]
- Moretti, C.; Trabalza, S.; Granieri, L.; Caballo-Ponce, E.; Devescovi, G.; Del Pino, A.M.; Ramos, C.; Venturi, V.; Van Den Burg, H.A.; Buonaurio, R.; et al. A Na+/Ca2+ exchanger of the olive pathogen Pseudomonas savastanoi pv. savastanoi is critical for its virulence. Mol. Plant Pathol. 2019, 20, 716–730. [Google Scholar] [CrossRef] [PubMed]
- Cerboneschi, M.; Decorosi, F.; Biancalani, C.; Ortenzi, M.V.; Macconi, S.; Giovannetti, L.; Viti, C.; Campanella, B.; Onor, M.; Bramanti, E.; et al. Indole-3-acetic acid in plant-pathogen interactions: A key molecule for in planta bacterial virulence and fitness. Res. Microbiol. 2016, 167, 774–787. [Google Scholar] [CrossRef] [PubMed]
- Añorga, M.; Pintado, A.; Ramos, C.; De Diego, N.; Ugena, L.; Novák, O.; Murillo, J. Genes ptz and idi, coding for cytokinin ciosynthesis enzymes, are essential for tumorogenesis and in planta growth by P. syringae pv. savastanoi NCPPB 3335. Front. Plant Sci. 2020, 11, 1294. [Google Scholar] [CrossRef] [PubMed]
- Castaǹeda-Ojeda, M.P.; Moreno-Pérez, A.; López-Solanilla, E.; Ramos, C. Suppression of plant immune responses by the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III effector tyrosine phosphatases HopAO1 and HopAO2. Front. Plant Sci. 2017, 5, 680. [Google Scholar] [CrossRef]
- Alfano, J.R.; Charkowski, A.O.; Deng, W.L.; Badel, J.L.; Petnicki-Ocwieja, T.; Van Dijk, K.; Collmer, A. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 2000, 97, 4856–4861. [Google Scholar] [CrossRef] [PubMed]
- Lelliot, R.A.; Billing, E.; Hayward, A.C. A Determinative Scheme for the Fluorescent Plant Pathogenic Pseudomonads. J. Appl. Bacteriol. 1966, 29, 470–489. [Google Scholar] [CrossRef] [PubMed]
- Turco, S.; Drais, M.I.; Rossini, L.; Chaboteaux, E.; Rahi, Y.J.; Balestra, G.M.; Iacobellis, N.S.; Mazzaglia, A. Complete genome assembly of the levan-positive strain PVFi1 of Pseudomonas savastanoi pv. savastanoi isolated from olive knots in Central Italy. Environ. Micriobiol. Rep. 2022, 14, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, M.; Ohta, K.; Tanaka, K.; Takikawa, Y. Comparison among Japanese isolates of Pseudomonas savastanoi pv. savastanoi, causal agent of olive knot disease. J. Gen. Plant Pathol. 2017, 83, 152–161. [Google Scholar] [CrossRef]
- Hall, B.H.; Cother, E.J.; Whattam, M.; Noble, D.; Luck, J.; Cartwright, D. First report of olive knot caused by Pseudomonas savastanoi pv. savastanoi on olives (Olea europaea) in Australia. Austral Plant Pathol. 2004, 33, 433–436. [Google Scholar] [CrossRef]
- Marchi, G.; Viti, C.; Giovannetti, L.; Surico, G. Spread of levan-positive populations of Pseudomonas savastanoi pv. savastanoi, the causal agent of olive knot, in central Italy. Eur. J. Plant Pathol. 2005, 112, 101–112. [Google Scholar] [CrossRef]
- Penyalver, R.; García, A.; Ferrer, A.; Bertolini, E.; López, M.M. Detection of Pseudomonas savastanoi pv. savastanoi in Olive Plants by Enrichment and PCR. Appl. Environ. Microbiol. 2000, 66, 2673–2677. [Google Scholar] [CrossRef] [PubMed]
- Tegli, S.; Cerboneschi, M.; Marsili Libelli, I.; Santilli, E. Development of a versatile tool for the simultaneous differential detection of Pseudomonas savastanoi pathovars by End Point and Real-Time PCR. BMC Microbiol. 2010, 10, 156. [Google Scholar] [CrossRef] [PubMed]
- Gori, A.; Cerboneschi, M.; Tegli, S. High-Resolution Melting Analysis as a powerful tool to discriminate and genotype Pseudomonas savastanoi pathovars and strains. PLoS ONE 2012, 7, e30199. [Google Scholar] [CrossRef] [PubMed]
- Baltrus, D.A.; Mccann, H.C.; Guttman, D.S. Evolution, genomics, and epidemiology of Pseudomonas syringae. Mol. Plant Pathol. 2017, 18, 152–168. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, J.R.; Varvaro, L.; Parisi, L.; Audergon, J.M.; Moris, C.E. Disease and frost damage of woody plants caused by Pseudomonas syringae: Seeing the forests for the trees. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press Inc.: San Diego, CA, USA, 2014; pp. 235–295. [Google Scholar] [CrossRef]
- Rahi, Y.J.; Turco, S.; Taratufolo, M.C.; Tatì, M.; Cerboneschi, M.; Tegli, S.; Valentini, F.; D’Onghia, A.; Iacobellis, N.S.; Balestra, G.M.; et al. Genetic diversity and population structure of Pseudomonas savastanoi, an endemic pathogen of the Mediterranean area, revealed up to strain level by the MLVA assay. J. Plant Pathol. 2020, 102, 1051–1064. [Google Scholar] [CrossRef]
- Moretti, C.; Ferrante, P.; Hosni, T.; Valentini, F.; D’Onghia, A.; Fatmi, M.; Buonaurio, R. Characterization of Pseudomonas savastanoi pv. savastanoi strains collected from olive trees in different countries. In Pseudomonas Syringae Pathovars and Related Pathogens—Identification, Epidemiology and Genomics; Fatmi, M.B., Collmer, A., Iacobellis, N.S., Mansfield, J.W., Murillo, J., Schaad, N.W., Ullrich, M., Eds.; Springer Science+ Business Media: Berlin, Germany, 2008; pp. 321–329. [Google Scholar] [CrossRef]
- Koprivnjak, O.; Majetić, V.; Brkić Bubola, K.; Kosić, U. Variability of phenolic and volatile compounds in virgin olive oil from Leccino and Istarska bjelica cultivars in relation to their fruit mixtures. Food Technol. Biotechnol. 2012, 50, 216–221. [Google Scholar]
- Kovačić, I.; Bilić, J.; Dudaš, S.; Poljuha, D. Phenolic content and antioxidant capacity of Istrian olive leaf infusion. Agriculture 2017, 23, 38–45. [Google Scholar] [CrossRef]
- Vuletin Selak, G.; Raboteg Božiković, M.; Abrouk, D.; Bolčić, M.; Žanić, K.; Perica, S.; Normand, P.; Pujić, P. Pseudomonas ST1 and Pantoea Paga Strains Cohabit in Olive Knots. Microorganisms 2022, 10, 1528. [Google Scholar] [CrossRef]
- Tolba, I.H.; Soliman, M.A. Efficacy of native antagonistic bacterial isolates in biological control of crown gall disease in Egypt. Ann. Agric. Sci. 2013, 58, 43–49. [Google Scholar] [CrossRef]
- Mina, D.; Pereira, J.A.; Lino-Neto, T.; Baptista, P. Screening the olive phyllosphere: Search and find potential antagonists against Pseudomonas savastanoi pv. savastanoi. Front. Microbiol. 2020, 11, 2051. [Google Scholar] [CrossRef] [PubMed]
- Zidarič, I. Olive Knot Disease (Pseudomonas syringae subsp. savastanoi Janse) Distribution in Slovenian Istria. Graduation Thesis, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia, 2016. Available online: http://www.digitalna-knjiznica.bf.uni-lj.si/agronomija/dn_zidaric_igor.pdf (accessed on 15 June 2022).
- Scortichini, M.; Rossi, M.P.; Salerno, M. Relationship of genetic structure of Pseudomonas savastanoi pv. savastanoi populations from Italian olive trees and patterns of host genetic diversity. Plant Pathol. 2004, 53, 491–497. [Google Scholar] [CrossRef]
- Pérez-Martinez, I.; Zhao, Y.; Murillo, J.; Sundin, G.W.; Ramos, C. Global genomic analysis of Pseudomonas savastanoi pv. savastanoi plasmids. J. Bacteriol. 2008, 190, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Surico, G.; Marchi, G. Olive knot disease: New insights in the ecology, physiology and epidemiology of Pseudomonas savastanoi pv. savastanoi. In Pseudomonas Syringae and Related Pathogens: Biology and Genetic; Iacobellis, N.S., Collmer, A., Utcheson, S.W., Mansfield, J.W., Morris, C.E., Murillo, J., Schaad, N.W., Stead, D.E., Surico, G., Ullrich, M.S., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2003; pp. 17–28. [Google Scholar]
- Mugnai, L.; Giovannetti, L.; Ventura, S.; Surico, G. The Grouping of Strains of Pseudomonas syringae subsp. savastanoi by DNA Restriction Fingerprinting. J. Phytopathol. 1994, 142, 209–218. [Google Scholar] [CrossRef]
- Bultreys, A.; Gheysen, I.; Maraite, H.; de Hoffman, E. Characterization of fluorescent and nonfluorescent peptide siderophores produced by Pseudomonas syringae strains and their potential use in strain identification. Appl. Environ. Microbiol. 2001, 67, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Krid, S.; Rhouma, A.; Quesada, J.M.; Penyalver, R.; Gargouri, A. Delineation of Pseudomonas savastanoi pv. savastanoi strains in Tunisia by random amplified polymorphic DNA analysis. J. Appl. Microbiol. 2009, 106, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Moretti, C.; Vinatzer, B.A.; Onofri, A.; Valentini, F.; Buonaurio, R. Genetic and phenotypic diversity of Mediterranean populations of the olive knot pathogen, Pseudomonas savastanoi pv. savastanoi. Plant Pathol. 2016, 66, 595–605. [Google Scholar] [CrossRef]
- Braun-Kiewnick, A.; Sands, D.C. Gram-negative bacteria. In Laboratory Guide for Identification of Plant Pathogenic Bacteria, 3rd ed.; Schaad, N.W., Jones, J.B., Chun, W., Eds.; APS Press: St. Paul, MN, USA, 2001; pp. 84–103. [Google Scholar]
- Spergser, J.; Hess, C.; Loncaric, I.; Ramírez, A.S. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Is a Superior Diagnostic Tool for the Identification and Differentiation of Mycoplasmas Isolated from Animals. J. Clin. Microbiol. 2019, 57, e00316-19. [Google Scholar] [CrossRef]
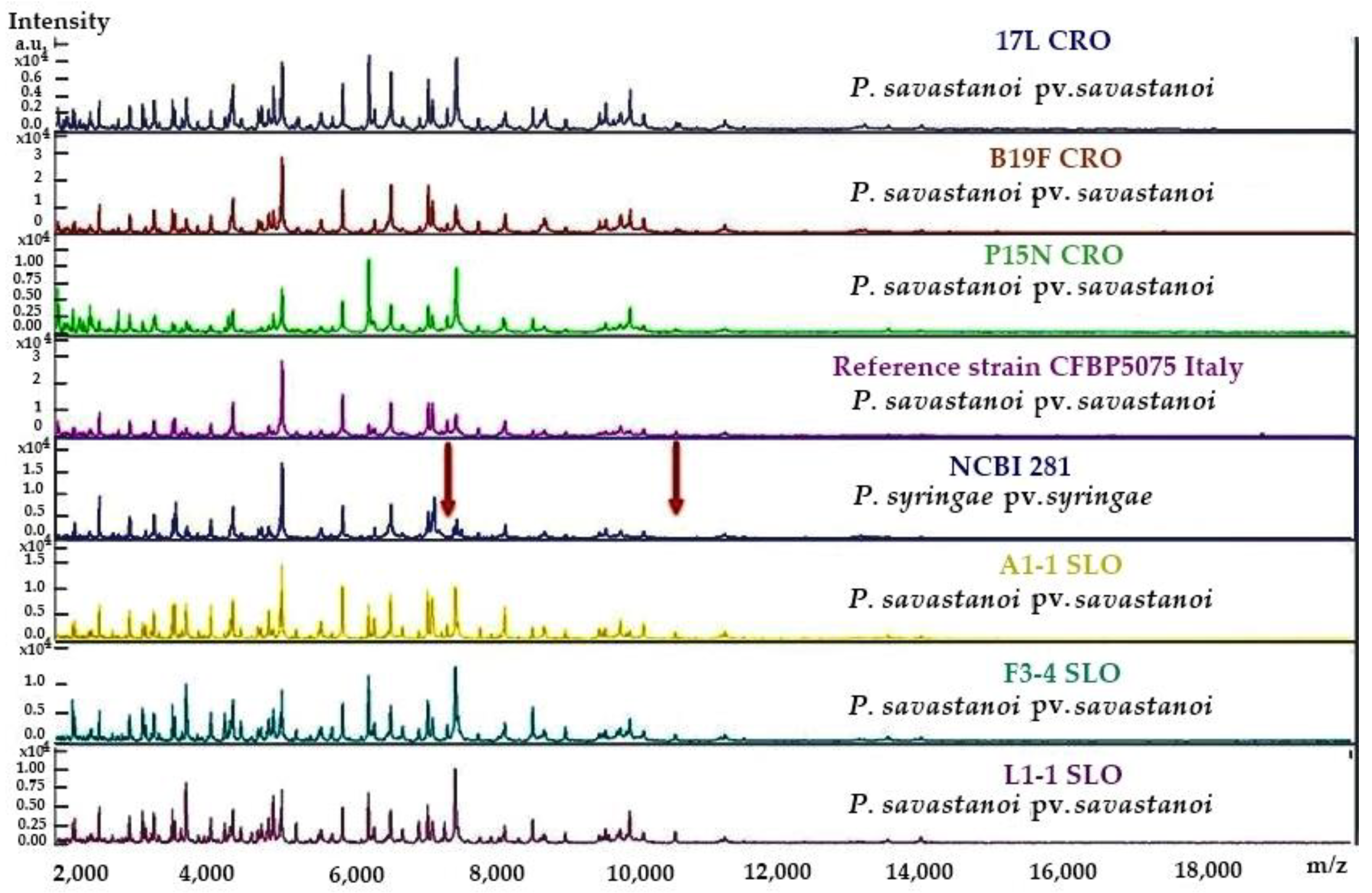
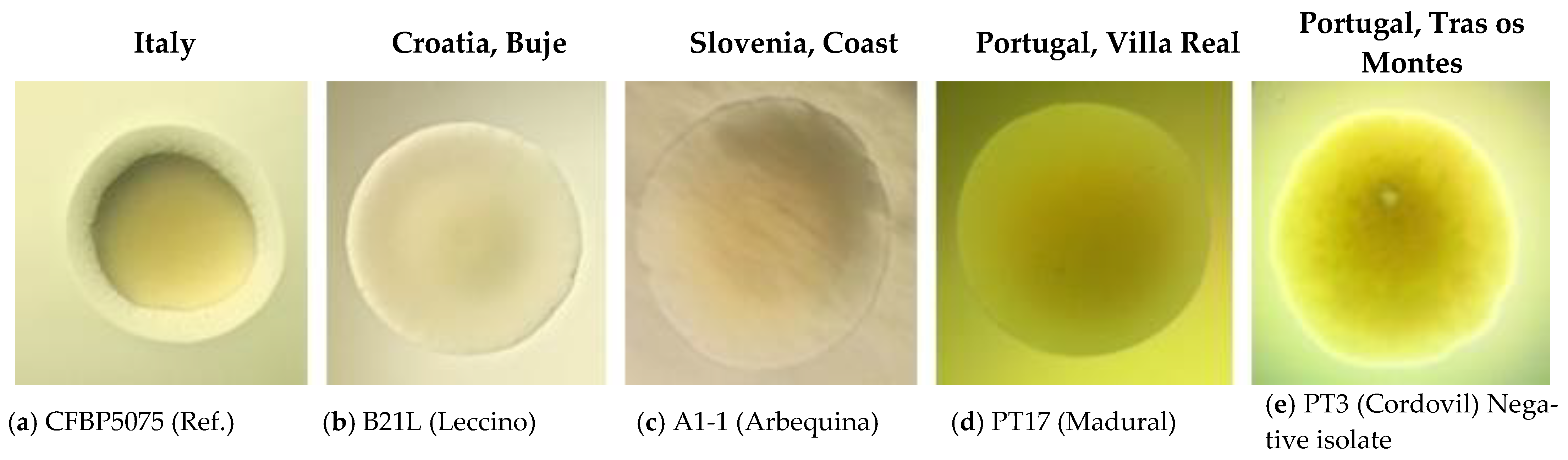
| Surveyed Country | Area (ha) | Olive Varieties (%) | Incidence * (%) | Severity § (cm) |
|---|---|---|---|---|
| Croatia | 600 | Leccino (82) | 59.5 | 0.97 ± 0.22 |
| Pendolino (69) | 48.4 | 1.04 ± 0.22 | ||
| Istarska bjelica (67) | 30.0 | 0.85 ± 0.17 | ||
| Frantoio (58) | 67.9 | 1.21 ± 0.33 | ||
| Buža (56) | 32.0 | 0.88 ± 0.20 | ||
| Maurino (18) | 50.0 | 0.88 ± 0.08 | ||
| Picholine (16) | 14.3 | 1.40 ± 0.20 | ||
| Moraiolo (13) | 33.3 | 0.75 ± 0.05 | ||
| Karbonaca (13) | 83.3 | 0.76 ± 0.08 | ||
| Rosinjola (11) | 20.0 | 0.80 ± 0.01 | ||
| Leccio del Corno (9) | 25.0 | 1.20 ± 0.40 | ||
| Ascolana tenera (9) | 0.00 | 0.00 ± 0.00 | ||
| Porečka rosulja (7) | 33.3 | 0.90 ± 0.10 | ||
| Italian rosinjola (3) | 100 | 0.80 ± 0.20 | ||
| Nocciara (3) | 100 | 0.80 ± 0.10 | ||
| Buža momjanska (3) | 100 | 0.60 ± 0.10 | ||
| Karbonera (3) | 100 | 1.00 ± 0.20 | ||
| Moražola (3) | 100 | 0.80 ± 0.01 | ||
| Cipressino (3) | 100 | 0.80 ± 0.01 | ||
| Pendolino piangente (3) | 100 | 1.10 ± 0.30 | ||
| Bianchera di Udine (3) | 100 | 0.90 ± 0.30 | ||
| Arbequina (3) | 100 | 0.70 ± 0.10 | ||
| Ascolana tenera (12.5) | 100 | 0.75 ± 0.25 | ||
| Slovenia | 2.40 | Frantoio (25) Ascolana tenera (12.5) | 100 100 | 1.97 ± 2.02 0.75 ± 0.25 |
| Pendolino (12.5) Frantoio (25) | 100 100 | 1.00 ± 0.00 1.97 ± 2.02 | ||
| Itrana (12.5) Pendolino (12.5) | 100 100 | 2.80 ± 0.40 1.00 ± 0.00 | ||
| Mata (12.5) Itrana (12.5) | 100 100 | 3.00 ± 0.00 2.80 ± 0.40 | ||
| Leccione (12.5) Mata (12.5) | 100 100 | 1.00 ± 0.00 3.00 ± 0.00 | ||
| Leccino (37.5) Leccione (12.5) | 66.6 100 | 2.14 ± 1.36 1.00 ± 0.00 | ||
| Arbequina (25) Leccino (37.5) | 50.0 66.6 | 1.00 ± 0.00 2.14 ± 1.36 | ||
| Istarska bjelica (75) Arbequina (25) | 00.0 50.0 | 0.00 ± 0.00 1.00 ± 0.00 | ||
| Istarska bjelica (75) | 00.0 | 0.00 ± 0.00 | ||
| Cobrançosa (40) | 100 | 0.83 ± 0.14 | ||
| Portugal | 3.25 | Cordovil (40) Cobrançosa (40) | 100 100 | 0.80 ± 0.00 0.83 ± 0.14 |
| Negruxa de Coimbra (20) Cordovil (40) | 100 100 | 0.89 ± 0.08 0.80 ± 0.00 | ||
| Negruxa (20) Negruxa de Coimbra (20) | 100 100 | 0.91 ± 0.06 0.89 ± 0.08 | ||
| Verdeal (60) Negruxa (20) | 100 100 | 0.95 ± 0.14 0.91 ± 0.06 | ||
| Madural (20) Verdeal (60) | 100 100 | 1.03 ± 0.13 0.95 ± 0.14 | ||
| Negruxa de Tras os Montes (20) Madural (20) | 100 100 | 0.96 ± 0.19 1.03 ± 0.13 | ||
| Xuinha/Azeitona de Coimbra (20) Negruxa de Tras os Montes (20) | 100 100 | 1.00 ± 0.00 0.96 ± 0.19 | ||
| Xuinha/Azeitona de Coimbra (20) | 100 | 1.00 ± 0.00 |
| Pss Isolate | Olive Variety | Country, Region, Municipality | Orchard Coordinates | Pss Specific Real-Time Assay * | PCR and MALDI–TOF |
|---|---|---|---|---|---|
| V4B | Buža | Croatia, Istria, Vodnjan | 44°56′99″ N; 13°51′81″ E | + | |
| V4 P | Pendolino | ||||
| V7 K | Karbonaca | Croatia, Istria, Vodnjan | 45°57′99″ N; 13°51′81″ E | + | |
| V9 B | Buža | Croatia, Istria, Vodnjan | 44°58′58″ N; 13°49′52″ E | + | |
| R12 L | Leccino | Croatia, Istria, Rovinj | 45°06′12″ N; 13°69′24″ E | + | |
| P15 TR | Italian rosinjola | Croatia, Istria, Poreč | 45°17′12″ N; 13°39′56″ E | + | |
| P15 N | Nocciara | ||||
| B17 L | Leccino | Croatia, Istria, Buje | 45°21′40″ N; 13°32′47″ E | + | |
| B19 F | Frantoio | Croatia, Istria, Buje | 45°24′50″ N; 13°35′14″ E | + | |
| B19 L | Leccino | ||||
| B19 P | Pendolino | ||||
| B21 L B21F | Leccino Frantoio | Croatia, Istria, Buje | 45°22′45″ N; 13°43′38″ E | + | |
| B23 F | Frantoio | Croatia, Istria, Buje | 45°20′20″ N; 13°33′31″ E | + | |
| I6 L, I7 L | Leccino | Croatia, Istria, Poreč | 45°22′34″ N; 13°60′25″ E | + | |
| NIN | Unknown | Croatia, Istria, Pula | 44°89’56’’ N; 13°81’82’’ E | + | |
| U26 F | Frantoio | Croatia, Istria, Umag | 45°24′27″ N; 13°33′24″ E | + | |
| U27 F | Frantoio | Croatia, Istria, Umag | 45°28′43″ N; 13°30′43″ E | + | |
| VS28 N | Unknown | Croatia, Istria, Višnjan | 45°15′58″ N; 13°43′20″ E | + | |
| VS28 L | Leccino | Croatia, Istria, Višnjan | 45°15′58″ N; 13°43′20″ E | + | |
| VS29 L | Leccino | Croatia, Istria, Višnjan | 45°17′53″ N; 13°42′50″ E | + | |
| VS31 L | Leccino | Croatia, Istria, Višnjan | 45°14′44″ N; 13°42′15″ E | + | |
| R34 F | Frantoio | Croatia, Istria, Rovinj | 45°02′53″ N; 13°44′10″ E | + | |
| BR37 L | Leccino | Croatia, Istria, Brtonigla | 45°22′48″ N; 13°36′28″ E | + | |
| N40 M | Maurino | Croatia, Istria, Novigrad | 44°21′46″ N; 13°33′46″ E | + | |
| N41 F | Frantoio | Croatia, Istria, Novigrad | 45°21′34″ N; 13°35′77″ E | + | |
| N42 F | Frantoio | Croatia, Istria, Novigrad | 45°21′06″ N; 13°35′01″ E | + | |
| N43 L N43 P | Leccino Pendolino | Croatia, Istria, Novigrad | 45°22′05″ N; 13°33′40″ E | + | |
| P44 F | Frantoio | Croatia, Istria, Poreč | 45°14′26″ N; 13°42′17″ E | + | |
| B45 C-PR | Porečka rosulja | Croatia, Istria, Buje | 45°24′42″ N; 13°30′10″ E | + | |
| F1-2, F1-3, F1-5F2-2, F2-5 A1-1, A1-3, A1-4 L1-1, L2-1, L2-3 AT1-2, AT2-6, AT2-4, AT1-5, AT1-4 | Frantoio Arbequina Leccino Ascolana tenera | Slovenia, Coast | 45°31′59″ N; 13°30′10″ E | + | |
| L6-2, L6-4 L5-3, L5-1, L5-4 L4-1, L4-4 L6-1 | Leccino | Slovenia, Goriška Brda | 46°0′11″ N; 13°34′36″ E | + | |
| F3-2, F3-4, F3-5 F4-2, F4-1, F4-4 | Frantoio | Slovenia, Goriška Brda | 45°58′56″ N; 13°30′29″ E | + | |
| PT15 PT 17 | Cordovil Madural | Portugal, Villa Real | 41°09′41″ N; 7°51′20″ W | + |
| Characteristic | Croatia (Istria) | Slovenia (Coast and Goriška Brda) | Portugal (Villa Real) |
|---|---|---|---|
| Gram-reaction | + | + | + |
| Fluorescence on KB | - | + | + |
| Levan production | - | - | - |
| Oxidase reaction | - | DP | - |
| Pectolytic activity | - | - | - |
| Arginine dihydrolase activity | - | - | - |
| Tobacco HR | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Košćak, L.; Lamovšek, J.; Đermić, E.; Tegli, S.; Gruntar, I.; Godena, S. Identification and Characterisation of Pseudomonas savastanoi pv. savastanoi as the Causal Agent of Olive Knot Disease in Croatian, Slovenian and Portuguese Olive (Olea europaea L.) Orchards. Plants 2023, 12, 307. https://doi.org/10.3390/plants12020307
Košćak L, Lamovšek J, Đermić E, Tegli S, Gruntar I, Godena S. Identification and Characterisation of Pseudomonas savastanoi pv. savastanoi as the Causal Agent of Olive Knot Disease in Croatian, Slovenian and Portuguese Olive (Olea europaea L.) Orchards. Plants. 2023; 12(2):307. https://doi.org/10.3390/plants12020307
Chicago/Turabian StyleKošćak, Laura, Janja Lamovšek, Edyta Đermić, Stefania Tegli, Igor Gruntar, and Sara Godena. 2023. "Identification and Characterisation of Pseudomonas savastanoi pv. savastanoi as the Causal Agent of Olive Knot Disease in Croatian, Slovenian and Portuguese Olive (Olea europaea L.) Orchards" Plants 12, no. 2: 307. https://doi.org/10.3390/plants12020307
APA StyleKošćak, L., Lamovšek, J., Đermić, E., Tegli, S., Gruntar, I., & Godena, S. (2023). Identification and Characterisation of Pseudomonas savastanoi pv. savastanoi as the Causal Agent of Olive Knot Disease in Croatian, Slovenian and Portuguese Olive (Olea europaea L.) Orchards. Plants, 12(2), 307. https://doi.org/10.3390/plants12020307










