Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review
Abstract
1. Introduction
2. Results
2.1. Research in Electronic Databases
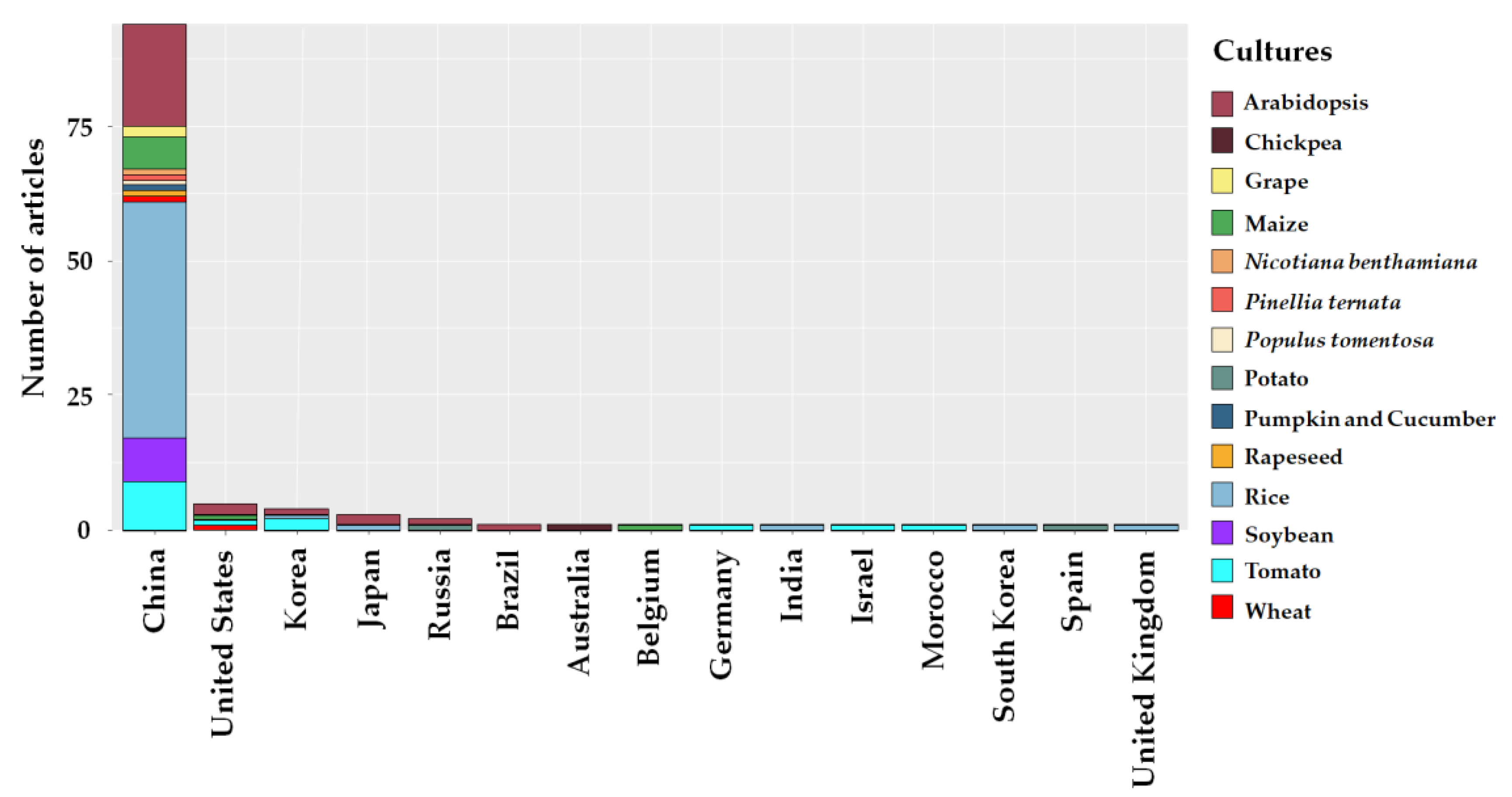
2.2. Study Sites
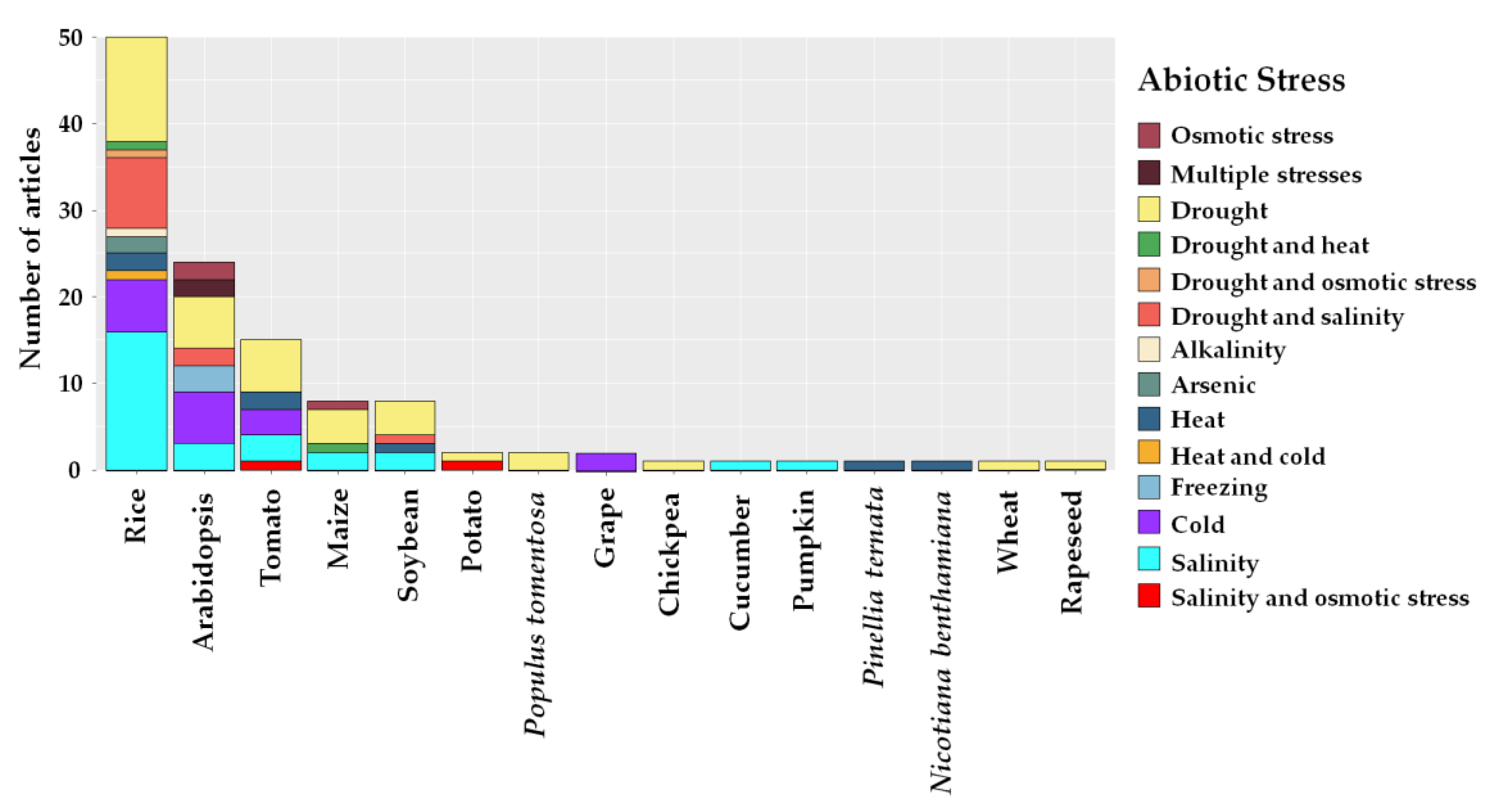
2.3. Edited Plant Crops and Abiotic Stresses
2.4. Types of Explants
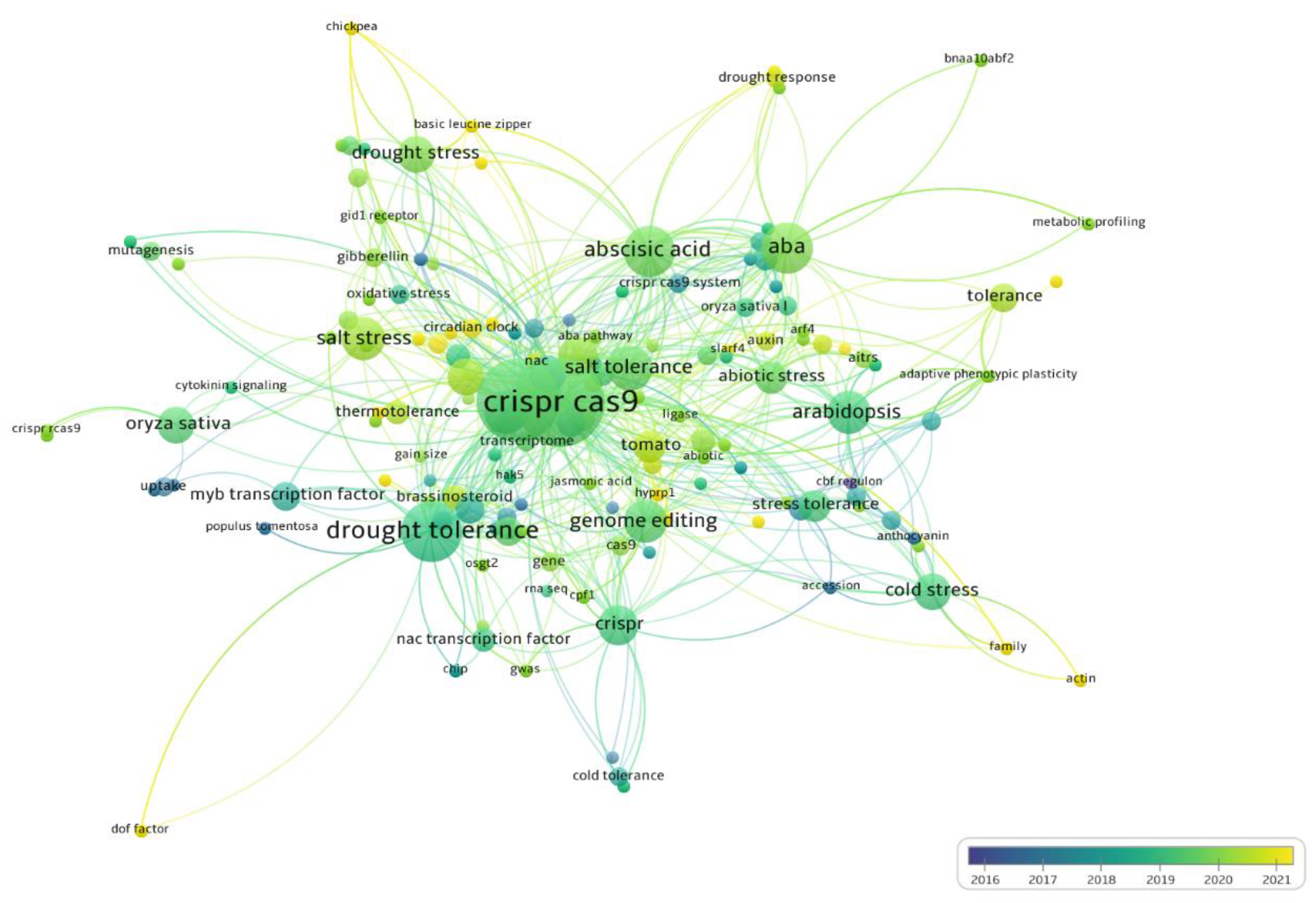
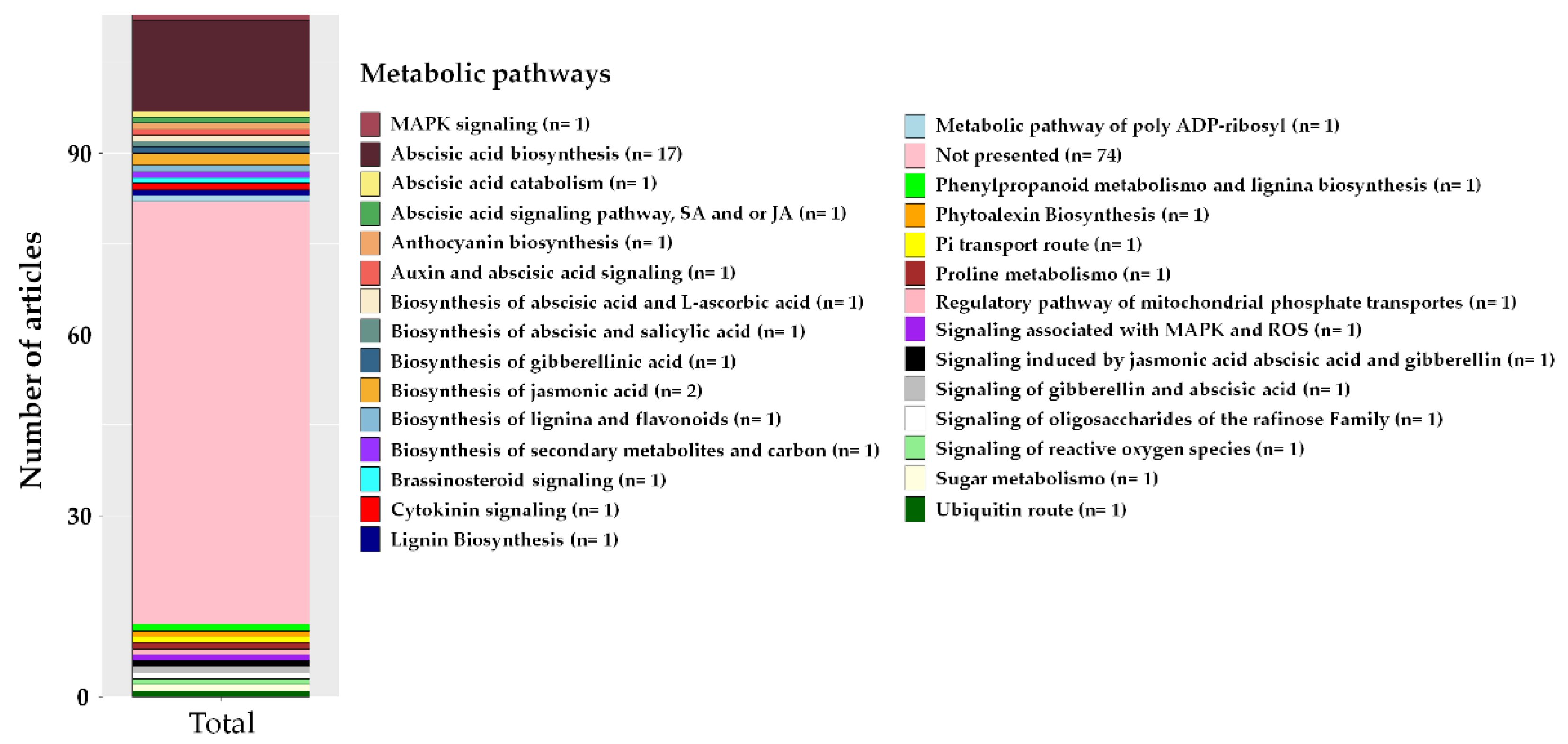
2.5. Main Genes and Metabolic Pathways
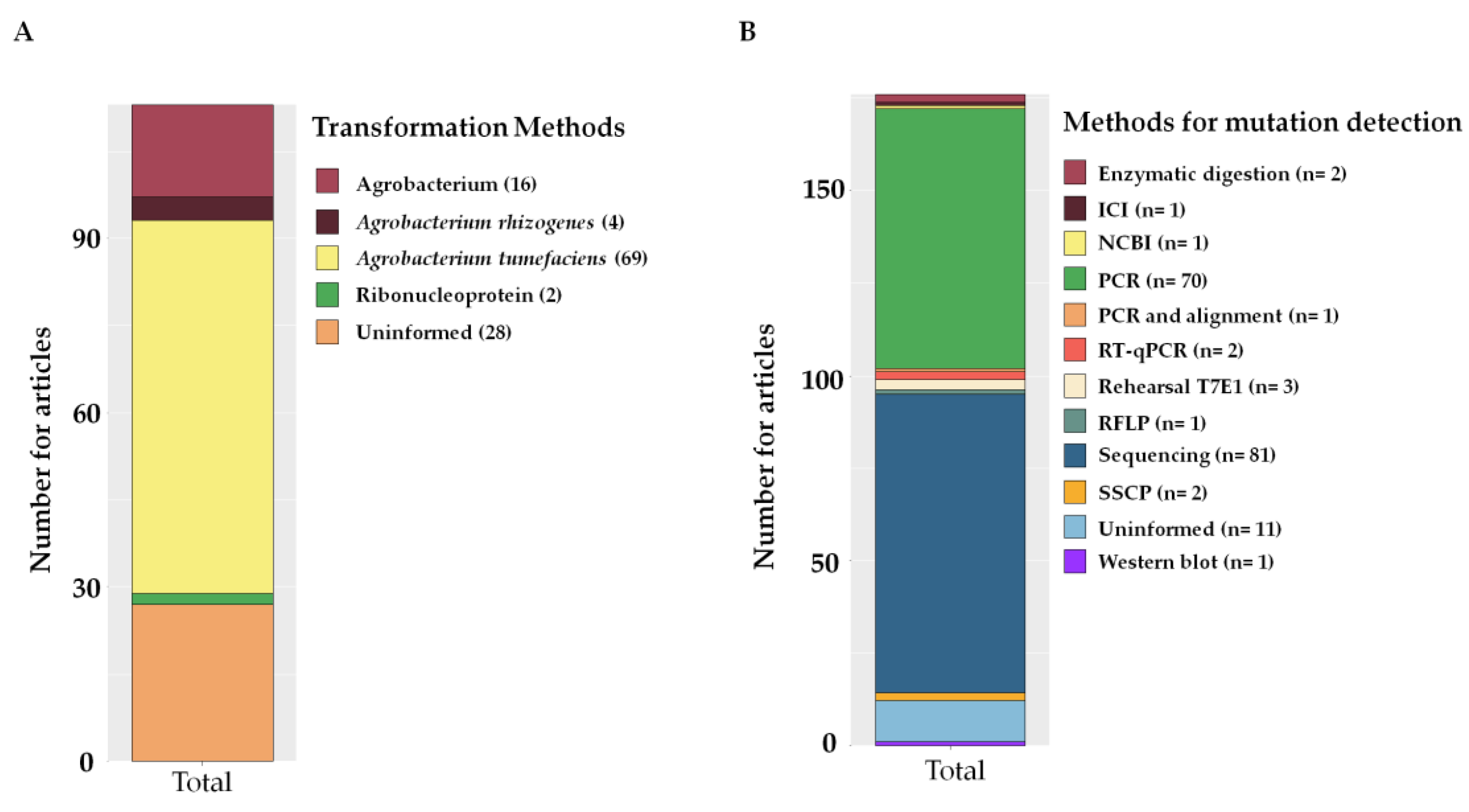
2.6. Methods of Editing with CRISPR
2.7. Auxiliary Methods
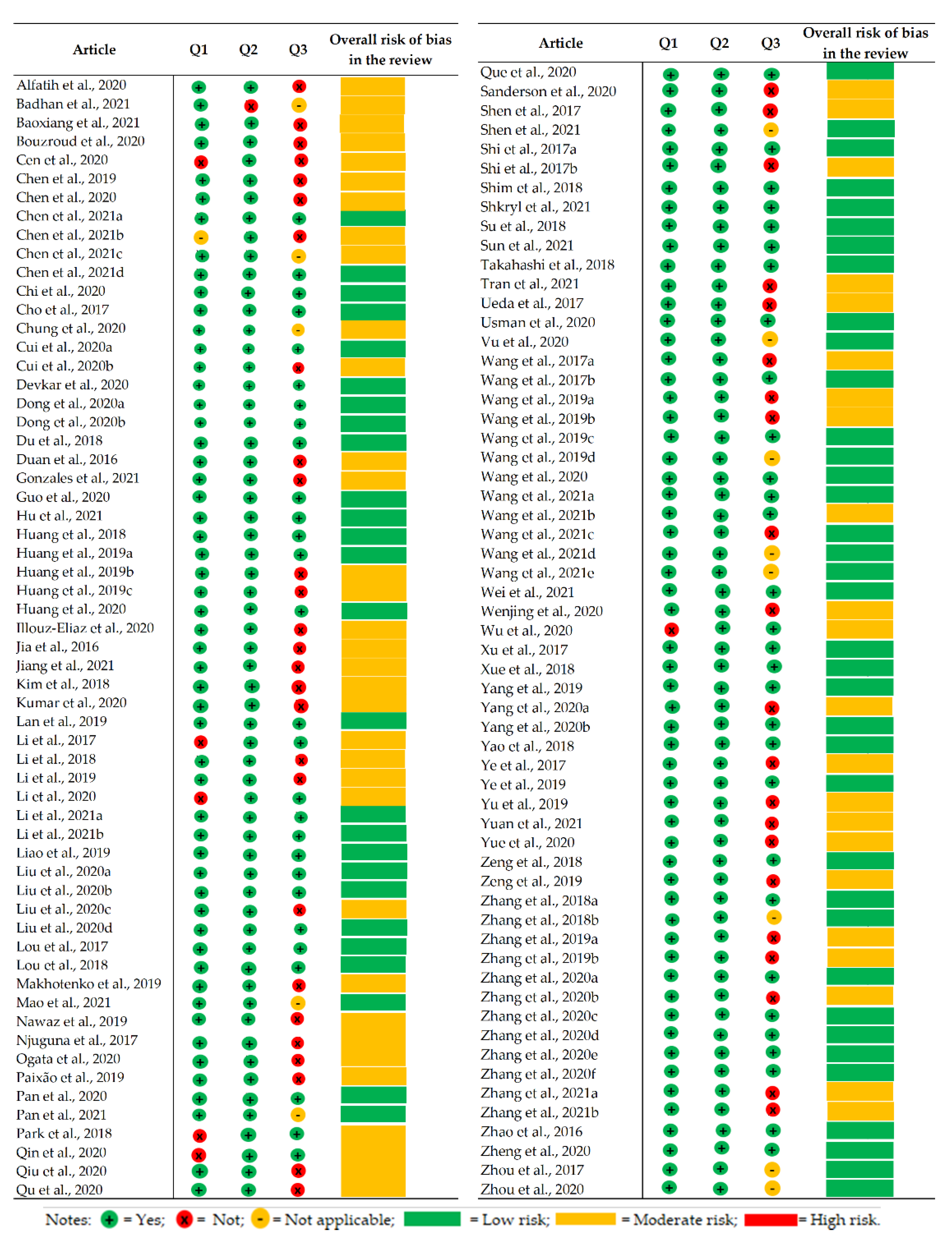
2.8. Risk of Bias Analysis
2.9. Literature Reviews
3. Discussion
3.1. Research in Electronic Databases
3.2. Study Sites, Plant Crops and Abiotic Stresses
3.3. Types of Explants
3.4. Genes and Metabolic Pathways
3.5. Methods for Editing with CRISPR/Cas
3.6. Auxiliary Methods
3.7. Risk of Bias
3.8. Final Considerations, Limitations, and Future Perspectives
4. Materials and Methods
4.1. Planning
4.2. Execution
4.3. Summarization
4.4. Risk of Bias Analysis
- Was off-target activity investigated?
- Was a phenotypic analysis performed?
- Is the identified protein studied?
4.5. Systematic Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The Future of Food and Agriculture: Trends and Challenges; FAO: Rome, Italy, 2017; ISBN 9789251095515. [Google Scholar]
- FAOSTAT. Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/faostat/en/#home (accessed on 13 March 2022).
- Onaga, G.; Wydra, K. Advances in plant tolerance to abiotic stresses. Plant Genom. 2016, 10, 229–272. [Google Scholar] [CrossRef]
- Pandey, P.; Irulappan, V.; Bagavathiannan, M.V.; Senthil-Kumar, M. Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front. Plant Sci. 2017, 8, 537. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, M.M.; Block, A.; Christensen, S.A.; Allen, L.H.; Schmelz, E.A. The effects of climate change associated abiotic stresses on maize phytochemical defenses. Phytochem. Rev. 2018, 17, 37–49. [Google Scholar] [CrossRef]
- Bhat, J.A.; Ali, S.; Salgotra, R.K.; Mir, Z.A.; Dutta, S.; Jadon, V.; Tyagi, A.; Mushtaq, M.; Jain, N.; Singh, G.P.; et al. Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 2016, 7, 221. [Google Scholar] [CrossRef]
- Raman, R. The impact of genetically modified (GM) crops in modern agriculture: A review. GM Crops Food 2017, 8, 195–208. [Google Scholar] [CrossRef]
- Mushtaq, M.; Bhat, J.A.; Mir, Z.A.; Sakina, A.; Ali, S.; Singh, A.K.; Tyagi, A.; Salgotra, R.K.; Dar, A.A.; Bhat, R. CRISPR/Cas approach: A new way of looking at plant-abiotic interactions. J. Plant Physiol. 2018, 224, 156–162. [Google Scholar] [CrossRef]
- Van Acker, R.; Rahman, M.M.; Cici, S.Z.H. Pros and cons of GMO crop farming. In Oxford Research Encyclopedia of Environmental Science; Oxford University Press: Oxford, UK, 2017; pp. 11–16. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 17, 816–821. [Google Scholar] [CrossRef]
- Khatodia, S.; Bhatotia, K.; Passricha, N.; Khurana, S.M.P.; Tuteja, N. The CRISPR/Cas Genome-Editing Tool: Application in Improvement of Crops. Front. Plant Sci. 2016, 7, 506. [Google Scholar] [CrossRef]
- Ghosh, S.; Dey, G. Biotic and abiotic stress tolerance through CRISPR-Cas mediated genome editing. J. Plant Biochem. Biotechnol. 2022, 31, 227–238. [Google Scholar] [CrossRef]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef]
- Bae, T.; Hur, J.W.; Kim, D.; Hur, J.K. Recent trends in CRISPR-Cas system: Genome, epigenome, and transcriptome editing and CRISPR delivery systems. Genes Genom. 2019, 41, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Karlson, C.K.S.; Mohd-Noor, S.N.; Nolte, N.; Tan, B.C. CRISPR/dCas9-based systems: Mechanisms and applications in plant sciences. Plants 2021, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Patron, N.J.; Nekrasov, V. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 2015, 32, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.J.; Nogoy, F.M.; Lee, S.K.; Cho, Y.G.; Kang, K.K. Application of ZFN for site directed mutagenesis of rice SSIVa gene. Biotechnol. Bioprocess Eng. 2018, 23, 108–115. [Google Scholar] [CrossRef]
- Chennakesavulu, K.; Singh, H.; Trivedi, P.K.; Jain, M.; Yadav, S.R. State-of-the-Art in CRISPR Technology and Engineering Drought, Salinity, and Thermo-tolerant crop plants. Plant Cell Rep. 2021, 41, 815–831. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Kim, M.J.; Kim, J.Y. CRISPR-mediated engineering across the central dogma in plant biology for basic research and crop improvement. Mol. Plant 2021, 14, 127–150. [Google Scholar] [CrossRef]
- Shelake, R.M.; Kadam, U.S.; Kumar, R.; Pramanik, D.; Singh, A.K.; Kim, J.Y. Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives. Plant Commun. 2022, 3, 100417. [Google Scholar] [CrossRef]
- Wenjing, W.; Chen, Q.; Singh, P.K.; Huang, Y.; Pei, D. CRISPR/Cas9 edited HSFA6a and HSFA6b of Arabidopsis thaliana offers ABA and osmotic stress insensitivity by modulation of ROS homeostasis. Plant Signal. Behav. 2020, 15, 1816321. [Google Scholar] [CrossRef]
- Tran, M.T.; Doan, D.T.H.; Kim, J.; Song, Y.J.; Sung, Y.W.; Das, S.; Kim, E.J.; Son, G.H.; Kim, S.H.; Vu, T.V.; et al. CRISPR/Cas9-based precise excision of SlHyPRP1 domain (s) to obtain salt stress-tolerant tomato. Plant Cell Rep. 2021, 40, 999–1011. [Google Scholar] [CrossRef]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. CRISPR/Cas9-induced mutagenesis of semi-rolled leaf1,2 confers curled leaf phenotype and drought tolerance by influencing protein expression patterns and ROS scavenging in rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef]
- Badhan, S.; Ball, A.S.; Mantri, N. First report of CRISPR/Cas9 mediated DNA-free editing of 4CL and RVE7 genes in chickpea protoplasts. Int. J. Mol. Sci. 2021, 22, 396. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xuan, H.; Yang, C.; Guo, N.; Wang, H.; Zhao, J.; Xing, H. GmHsp90A2 is involved in soybean heat stress as a positive regulator. Plant Sci. 2019, 285, 26–33. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Tong, Q.; Xu, G.; Xu, M.; Li, H.; Fan, P.; Li, S.; Liang, Z. Transcriptomic analysis of grapevine Dof transcription factor gene family in response to cold stress and functional analyses of the VaDof17d gene. Planta 2021, 253, 55. [Google Scholar] [CrossRef]
- Zulfiqar, M.; Qamar, I.; Murtaza, M.; Haider, Z.; Javed, A.; Abbas, A.; Haider, A.; Masood, M.U.; Iftikhar, A. A novel plant genome editing CRISPR/Cas9 System: To modify stress tolerance responses in plants. Am. J. Biotechnol. Biosci. 2019, 3, 16. [Google Scholar]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N. Application of genetic modification and genome editing for developing climate-smart banana. Food Energy Secur. 2019, 8, e00168. [Google Scholar] [CrossRef]
- Farhat, S.; Jain, N.; Singh, N.; Sreevathsa, R.; Dash, P.K.; Rai, R.; Yadav, S.; Kumar, P.; Sarkar, A.K.; Jain, A.; et al. CRISPR-Cas9 directed genome engineering for enhancing salt stress tolerance in rice. Semin. Cell Dev. Biol. 2019, 96, 91–99. [Google Scholar] [CrossRef]
- Hyun, T.K. CRISPR/Cas-based genome editing to improve abiotic stress tolerance in plants. Bot. Serb. 2020, 44, 121–127. [Google Scholar] [CrossRef]
- Biswas, D.; Saha, S.C.; Dey, A. CRISPR-Cas genome-editing tool in plant abiotic stress-tolerance. Plant Gene 2021, 26, 100286. [Google Scholar] [CrossRef]
- Li, X.; Xu, S.; Fuhrmann-Aoyagi, M.B.; Yuan, S.; Iwama, T.; Kobayashi, M.; Miura, K. CRISPR/Cas9 Technique for Temperature, Drought, and Salinity Stress Responses. Curr. Issues Mol. Biol. 2022, 44, 2664–2682. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, X.; Zhang, Y.; Zhang, Q. CRISPR/Cas genome editing improves abiotic and biotic stress tolerance of crops. Front. Genome Ed. 2022, 4, 987817. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, M.F.; Karlson, C.K.S.; Teoh, E.Y.; Lau, S.-E.; Tan, B.C. Genome Editing for Sustainable Crop Improvement and Mitigation of Biotic and Abiotic Stresses. Plants 2022, 11, 2625. [Google Scholar] [CrossRef] [PubMed]
- Modrzejewski, D.; Hartung, F.; Lehnert, H.; Sprink, T.; Kohl, C.; Keilwagen, J.; Wilhelm, R. Which factors affect the occurrence of off-target effects caused by the use of CRISPR/Cas: A systematic review in plants. Front. Plant Sci. 2020, 11, 574959. [Google Scholar] [CrossRef] [PubMed]
- Modrzejewski, D.; Hartung, F.; Sprink, T.; Krause, D.; Kohl, C.; Wilhelm, R. What is the available evidence for the range of applications of genome-editing as a new tool for plant trait modification and the potential occurrence of associated off-target effects: A systematic map. Environ. Evid. 2019, 8, 27. [Google Scholar] [CrossRef]
- Pandey, V.K.; Tripathi, A.; Bhushan, R.; Ali, A.; Dubey, P.K.; Therapy, G. Application of CRISPR/Cas9 genome editing in genetic disorders: A systematic review up to date. J. Genet. Syndr. Gene Ther. 2017, 8, 1000321. [Google Scholar] [CrossRef]
- Babačić, H.; Mehta, A.; Merkel, O.; Schoser, B. CRISPR-cas gene-editing as plausible treatment of neuromuscular and nucleotide-repeat-expansion diseases: A systematic review. PLoS ONE 2019, 14, e0212198. [Google Scholar] [CrossRef]
- Bhatt, A.; Bumbrah, G.S.; Ruwali, M.; Hameed, S.; Fatima, Z. Diagnostic efficiency of RT-LAMP integrated CRISPR-Cas technique for COVID-19: A systematic review and meta-analysis. Pathog. Glob. Health 2022, 116, 410–420. [Google Scholar] [CrossRef]
- Maganti, H.B.; Kirkham, A.M.; Bailey, A.J.; Shorr, R.; Kekre, N.; Pineault, N.; Allan, D.S. Use of CRISPR/Cas9 gene editing to improve chimeric antigen-receptor T cell therapy: A systematic review and meta-analysis of preclinical studies. Cytotherapy 2022, 24, 405–412. [Google Scholar] [CrossRef]
- Silva, F.A.F.; Brito, B.B.; Santos, M.L.C.; Marques, H.S.; Silva, R.T.; Carvalho, L.S.D., Jr.; Vieira, E.S.; Oliveira, M.V.; Melo, F.F. COVID-19 gastrointestinal manifestations: A systematic review. Rev. Soc. Bras. Med. Trop. 2020, 53, e20200714. [Google Scholar] [CrossRef]
- Rocha, A.D.J.; Soares, J.M.D.S.; Nascimento, F.D.S.; Santos, A.S.; Amorim, V.B.D.O.; Ferreira, C.F.; Haddad, F.; Santos-Serejo, J.A.; Amorim, E.P. Improvements in the resistance of the banana species to Fusarium wilt: A systematic review of methods and perspectives. J. Fungi 2021, 7, 249. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.M.; Rocha, A.J.; Nascimento, F.S.; Santos, A.S.; Miller, R.N.; Ferreira, C.F.; Haddad, F.; Amorim, V.B.O.; Amorim, E.P. Genetic improvement for resistance to black Sigatoka in bananas: A systematic review. Front. Plant Sci. 2021, 12, 657916. [Google Scholar] [CrossRef] [PubMed]
- Benevenuto, R.F.; Venter, H.J.; Zanatta, C.B.; Nodari, R.O.; Agapito-Tenfen, S.Z. Alterations in genetically modified crops assessed by omics studies: Systematic review and meta-analysis. Trends Food Sci. Technol. 2022, 120, 325–337. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Xing, H.-L.; Dong, L.; Wang, Z.; Zhang, H.-Y.; Han, C.-Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.; Al-Sadi, A.M.; Pour-Aboughadareh, A.; Burritt, D.J.; Tran, L.S.P. Genome editing using CRISPR/Cas9–targeted mutagenesis: An opportunity for yield improvements of crop plants grown under environmental stresses. Plant Physiol. Biochem. 2018, 131, 31–36. [Google Scholar] [CrossRef]
- Bhat, M.A.; Mir, R.A.; Kumar, V.; Shah, A.A.; Zargar, S.M.; Rahman, S.; Jan, A.T. Mechanistic insights of CRISPR/Cas-mediated genome editing towards enhancing abiotic stress tolerance in plants. Physiol. Plant. 2021, 172, 1255–1268. [Google Scholar] [CrossRef]
- Biswal, A.K.; Mangrauthia, S.K.; Reddy, M.R.; Yugandhar, P. CRISPR mediated genome engineering to develop climate smart rice: Challenges and opportunities. Semin. Cell Dev. Biol. 2019, 96, 100–106. [Google Scholar] [CrossRef]
- Debbarma, J.; Sarki, Y.N.; Saikia, B.; Boruah, H.P.D.; Singha, D.L.; Chikkaputtaiah, C. Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR–Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: A review. Mol. Biotechnol. 2019, 61, 153–172. [Google Scholar] [CrossRef]
- Gentzel, I.N.; Ohlson, E.W.; Redinbaugh, M.G.; Wang, G.L. VIGE: Virus-induced genome editing for improving abiotic and biotic stress traits in plants. Stress Biol. 2022, 2, 2. [Google Scholar] [CrossRef]
- Jain, M. Function genomics of abiotic stress tolerance in plants: A CRISPR approach. Front. Plant Sci. 2015, 6, 375. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.K.; Bharat, S.S.; Mishra, R. Engineering drought tolerance in plants through CRISPR/Cas genome editing. 3 Biotech 2020, 10, 400. [Google Scholar] [CrossRef] [PubMed]
- Khalid, T.; Hassan, S.; Ashraf, S.; Naseem, H.; Nasim, F.; Batool, Y. Functional Characterization of CNGC19 and CNGC20 of Arabidopsis through CRISPR-Cas9. Sch. Int. J. Biochem. 2020, 3, 136–142. [Google Scholar] [CrossRef]
- Ma, L.; Liang, Z. CRISPR technology for abiotic stress resistant crop breeding. Plant Growth Regul. 2021, 94, 115–129. [Google Scholar] [CrossRef]
- Razzaq, M.K.; Aleem, M.; Mansoor, S.; Khan, M.A.; Rauf, S.; Iqbal, S.; Siddique, K.H.M. Omics and CRISPR-Cas9 approaches for molecular insight, functional gene analysis, and stress tolerance development in crops. Int. J. Mol. Sci. 2021, 22, 1292. [Google Scholar] [CrossRef] [PubMed]
- Saikia, B.; Singh, S.; Debbarma, J.; Velmurugan, N.; Dekaboruah, H.; Arunkumar, K.P.; Chikkaputtaiah, C. Multigene CRISPR/Cas9 genome editing of hybrid proline rich proteins (HyPRPs) for sustainable multi-stress tolerance in crops: The review of a promising approach. Physiol. Mol. Biol. Plants 2020, 26, 857–869. [Google Scholar] [CrossRef]
- Sami, A.; Xue, Z.; Tazein, S.; Arshad, A.; He Zhu, Z.; Ping Chen, Y.; Hong, Y.; Zhu, X.T.; Zhou, K.J. CRISPR–Cas9-based genetic engineering for crop improvement under drought stress. Bioengineered 2021, 12, 5814–5829. [Google Scholar] [CrossRef]
- Singh, J.; Sharma, D.; Brar, G.S.; Sandhu, K.S.; Wani, S.H.; Kashyap, R.; Kour, A.; Singh, S. CRISPR/Cas Tool Designs for Multiplex Genome Editing and Its Applications in Developing Biotic and Abiotic Stress Resistant Crop Plants. Mol. Biol. Rep. 2022, 49, 11443–11467. [Google Scholar] [CrossRef]
- Zafar, S.A.; Zaidi, S.S.E.A.; Gaba, Y.; Singla-Pareek, S.L.; Dhankher, O.P.; Li, X.; Xueyong, L.; Mansoor, S.; Pareek, A. Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J. Exp. Bot. 2020, 71, 470–479. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Li, J.F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Liu, J.; Xi, J.J.; Qiu, J.L.; Gao, C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Klimek-Chodacka, M.; Oleszkiewicz, T.; Lowder, L.G.; Qi, Y.; Baranski, R. Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 2018, 37, 575–586. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Q.; Li, Q.; Men, K.; He, Z.; Deng, H.; Ji, W.; Wei, Y. Recent advances in therapeutic genome editing in China. Hum. Gene Ther. 2018, 29, 136–145. [Google Scholar] [CrossRef]
- Gao, W.; Xu, W.T.; Huang, K.L.; Guo, M.Z.; Luo, Y.B. Risk analysis for genome editing-derived food safety in China. Food Control 2018, 84, 128–137. [Google Scholar] [CrossRef]
- Sun, W.; Wang, H. Recent advances of genome editing and related technologies in China. Gene Ther. 2020, 27, 312–320. [Google Scholar] [CrossRef]
- Cohen, J. To Feed Its 1.4 Billion, China Bets Big on Genome Editing of Crops. 2019. Available online: https://www.science.org/content/article/feed-its-14-billion-china-bets-big-genome-editing-crops (accessed on 21 March 2022).
- Provart, N.J.; Alonso, J.; Assmann, S.M.; Bergmann, D.; Brady, S.M.; Brkljacic, J.; Browse, J.; Chapple, C.; Colot, V.; Cutler, S.; et al. 50 years of Arabidopsis research: Highlights and future directions. New Phytol. 2016, 209, 921–944. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Rensink, W.A.; Buell, C.R. Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species. Plant Physiol. 2004, 135, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liu, C.; Zhao, R.; Wang, L.; Chen, L.; Yu, W.; Zhang, S.; Sheng, J.; Shen, L. CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019, 19, 38. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Ma, N.; Wang, C.; Fan, H.; Wang, M.; Zhang, J.; Cao, J.; Wang, D. A Golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front. Plant Sci. 2021, 12, 638340. [Google Scholar] [CrossRef]
- Shi, Y.; Gao, X.; Xu, Y.; Giorgi, F.; Chen, D. Effects of climate change on heating and cooling degree days and potential energy demand in the household sector of China. Clim. Res. 2016, 67, 135–149. [Google Scholar] [CrossRef]
- Chakraborty, N.; Chakraborty, P.; Sen, M.; Bandopadhyay, R. Choice of Explant for Plant Genetic Transformation. In Biolistic DNA Delivery in Plants; Rustgi, S., Luo, H., Eds.; Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2124, pp. 107–123. [Google Scholar] [CrossRef]
- Cho, S.; Yu, S.I.; Park, J.; Mao, Y.; Zhu, J.K.; Yun, D.J.; Lee, B. Accession-dependent CBF gene deletion by CRISPR/Cas system in Arabidopsis. Front. Plant Sci. 2017, 8, 1910. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, N.; Zhang, Q.; Zhou, G.; Tian, H.; Hussain, S.; Ahmed, S.; Wang, T.; Wang, S. Genome editing to integrate seed size and abiotic stress tolerance traits in Arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int. J. Mol. Sci. 2019, 20, 2695. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Li, H.; Ding, Y.; Shi, Y.; Song, C.; Gong, Z.; Yang, S. BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell 2019, 31, 2682–2696. [Google Scholar] [CrossRef]
- Sanderson, B.J.; Park, S.; Jameel, M.I.; Kraft, J.C.; Thomashow, M.F.; Schemske, D.W.; Oakley, C.G. Genetic and physiological mechanisms of freezing tolerance in locally adapted populations of a winter annual. Am. J. Bot. 2020, 107, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, T.; Rehman, A.U.; Wang, Y.; Qi, J.; Li, Z.; Song, C.; Wang, B.; Yang, S.; Gong, Z. Arabidopsis U-box E3 ubiquitin ligase PUB11 negatively regulates drought tolerance by degrading the receptor-like protein kinases LRR1 and KIN7. J. Integr. Plant Biol. 2021, 63, 494–509. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Cao, Y.; Ku, L.; Yao, W.; Cao, Y.; Ren, Z.; Dou, D.; Wang, H.; Ren, Z.; Liu, H.; et al. Dual functions of ZmNF-YA3 in photoperiod-dependent flowering and abiotic stress responses in maize. J. Exp. Bot. 2018, 69, 5177–5189. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, C.; Wang, H.; Wang, S.; Yang, S.; Liu, X.; Yan, J.; Li, B.; Beatty, M.; Zastrow-Hayes, G.; et al. Mapping regulatory variants controlling gene expression in drought response and tolerance in maize. Genome Biol. 2020, 21, 163. [Google Scholar] [CrossRef] [PubMed]
- Ogata, T.; Ishizaki, T.; Fujita, M.; Fujita, Y. CRISPR/Cas9-targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS ONE 2020, 15, e0243376. [Google Scholar] [CrossRef]
- Pan, Z.; Liu, M.; Zhao, H.; Tan, Z.; Liang, K.; Sun, Q.; Gong, D.; He, H.; Zhou, W.; Qiu, F. ZmSRL5 is involved in drought tolerance by maintaining cuticular wax structure in maize. J. Integr. Plant Biol. 2020, 62, 1895–1909. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; La Rota, M.; Hoerster, G.; Hastings, C.; Wang, N.; Chamberlin, M.; Wu, E.; Jones, T.; Gordon-Kamm, W. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol.-Plant 2018, 54, 240–252. [Google Scholar] [CrossRef]
- Qu, M.; Essemine, J.; Xu, J.; Ablat, G.; Perveen, S.; Wang, H.; Chen, K.; Zhao, Y.; Chen, G.; Chu, C.; et al. Alterations in stomatal response to fluctuating light increase biomass and yield of rice under drought conditions. Plant J. 2020, 104, 1334–1347. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Chen, S.; Ma, X.; Wei, H.; Chen, C.; Gao, N.; Zou, Y.; Kong, D.; Li, T.; et al. An APETALA2/ethylene responsive factor, OsEBP89 knockout enhances adaptation to direct-seeding on wet land and tolerance to drought stress in rice. Mol. Genet. Genom. 2020, 295, 941–956. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Kanchiswamy, C.N. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Crop J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Reduction of tomato-plant chilling tolerance by CRISPR–Cas9-mediated SlCBF1 mutagenesis. J. Agric. Food Chem. 2018, 66, 9042–9051. [Google Scholar] [CrossRef]
- Park, S.; Gilmour, S.J.; Grumet, R.; Thomashow, M.F. CBF-dependent and CBF-independent regulatory pathways contribute to the differences in freezing tolerance and cold-regulated gene expression of two Arabidopsis ecotypes locally adapted to sites in Sweden and Italy. PLoS ONE 2018, 13, e0207723. [Google Scholar] [CrossRef]
- Wang, F.; Wang, X.; Zhang, Y.; Yan, J.; Ahammed, G.J.; Bu, X.; Sun, X.; Liu, Y.; Xu, T.; Qi, H.; et al. SlFHY3 and SlHY5 act compliantly to enhance cold tolerance through the integration of myo-inositol and light signaling in tomato. New Phytol. 2021, 233, 2127–2143. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef]
- Yu, W.; Wang, L.; Zhao, R.; Sheng, J.; Zhang, S.; Li, R.; Shen, L. Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol. 2019, 19, 354. [Google Scholar] [CrossRef]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Chen, L.; Yang, H.; Fang, Y.; Guo, W.; Chen, H.; Zhang, X.; Dai, W.; Chen, S.; Hao, Q.; Yuan, S.; et al. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 2021, 19, 702–716. [Google Scholar] [CrossRef]
- Mao, H.; Jian, C.; Cheng, X.; Chen, B.; Mei, F.; Li, F.; Zhang, Y.; Li, S.; Du, L.; Li, T.; et al. The wheat ABA receptor gene TaPYL1-1B contributes to drought tolerance and grain yield by increasing water-use efficiency. Plant Biotechnol. J. 2021, 20, 846–861. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Que, Z.; Xia, Y.; Tang, N.; Li, D.; He, R.; Cao, M. Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol. 2017, 60, 539–547. [Google Scholar] [CrossRef]
- Que, Z.; Lu, Q.; Liu, T.; Li, S.; Zou, J.; Chen, G. The rice annexin gene OsAnn5 is a positive regulator of cold stress tolerance at the seedling stage. Res. Sq. 2020, 1, 1–14. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.; Wang, H.; Yu, D. The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol. 2018, 18, 203. [Google Scholar] [CrossRef]
- Yang, C.; Huang, Y.; Lv, W.; Zhang, Y.; Bhat, J.A.; Kong, J.; Xing, H.; Zhao, J.; Zhao, T. GmNAC8 acts as a positive regulator in soybean drought stress. Plant Sci. 2020, 293, 110442. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef]
- Shim, J.S.; Oh, N.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Kim, J.K. Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 2018, 9, 310. [Google Scholar] [CrossRef]
- Wang, B.; Zhong, Z.; Wang, X.; Han, X.; Yu, D.; Wang, C.; Song, W.; Zheng, X.; Chen, C.; Zhang, Y. Knockout of the OsNAC006 transcription factor causes drought and heat sensitivity in rice. Int. J. Mol. Sci. 2020, 21, 2288. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Huang, J.; Xia, J. OsNAC45 is involved in ABA response and salt tolerance in rice. Rice 2020, 13, 79. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Bendaou, N.; Bouzayen, M.; Zsögön, A.; Smouni, A.; et al. Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xun, H.; Wang, W.; Ding, X.; Tian, H.; Hussain, S.; Dong, Q.; Li, Y.; Cheng, Y.; Wang, C.; et al. Mutation of GmAITR genes by CRISPR/Cas9 genome editing results in enhanced salinity stress tolerance in soybean. Front. Plant Sci. 2021, 12, 779598. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, N.; Zhou, G.; Hussain, S.; Ahmed, S.; Tian, H.; Wang, S. Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 2021, 21, 137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Z.; Xiong, Y.; Shi, J.; Chen, C.; Pan, Y.; Xue, T.; Xue, J.; Duan, Y. Stearic acid desaturase gene negatively regulates the thermotolerance of Pinellia ternata by modifying the saturated levels of fatty acids. Ind. Crops Prod. 2021, 166, 113490. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Luo, Y.; Zhang, L.; Yao, Y.; Han, L.; Chen, Z.; Wang, L.; Li, Y. OsABA8ox2, an ABA catabolic gene, suppresses root elongation of rice seedlings and contributes to drought response. Crop J. 2020, 8, 480–491. [Google Scholar] [CrossRef]
- Yang, W.; Chen, S.; Cheng, Y.; Zhang, N.; Ma, Y.; Wang, W.; Tian, H.; Li, Y.; Hussain, S.; Wang, S. Cell wall/vacuolar inhibitor of fructosidase 1 regulates ABA response and salt tolerance in Arabidopsis. Plant Signal. Behav. 2020, 15, 1744293. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, X.; Dong, S.; Jiang, X.; Wang, L.; Yu, J.; Zhou, Y. Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato. Plant Biotechnol. J. 2019, 18, 1041–1055. [Google Scholar] [CrossRef]
- Pan, C.; Yang, D.; Zhao, X.; Liu, Y.; Li, M.; Ye, L.; Ali, M.; Yu, F.; Lamin-Samu, A.T.; Fei, Z.; et al. PIF4 negatively modulates cold tolerance in tomato anthers via temperature-dependent regulation of tapetal cell death. Plant Cell 2021, 33, 2320–2339. [Google Scholar] [CrossRef]
- Shen, C.; Zhang, Y.; Li, Q.; Liu, S.; He, F.; An, Y.; Zhou, Y.; Liu, C.; Yin, W.; Xia, X. PdGNC confers drought tolerance by mediating stomatal closure resulting from NO and H2O2 production via the direct regulation of PdHXK1 expression in Populus. New Phytol. 2021, 230, 1868–1882. [Google Scholar] [CrossRef]
- Wang, F.Z.; Chen, M.X.; Yu, L.J.; Xie, L.J.; Yuan, L.B.; Qi, H.; Xiao, M.; Guo, W.; Chen, Z.; Yi, K.; et al. OsARM1, an R2R3 MYB transcription factor, is involved in regulation of the response to arsenic stress in rice. Front. Plant Sci. 2017, 8, 1868. [Google Scholar] [CrossRef]
- Ye, Y.; Li, P.; Xu, T.; Zeng, L.; Cheng, D.; Yang, M.; Luo, J.; Lian, X. OsPT4 contributes to arsenate uptake and transport in rice. Front. Plant Sci. 2017, 8, 2197. [Google Scholar] [CrossRef]
- Paixão, J.F.R.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Melo, B.P.; Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 8080. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.P.; Deng, K.; Cheng, Y.; Zhong, Z.; Tian, L.; Tang, X.; Tang, A.; Zheng, X.; Zhang, T.; Qi, Y.; et al. CRISPR-Cas9 Based Genome Editing Reveals New Insights into MicroRNA Function and Regulation in Rice. Front. Plant Sci. 2017, 8, 1598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, H.; Srivastava, A.K.; Pan, Y.; Bai, J.; Fang, J.; Shi, H.; Zhu, J.K. Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 2018, 176, 2082–2094. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.J.; Chung, H.; Oh, N.; Choi, J.; Bang, S.W.; Jung, S.E.; Jung, H.; Shim, J.S.; Kim, J. Efficiency of recombinant CRISPR/rCas9-mediated miRNA gene editing in rice. Int. J. Mol. Sci. 2020, 21, 9606. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, W.; Li, X.; Sun, D.; Xu, K.; Feng, C.; Foka, I.C.K.; Ketehouli, T.; Gao, H.; Wang, N.; et al. Integration of sRNA, degradome, transcriptome analysis and functional investigation reveals gma-miR398c negatively regulates drought tolerance via GmCSDs and GmCCS in transgenic Arabidopsis and soybean. BMC Plant Biol. 2020, 20, 190. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Zhang, D.P. Abscisic Acid: Metabolism, Transport and Signaling; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019, 287, 110188. [Google Scholar] [CrossRef]
- Usman, B.; Nawaz, G.; Zhao, N.; Liao, S.; Liu, Y.; Li, R. Precise editing of the OSPYL9 gene by RNA-guided cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza sativa L.) by regulating circadian rhythm and abiotic stress responsive proteins. Int. J. Mol. Sci. 2020, 21, 7854. [Google Scholar] [CrossRef]
- Yuan, L.; Xie, G.Z.; Zhang, S.; Li, B.; Wang, X.; Li, Y.; Liu, T.; Xu, X. GmLCLs negatively regulate ABA perception and signalling genes in soybean leaf dehydration response. Plant Cell Environ. 2021, 44, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wong, D.C.J.; Wang, Y.; Xu, G.; Ren, C.; Liu, Y.; Kuang, Y.; Fan, P.; Li, S.; Xin, H.; et al. GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 2021, 186, 1660–1678. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Guo, L.; Wang, Z.; Zhang, X.; Zhao, B.; Guo, Y.D.; Zhanga, N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020, 301, 110683. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef]
- Chi, C.; Li, X.; Fang, P.; Xia, X.; Shi, K.; Zhou, Y.; Zhou, J.; Yu, J. Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J. Exp. Bot. 2020, 71, 1092–1106. [Google Scholar] [CrossRef]
- Qin, Q.; Wang, Y.; Huang, L.; Du, F.; Zhao, X.; Li, Z.; Wang, W.; Fu, B. A U-box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 2020, 102, 89–107. [Google Scholar] [CrossRef]
- Aquino, S.O.; Carneiro, F.A.; Rêgo, E.C.S.; Alves, G.S.C.; Andrade, A.C.; Marraccini, P. Functional analysis of different promoter haplotypes of the coffee (Coffea canephora) CcDREB1D gene through genetic transformation of Nicotiana tabacum. Plant Cell Tissue Organ Cult. 2017, 132, 279–294. [Google Scholar] [CrossRef]
- El-Mounadi, K.; Morales-Floriano, M.L.; Garcia-Ruiz, H. Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front. Plant Sci. 2020, 11, 56. [Google Scholar] [CrossRef]
- Makhotenko, A.V.; Khromov, A.V.; Snigir, E.A.; Makarova, S.S.; Makarov, V.V.; Suprunova, T.P.; Kalinina, N.O.; Taliansky, M.E. Functional analysis of coilin in virus resistance and stress tolerance of potato Solanum tuberosum using CRISPR-Cas9 editing. Dokl. Biochem. Biophys. 2019, 484, 88–91. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 234. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, J.; Sun, T.; Wang, X.; Zhu, C.; Ai, Y.; Gu, H. The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integr. Plant Biol. 2017, 59, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Jiang, N.; Xu, Z.; Xu, Q. Heterotrimeric G protein are involved in the regulation of multiple agronomic traits and stress tolerance in rice. BMC Plant Biol. 2020, 20, 90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xiang, Y.; He, N.; Liu, X.; Liu, H.; Fang, L.; Zhang, F.; Sun, X.; Zhang, D.; Li, X.; et al. Enhanced vitamin C production mediated by an ABA-induced PTP-like nucleotidase improves plant drought tolerance in Arabidopsis and maize. Mol. Plant 2020, 13, 760–776. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.E.A.; Mahas, A.; Vanderschuren, H.; Mahfouz, M.M. Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 2020, 21, 289. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.V.; Sivankalyani, V.; Kim, E.J.; Doan, D.T.H.; Tran, M.T.; Kim, J.; Sung, Y.W.; Park, M.; Kang, Y.J.; Kim, J.Y. Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato. Plant Biotechnol. J. 2020, 18, 2133–2143. [Google Scholar] [CrossRef]
- Zetsche, B.; Heidenreich, M.; Mohanraju, P.; Fedorova, I.; Kneppers, J.; DeGennaro, E.M.; Winblad, N.; Choudhury, S.R.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 2017, 35, 31–34. [Google Scholar] [CrossRef]
- Zaidi, S.S.E.A.; Mahfouz, M.M.; Mansoor, S. CRISPR-Cpf1: A new tool for plant genome editing. Trends Plant Sci. 2017, 22, 550–553. [Google Scholar] [CrossRef]
- Ruduś, I.; Kępczyński, J. Reference gene selection for molecular studies of dormancy in wild oat (Avena fatua L.) caryopses by RT-qPCR method. PLoS ONE 2018, 13, e0192343. [Google Scholar] [CrossRef]
- Cai, J.; Li, P.; Luo, X.; Chang, T.; Li, J.; Zhao, Y.; Xu, Y. Selection of appropriate reference genes for the detection of rhythmic gene expression via quantitative real-time PCR in Tibetan hulless barley. PLoS ONE 2018, 13, e0190559. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhang, X.; Liu, J.; Hou, L.; Liu, H.; Zhao, X. OsProDH negatively regulates thermotolerance in rice by modulating proline metabolism and reactive oxygen species scavenging. Rice 2020, 13, 61. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.C.; Lin, T.C.; Kieber, J.; Tsai, Y.C. Response regulators 9 and 10 negatively regulate salinity tolerance in rice. Plant Cell Physiol. 2019, 60, 2549–2563. [Google Scholar] [CrossRef]
- Cui, Y.; Lu, S.; Li, Z.; Cheng, J.; Hu, P.; Zhu, T.; Wang, X.; Jin, M.; Wang, X.; Li, L.; et al. CYCLIC NUCLEOTIDE-GATED ION CHANNELs 14 and 16 promote tolerance to heat and chilling in rice. Plant Physiol. 2020, 183, 1794–1808. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, X.; He, Y.; Xu, H.; Wang, L. Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1-mediated sodium homeostasis. EMBO J. 2021, 40, e105086. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.; Altman, D.G.; Gøtzsche, P.C.; Jüni, P.; Moher, D.; Oxman, A.D.; Savović, J.; Schulz, K.F.; Weeks, L.; Sterne, J.A.C. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. BMJ 2011, 343, d5928. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. GABA-enriched tomato is first CRISPR-edited food to enter market. Nat. Biotechnol. 2022, 40, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.M.D.C.; Pimenta, C.A.D.M.; Nobre, M.R.C. The PICO strategy for the research question construction and evidence search. Rev. Lat.-Am. Enferm. 2007, 15, 508–511. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016; Available online: https://www.R-project.org (accessed on 10 March 2022).
- Van Eck, N.; Waltman, L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 2010, 84, 523–538. [Google Scholar] [CrossRef]
- Alfatih, A.; Wu, J.; Jan, S.U.; Zhang, Z.S.; Xia, J.Q.; Xiang, C.B. Loss of rice PARAQUAT TOLERANCE 3 confers enhanced resistance to abiotic stresses and increases grain yield in field. Plant Cell Environ. 2020, 43, 2743–2754. [Google Scholar] [CrossRef]
- Baoxiang, W.; Yan, L.; Yifeng, W.; Jingfang, L.; Zhiguang, S.; Ming, C.; Yungao, X.; Bo, X.; Bo, Y.; Jian, L.; et al. OsbZIP72 Is Involved in Transcriptional Gene-Regulation Pathway of Abscisic Acid Signal Transduction by Activating Rice High-Affinity Potassium Transporter OsHKT1;1. Rice Sci. 2021, 28, 257–267. [Google Scholar] [CrossRef]
- Cen, W.; Zhao, W.; Ma, M.; Lu, S.; Liu, J.; Cao, Y.; Zeng, Z.; Wei, H.; Wang, S.; Li, R.; et al. The wild rice locus CTS-12 mediates ABA-dependent stomatal opening modulation to limit water loss under severe chilling stress. Front. Plant Sci. 2020, 11, 1711. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, J.; Dong, L.; Zeng, D.; Guo, L.; Zhang, G.; Zhu, L.; Qian, Q. The tolerance of salinity in rice requires the presence of a functional copy of FLN2. Biomolecules 2020, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhu, X.; Liu, X.; Wu, C.; Yu, C.; Hu, G.; Chen, L.; Chen, R.; Bouzayen, M.; Zouine, M.; et al. Knockout of auxin response factor SlARF4 improves tomato resistance to water deficit. Int. J. Mol. Sci. 2021, 22, 3347. [Google Scholar] [CrossRef] [PubMed]
- Devkar, V.; Thirumalaikumar, V.P.; Xue, G.P.; Vallarino, J.G.; Turečková, V.; Strnad, M.; Fernie, A.R.; Hoefgen, R.; Mueller-Roeber, B.; Balazadeh, S. Multifaceted regulatory function of tomato SlTAF1 in the response to salinity stress. New Phytol. 2020, 225, 1681–1698. [Google Scholar] [CrossRef]
- Dong, N.; Yin, W.; Liu, D.; Zhang, X.; Yu, Z.; Huang, W.; Liu, J.; Yang, Y.; Meng, M.; Niu, M.; et al. Regulation of brassinosteroid signaling and salt resistance by SERK2 and potential utilization for crop improvement in rice. Front. Plant Sci. 2020, 11, 621859. [Google Scholar] [CrossRef]
- Dong, N.Q.; Sun, Y.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.-H.; Zhang, H.; Yang, Y.-B.; Li, Y.-C.; et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef]
- Gonzales, L.; Shi, L.; Bergonzi, S.B.; Oortwijn, M.; Franco-Zorrilla, J.M.; Solano-Tavira, R.; Visser, R.G.F.; Abelenda, J.A.; Bachem, C.W.B. Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 2021, 105, 855–869. [Google Scholar] [CrossRef]
- Hu, Z.; Li, J.; Ding, S.; Cheng, F.; Li, X.; Jiang, Y.; Yu, J.; Foyer, C.H.; Shi, K. The protein kinase CPK28 phosphorylates ascorbate peroxidase and enhances thermotolerance in tomato. Plant Physiol. 2021, 186, 1302–1317. [Google Scholar] [CrossRef]
- Huang, Y.; Cao, H.; Yang, L.; Chen, C.; Shabala, L.; Xiong, M.; Niu, M.; Liu, J.; Zheng, Z.; Zhou, L.; et al. Tissue-specific respiratory burst oxidase homolog-dependent H2O2 signaling to the plasma membrane H+-ATPase confers potassium uptake and salinity tolerance in Cucurbitaceae. J. Exp. Bot. 2019, 70, 5879–5893. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Xin, S.; Xie, G.; Han, J.; Liu, Z.; Wang, B.; Zhang, S.; Wu, Q.; Cheng, X. Mutagenesis reveals that the rice OsMPT3 gene is an important osmotic regulatory factor. Crop J. 2020, 8, 465–479. [Google Scholar] [CrossRef]
- Illouz-Eliaz, N.; Nissan, I.; Nir, I.; Ramon, U.; Shohat, H.; Weiss, D. Mutations in the tomato gibberellin receptors suppress xylem proliferation and reduce water loss under water-deficit conditions. J. Exp. Bot. 2020, 71, 3603–3612. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Liu, Y.; Li, R.; Li, S.; Tan, Y.; Huang, J.; Shu, Q. An inositol 1, 3, 4, 5, 6-pentakisphosphate 2-kinase 1 mutant with a 33-nt deletion showed enhanced tolerance to salt and drought stress in rice. Plants 2021, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.V.S.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, a SBP-box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.). G3 Genes Genomes Genet. 2019, 9, 4107–4114. [Google Scholar] [CrossRef]
- Li, S.; Zhang, J.; Liu, L.; Wang, Z.; Li, Y.; Guo, L.; Li, Y.; Zhang, X.; Ren, S.; Zhao, B.; et al. SlTLFP8 reduces water loss to improve water-use efficiency by modulating cell size and stomatal density via endoreduplication. Plant Cell Environ. 2020, 43, 2666–2679. [Google Scholar] [CrossRef]
- Li, J.; Zhang, M.; Yang, L.; Mao, X.; Li, J.; Li, L.; Wang, J.; Liu, H.; Zheng, H.; Li, L.; et al. OsADR3 increases drought stress tolerance by inducing antioxidant defense mechanisms and regulating OsGPX1 in rice (Oryza sativa L.). Crop J. 2021, 9, 1003–1017. [Google Scholar] [CrossRef]
- Liu, X.H.; Lyu, Y.S.; Yang, W.; Yang, Z.T.; Lu, S.J.; Liu, J.X. A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol. J. 2020, 18, 1317–1329. [Google Scholar] [CrossRef]
- Liu, X.; Wu, D.; Shan, T.; Xu, S.; Qin, R.; Li, H.; Negm, M.; Wu, D.; Li, J. The trihelix transcription factor OsGTγ-2 is involved adaption to salt stress in rice. Plant Mol. Biol. 2020, 103, 545–560. [Google Scholar] [CrossRef]
- Nawaz, G.; Han, Y.; Usman, B.; Liu, F.; Qin, B.; Li, R. Knockout of OsPRP1, a gene encoding proline-rich protein, confers enhanced cold sensitivity in rice (Oryza sativa L.) at the seedling stage. 3 Biotech 2019, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Njuguna, E.; Coussens, G.; Aesaert, S.; Neyt, P.; Anami, S.; Van Lijsebettens, M. Modulation of energy homeostasis in maize and Arabidopsis to develop lines tolerant to drought, genotoxic and oxidative stresses. Afr. Focus 2017, 30, 66–76. [Google Scholar] [CrossRef]
- Qiu, T.; Qi, M.; Ding, X.; Zheng, Y.; Zhou, T.; Chen, Y.; Han, N.; Zhu, M.; Bian, H.; Wang, J. The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings. Ann. Bot. 2020, 125, 805–819. [Google Scholar] [CrossRef]
- Shkryl, Y.; Yugay, Y.; Avramenko, T.; Grigorchuk, V.; Gorpenchenko, T.; Grischenko, O.; Bulgakov, V. CRISPR/Cas9-Mediated Knockout of HOS1 Reveals Its Role in the Regulation of Secondary Metabolism in Arabidopsis thaliana. Plants 2021, 10, 104. [Google Scholar] [CrossRef]
- Ueda, M.; Matsui, A.; Tanaka, M.; Nakamura, T.; Abe, T.; Sako, K.; Sasaki, T.; Kim, J.M.; Ito, A.; Nishino, N.; et al. The distinct roles of class I and II RPD3-like histone deacetylases in salinity stress response. Plant Physiol. 2017, 175, 1760–1773. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Xie, G.; Liu, Z.; He, R.; Han, J.; Huang, S.; Liu, L.; Cheng, X. Mutagenesis reveals that the OsPPa6 gene is required for enhancing the alkaline tolerance in rice. Front. Plant Sci. 2019, 10, 759. [Google Scholar] [CrossRef]
- Wang, B.; Zhaohui, Z.; Huanhuan, Z.; Xia, W.; Binglin, L.; Lijia, Y.; Xiangyan, H.; Deshui, Y.; Xuelian, Z.; Chunguo, W.; et al. Targeted mutagenesis of NAC transcription factor gene, OsNAC041, leading to salt sensitivity in rice. Rice Sci. 2019, 26, 98–108. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y.; et al. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yan, G.; Duan, Z.; Wang, Z.; Kang, C.; Guo, L.; Liu, K.; Tu, J.; Shen, J.; Yi, B.; et al. Roles of the Brassica napus DELLA protein BnaA6. RGA, in modulating drought tolerance by interacting with the ABA signaling component BnaA10.ABF2. Front. Plant Sci. 2020, 11, 577. [Google Scholar] [CrossRef]
- Xu, C.; Fu, X.; Liu, R.; Guo, L.; Ran, L.; Li, C.; Tian, Q.; Jiao, B.; Wang, B.; Luo, K. PtoMYB170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis. Tree Physiol. 2017, 37, 1713–1726. [Google Scholar] [CrossRef]
- Xue, Y.; Yang, Y.; Yang, Z.; Wang, X.; Guo, Y. VAMP711 is required for abscisic acid-mediated inhibition of plasma membrane H+-ATPase activity. Plant Physiol. 2018, 178, 1332–1343. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Xu, K.; Chen, S.; Li, T.; Xia, H.; Chen, L.; Liu, H.; Luo, L. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 2019, 19, 260. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Cheng, X.; Gu, Z.; Huang, W.; Li, S.; Wang, L.; Wang, Y.F.; Xu, P.; Ma, H.; Ge, X. The AWPM-19 family protein OsPM1 mediates abscisic acid influx and drought response in rice. Plant Cell 2018, 30, 1258–1276. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.; Cao, H.; Liu, B. OsmiR535, a potential genetic editing target for drought and salinity stress tolerance in Oryza sativa. Plants 2020, 9, 1337. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.D.; Yang, C.C.; Qin, R.; Alamin, M.; Yue, E.K.; Jin, X.L.; Shi, S.H. A guanine insert in OsBBS1 leads to early leaf senescence and salt stress sensitivity in rice (Oryza sativa L.). Plant Cell Rep. 2018, 37, 933–946. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR–Cas9 system. Front. Plant Sci. 2019, 10, 1663. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef]
- Zhang, C.; Srivastava, A.K.; Sadanandom, A. Targeted mutagenesis of the SUMO protease, Overly Tolerant to Salt1 in rice through CRISPR/Cas9-mediated genome editing reveals a major role of this SUMO protease in salt tolerance. bioRxiv 2019, 555706. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Xing, J.; Wan, J.; Wang, X.; Zhang, J.; Wang, X.; Li, Z.; Zhang, M. Copalyl diphosphate synthase mutation improved salt tolerance in maize (Zea mays L.) via enhancing vacuolar Na+ sequestration and maintaining ROS homeostasis. Front. Plant Sci. 2020, 11, 457. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, X.; Liu, Q.; Ahammed, G.J.; Lin, R.; Wang, L.; Shao, S.; Yu, J.; Zhou, Y. The HY5 and MYB15 transcription factors positively regulate cold tolerance in tomato via the CBF pathway. Plant Cell Environ. 2020, 43, 2712–2726. [Google Scholar] [CrossRef]
- Zhang, P.; Qian, D.; Luo, C.; Niu, Y.; Li, T.; Li, C.; Xiang, Y.; Wang, X.; Niu, Y. Arabidopsis ADF5 acts as a downstream target gene of CBFs in response to low-temperature stress. Front. Cell Dev. Biol. 2021, 9, 635533. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Zhou, J.; Zhang, F.; Yin, J.; Zhou, G.; Li, Y.; Chen, F.; Xie, X. OsABAR1, a novel GRAM domain-containing protein, confers drought and salt tolerance via an ABA-dependent pathway in rice. Plant Physiol. Biochem. 2020, 152, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, C.; Fu, Y.; Liu, Q.; Jiao, X.; Wang, K. Expanding the range of CRISPR/Cas9 genome editing in rice. Mol. Plant 2016, 9, 943–945. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.P.; Xing, H.L.; Dong, L.; Zhang, H.Y.; Han, C.Y.; Wang, X.C.; Chen, Q.J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Kang, S.; He, L.; Zhao, J.; Zhang, S.; Hu, J.; Zeng, D.; Zhang, G.; Dong, G.; Gao, Z.; et al. The newly identified heat-stress sensitive albino 1 gene affects chloroplast development in rice. Plant Sci. 2018, 267, 168–179. [Google Scholar] [CrossRef]
- Chen, G.; Liu, C.; Gao, Z.; Zhang, Y.; Zhu, L.; Hu, J.; Ren, D.; Xu, G.; Qian, Q. Driving the expression of RAA1 with a drought-responsive promoter enhances root growth in rice, its accumulation of potassium and its tolerance to moisture stress. Environ. Exp. Bot. 2018, 147, 147–156. [Google Scholar] [CrossRef]
- Bao, A.L.; Chen, H.F.; Chen, L.M.; Chen, S.L.; Hao, Q.N.; Guo, W.; Qiu, D.; Shan, Z.; Yang, Z.; Yuan, S.; et al. CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 2019, 19, 131. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Qin, L.; Liu, X.; He, Y.; Wang, J.; Chen, L.; Lu, G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep. 2016, 6, 24765. [Google Scholar] [CrossRef]
- Liu, W.S.; Zhu, X.H.; Lei, M.G.; Xia, Q.Y.; Botella, J.R.; Zhu, J.K.; Mao, Y. A detailed procedure for CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana. Sci. Bull. 2015, 60, 1332–1347. [Google Scholar] [CrossRef]
- Cui, Y.; Zhu, M.; Xu, Z.; Xu, Q. Assessment of the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system. Appl. Genet. 2019, 132, 1887–1896. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.M.; Ye, X.; Guo, R.M.; Huang, J.; Wang, W.; Tang, J.Y.; Tan, L.; Zhu, J.K.; Chu, C.; Qian, Y. Genome-wide Targeted Mutagenesis in Rice Using the CRISPR/Cas9 System. Mol. Plant 2017, 10, 1242–1245. [Google Scholar] [CrossRef]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 2015, 5, 10342. [Google Scholar] [CrossRef]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, J.L. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef]
- Hu, B.; Li, D.; Liu, X.; Qi, J.; Gao, D.; Zhao, S.; Huang, S.; Sun, J.; Yang, L. Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol. Plant 2017, 10, 1575–1578. [Google Scholar] [CrossRef]
- Kirchner, T.W.; Niehaus, M.; Debener, T.; Schenk, M.K.; Herde, M. Efficient generation of mutations mediated by CRISPR/Cas9 in the hairy root transformation system of Brassica carinata. PLoS ONE 2017, 12, e0185429. [Google Scholar] [CrossRef]
- Illouz-Eliaz, N.; Ramon, U.; Shohat, H.; Blum, S.; Livne, S.; Mendelson, D. Multiple gibberellin receptors contribute to phenotypic stability under changing environments. Plant Cell 2019, 31, 1506–1519. [Google Scholar] [CrossRef]
- Lu, H.P.; Liu, S.M.; Xu, S.L.; Chen, W.Y.; Zhou, X.; Tan, Y.Y.; Huang, J.Z.; Shu, Q.Y. CRISPR-S: An active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol. J. 2017, 15, 1371–1373. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.F.; Li, H.; Qin, R.Y.; Li, J.; Qiu, C.H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 2015, 5, 11491. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Zhang, H.; Xu, N.; Zhang, B.; Gou, F.; Zhu, J.K. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 2013, 6, 2008–2011. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef]
- Yan, L.; Wei, S.; Wu, Y.; Hu, R.; Li, H.; Yang, W.; Xie, Q. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 2015, 8, 1820–1823. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhang, H.M.; Lou, D.J.; Yu, D.Q. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci. Rep. 2016, 6, 21451. [Google Scholar] [CrossRef]
- Liu, W.; Xie, X.; Ma, X.; Li, J.; Chen, J.; Liu, Y.G. DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant 2015, 8, 1431–1433. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Zhang, R.; Gao, J.; Qi, Y.; Song, G.; Li, W.; Li, Y.; Li, G. Efficient multiplex genome editing by CRISPR/Cas9 in common wheat. Plant Biotechnol. J. 2021, 19, 427. [Google Scholar] [CrossRef]
- Huang, C.; Sun, H.; Xu, D.; Chen, Q.; Liang, Y.; Wang, X.; Xu, G.; Tian, J.; Wang, C.; Li, D.; et al. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA 2017, 115, E334–E341. [Google Scholar] [CrossRef]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigenerational analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Zhou, Y.; Han, X.; Shen, C.; Wang, S.; Liu, C.; Yin, W.; Xia, X. The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 2020, 71, 1969–1984. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norvilleand, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted mutagenesis, precise gene editing and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015, 2, 931–945. [Google Scholar] [CrossRef]
- Xie, K.; Zhang, J.; Yang, Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR–Cas9-mediated genome editing in model plants and major crops. Mol. Plant 2014, 7, 923–926. [Google Scholar] [CrossRef]
- Osakabe, Y.; Watanabe, T.; Sugano, S.S.; Ueta, R.; Ishihara, R.; Shinozaki, K.; Osakabe, K. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 2016, 6, 26685. [Google Scholar] [CrossRef]
- Schiml, S.; Fauser, F.; Puchta, H. The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 2014, 80, 1139–1150. [Google Scholar] [CrossRef]
- Weber, E.; Engler, C.; Gruetzner, R.; Werner, S.; Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 2011, 6, e16765. [Google Scholar] [CrossRef]
- Engler, C.; Youles, M.; Gruetzner, R.; Ehnert, T.M.; Werner, S.; Jones, J.D.; Patron, N.J.; Marillonnet, S. A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 2014, 3, 839–843. [Google Scholar] [CrossRef]
- Čermák, T.; Baltes, N.J.; Cegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef]
- Tang, X.; Zheng, X.; Qi, Y.; Zhang, D.; Cheng, Y.; Tang, A.; Voytas, D.F.; Zhang, Y. Single Transcript CRISPR-Cas9 System for Efficient Genome Editing in Plants. Mol. Plant 2016, 9, 1088–1091. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.H.; Zhang, Y.; You, Q.; Tang, X.; Ren, Q.; Liu, S.; Yang, L.; Wang, Y.; Liu, X.; Liu, B.; et al. Plant Genome Editing Using FnCpf1 and LbCpf1 Nucleases at Redefined and Altered PAM Sites. Mol. Plant 2018, 11, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wu, N.; Zhang, L.Y.; Ahammed, G.J.; Chen, X.X.; Xiang, X.; Zhou, J.; Xia, X.; Shi, K.; Yu, J.; et al. Light signaling-dependent regulation of photoinhibition and photoprotection in tomato. Plant Physiol. 2018, 176, 1311–1326. [Google Scholar] [CrossRef]
- Ren, C.; Liu, X.; Zhang, Z.; Wang, Y.; Duan, W.; Li, S.; Liang, Z. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 2016, 6, 32289. [Google Scholar] [CrossRef]
- Yang, H.; Wu, J.J.; Tang, T.; Liu, K.D.; Dai, C. CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 2017, 7, 7489. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Liu, T.; Li, C.; Jiao, B.; Li, S.; Hou, Y.; Luo, K. Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 2015, 5, 12217. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, L.; Su, T.; Wang, Q.; Gao, Y.; Zhang, S.; Jia, Q.; Yu, G.; Fu, Y.; Cheng, Q.; et al. Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ. 2020, 43, 637–648. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.G. CRISPR/Cas9-based multiplex genome editing in monocot and dicot plants. Curr. Protoc. Mol. Biol. 2016, 115, 31–36. [Google Scholar] [CrossRef]
- Tang, G.; Yan, J.; Gu, Y.; Qiao, M.; Fan, R.; Mao, Y.; Tang, X. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 2012, 58, 118–125. [Google Scholar] [CrossRef]
- Xu, R.; Wei, P.; Yang, J. Use of CRISPR/Cas Genome Editing Technology for Targeted Mutagenesis in Rice. In In Vitro Mutagenesis; Reeves, A., Ed.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2016; Volume 1498. [Google Scholar] [CrossRef]
- Liu, Z.; Jia, Y.; Ding, Y.; Shi, Y.; Li, Z.; Guo, Y.; Gong, F.; Yang, S. Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell. 2017, 66, 117–128.e5. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhao, F.; Li, Q.; Shen, S.; Li, Z.; Teng, J.; Sheng, W.; Zhang, A.; Xue, J. High efficiency Agrobacterium-mediated transformation of Pinellia ternata using petiole explants from submerged cultures. Biologia 2015, 70, 1351–1358. [Google Scholar] [CrossRef]
- Zhang, Z.; Mao, Y.; Ha, S.; Liu, W.; Botella, J.R.; Zhu, J.K. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2015, 35, 1519–1533. [Google Scholar] [CrossRef] [PubMed]




| Article | Culture | Objective of the Review |
|---|---|---|
| [49] | Cultures in general | Applications of CRISPR/Cas9-mediated gene editing to produce plants grown under stressful environmental conditions. |
| [50] | Cultures in general | Development of crops with high yields and tolerance to abiotic stresses. |
| [51] | Rice | CRISPR/Cas9 to develop heat-tolerant rice and tolerance to water deficits and floods. |
| [33] | Cultures in general | CRISPR/Cas9 for tolerance to abiotic stress. |
| [19] | Cultures in general | Comprehensive overview of CRISPR/Cas technology to improve tolerance to abiotic stress. |
| [52] | Cultures in general | CRISPR/Cas9 and ERFs * used for tolerance to abiotic stress. |
| [31] | Rice | Rice generated to be capable of sustaining growth under conditions of high salinity, using CRISPR/Cas. |
| [53] | Cultures in general | Application of CRISPR/Cas technology for the elimination/deactivation of genes associated with abiotic stresses. |
| [12] | Cultures in general | Biotic and abiotic factors. |
| [32] | Cultures in general | Application of CRISPR/Cas for tolerance to abiotic stress. |
| [54] | Cultures in general | CRISPR/Cas9 to understand tolerance to abiotic stress. |
| [55] | Cultures in general | Tolerance to water stress. |
| [56] | Arabidopsis | Functional role of CNGC19 and CNGC20 * in Arabidopsis using CRISPR/Cas9. |
| [57] | Cultures in general | Genome editing approaches based on CRISPR/Cas that have been used in plants for tolerance to abiotic stress. |
| [8] | Cultures in general | CRISPR/Cas approaches and their efficiency to improve plant growth and responses to abiotic stress. |
| [58] | Cultures in general | Applications of omics and CRISPR/Cas9 for the development of stress-tolerant cultures. |
| [59] | Cultures in general | Genome editing based on CRISPR/Cas9 in targeting HyPRPs* for tolerance to multiple stresses. |
| [60] | Cultures in general | Tolerance to drought, yield, and domestication. |
| [61] | Cultures in general | Abiotic and biotic factors. |
| [30] | Banana | Recent and prospective advances in the application of genetic modification and genome editing for the development of bananas resistant to high temperatures and water deficits. |
| [62] | Cultures in general | Evaluation of available tools and target genes to obtain plants with greater tolerances to abiotic stresses. |
| [29] | Cultivation plants | Production of multiple stress-tolerant crops using CRISPR/Cas9. |
| Questions |
|---|
| Q1. Which cultures have been edited using the CRISPR/Cas technique? |
| Q2. Which genes have been edited using the CRISPR/Cas technique? |
| Q3. What metabolic pathways are reported in studies with CRISPR/Cas? |
| Q4. Which countries or continents most widely use the CRISPR/Cas technique for tolerance to abiotic stresses? |
| Q5. Which enzymes other than Cas9 are used in CRISPR? |
| Q6. What protocols are proposed for editing with CRISPR/Cas? |
| Q7. Which explants are most used for gene editing with CRISPR/Cas? |
| Q8. What type of vector and bacteria are most reported as being used with CRISPR/Cas? |
| Q9. What methods are used to confirm the efficiency of the CRISPR/Cas technique? |
| Q10. Which abiotic factors is CRISPR/Cas used to modify? |
| Q11. What auxiliary methods to CRISPR/Cas for tolerance to abiotic stresses are used? |
| Description | Abbreviation | Components of the Question |
|---|---|---|
| Population | P | Agricultural crops with abiotic stresses. |
| Interest/Intervention | I | Gene editing based on CRISPR/Cas technology for plant breeding. |
| Comparison | C | Methods of plant breeding that do not include editing genes with CRISPR/Cas. |
| Outcome | O | Editing genes that confer tolerance to abiotic stresses in plants. |
| Study type | S | Scientific articles and literature reviews. |
| Database | Keyword Variations |
|---|---|
| Google Scholar | (“abiotic factors” OR “water deficit” OR “drought tolerance” OR “salinity tolerance” OR “cold tolerance” OR “heat tolerance”) AND (“CRISPR/Cas9” OR CRISPR-Cas9 OR “CRISPR-Cas in plants”). |
| Springer | |
| CAPES Journal Portal | |
| CABI Direct | |
| Web of Science | (crop OR crops OR plant OR plants OR seed OR seeds OR Arabidopsis OR Tobacco OR Nicotiana OR “zea mays” OR maize OR wheat OR Triticum OR barley OR hordeum OR rice OR oryza OR soybean OR “Glycine max” OR potato OR Solanum OR “sweet potato” OR “Ipomoea batatas” OR “sugar beet” OR “sugar-beet” OR “fodder beet” OR “beta vulgaris” OR tomato OR cucumber OR cucumis OR onion OR allium OR apple OR apples OR malus OR orange OR “Citrus sinensis” OR banana OR musa OR manihot OR cassava OR “Manihot esculenta” OR sugarcane OR “Saccharum officinarum” OR cotton OR “Gossypium hirsutum” “oil palm” OR “Elaeis guineensis” OR watermelon OR Citrullus) AND (“genome edit*” OR “genome-edit*” OR “genome editing in plants” OR CRISPR/Cas9 OR CRISPR-Cas9 OR “CRISPR/Cas9-targeted mutagenesis” OR “targeted mutagenesis” OR “genome editing technology”) AND (“abiotic factors” OR “Abiotic stress” OR “water deficit” OR “drought tolerance” OR “salinity tolerance” OR “cold tolerance” OR “heat tolerance”) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nascimento, F.d.S.; Rocha, A.d.J.; Soares, J.M.d.S.; Mascarenhas, M.S.; Ferreira, M.d.S.; Morais Lino, L.S.; Ramos, A.P.d.S.; Diniz, L.E.C.; Mendes, T.A.d.O.; Ferreira, C.F.; et al. Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review. Plants 2023, 12, 305. https://doi.org/10.3390/plants12020305
Nascimento FdS, Rocha AdJ, Soares JMdS, Mascarenhas MS, Ferreira MdS, Morais Lino LS, Ramos APdS, Diniz LEC, Mendes TAdO, Ferreira CF, et al. Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review. Plants. 2023; 12(2):305. https://doi.org/10.3390/plants12020305
Chicago/Turabian StyleNascimento, Fernanda dos Santos, Anelita de Jesus Rocha, Julianna Matos da Silva Soares, Marcelly Santana Mascarenhas, Mileide dos Santos Ferreira, Lucymeire Souza Morais Lino, Andresa Priscila de Souza Ramos, Leandro Eugenio Cardamone Diniz, Tiago Antônio de Oliveira Mendes, Claudia Fortes Ferreira, and et al. 2023. "Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review" Plants 12, no. 2: 305. https://doi.org/10.3390/plants12020305
APA StyleNascimento, F. d. S., Rocha, A. d. J., Soares, J. M. d. S., Mascarenhas, M. S., Ferreira, M. d. S., Morais Lino, L. S., Ramos, A. P. d. S., Diniz, L. E. C., Mendes, T. A. d. O., Ferreira, C. F., Santos-Serejo, J. A. d., & Amorim, E. P. (2023). Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review. Plants, 12(2), 305. https://doi.org/10.3390/plants12020305








