Floral Development on Vitis vinifera Is Associated with MADS-Box Transcription Factors through the Transcriptional Regulation of VviZIP3
Abstract
:1. Introduction
2. Results
2.1. Identification of MADS-Type Response Elements in VviZIP3 Promoter
2.2. Isolation of Promoter Region of VviZIP3 in Carmenérè
2.3. The MADS AGAMOUS and APETALA1 Proteins Belonging to V. vinifera Are Similar to Their Orthologs in A. thaliana
2.4. Functionality of CArG Boxes Present in VviZIP3 Promoter
2.5. The Functionality of CArG Boxes Present in VviZIP3 Promoter Is Carried out by MADS Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Purification of Genomic DNA from V. vinifera cv. Carménère
4.3. In Silico Analysis of the Cis-Regulatory Regions of the VviZIP3 Promoter Region
4.4. VviZIP3 Promoter Region Isolation
4.5. Construction of VviZIP3 Reporter Gene-Promoter Fusions
4.6. Transformation of PZIP3 Fragments in A. thaliana
4.7. Histochemical GUS Staining
4.8. Analysis of Reporter Gene Expression via Quantitative RT-PCR
4.9. VviZIP3 Promoter Site-Directed Deletions
4.10. Analysis of Reporter Gene Expression in Fragments Obtained from Site-Directed Deletion
4.11. Transformation of Mutant Lines of A. thaliana
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barrett, A.J. Enzyme Nomenclature. Recommendations 1992. Eur. J. Biochem. 1995, 232, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.A.T.; Kyriacou, B.; Callahan, D.L.; Carruthers, L.; Stangoulis, J.; Lombi, E.; Tester, M. Constitutive Overexpression of the OsNAS Gene Family Reveals Single-Gene Strategies for Effective Iron- and Zinc-Biofortification of Rice Endosperm. PLoS ONE 2011, 6, e24476. [Google Scholar] [CrossRef]
- Lee, I.; Seo, Y.-S.; Coltrane, D.; Hwang, S.; Oh, T.; Marcotte, E.M.; Ronald, P.C. Genetic Dissection of the Biotic Stress Response Using a Genome-Scale Gene Network for Rice. Proc. Natl. Acad. Sci. USA 2011, 108, 18548–18553. [Google Scholar] [CrossRef]
- Cakmak, I. Enrichment of Cereal Grains with Zinc: Agronomic or Genetic Biofortification? Plant Soil 2008, 302, 1–17. [Google Scholar] [CrossRef]
- Alloway, B.J. Soil Factors Associated with Zinc Deficiency in Crops and Humans. Environ. Geochem. Health 2009, 31, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Hacisalihoglu, G.; Hart, J.J.; Wang, Y.-H.; Cakmak, I.; Kochian, L.V. Zinc Efficiency Is Correlated with Enhanced Expression and Activity of Zinc-Requiring Enzymes in Wheat. Plant Physiol. 2003, 131, 595–602. [Google Scholar] [CrossRef]
- Marschner, H. Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Academic Press: London, UK, 2011; ISBN 978-0-12-384905-2. [Google Scholar]
- Jackson, R. Wine Science. In Principles and Applications, 3rd ed.; Elsevier; Academic Press: Amsterdam, The Netherlands, 2008; ISBN 978-0-12-373646-8. [Google Scholar]
- Alva, O.; Roa-Roco, R.N.; Pérez-Díaz, R.; Yáñez, M.; Tapia, J.; Moreno, Y.; Ruiz-Lara, S.; González, E. Pollen Morphology and Boron Concentration in Floral Tissues as Factors Triggering Natural and GA-Induced Parthenocarpic Fruit Development in Grapevine. PLoS ONE 2015, 10, e0139503. [Google Scholar] [CrossRef]
- Eide, D.J. Homeostatic and Adaptive Responses to Zinc Deficiency in Saccharomyces Cerevisiae. J. Biol. Chem. 2009, 284, 18565–18569. [Google Scholar] [CrossRef]
- Ajeesh Krishna, T.P.; Antony Ceasar, S.; Maharajan, T.; Ramakrishnan, M.; Duraipandiyan, V.; Al-Dhabi, N.A.; Ignacimuthu, S. Improving the Zinc-Use Efficiency in Plants: A Review. Sabrao J. Breed. Genet. 2017, 49, 211–230. [Google Scholar]
- Sutherland, M.W. The Generation of Oxygen Radicals during Host Plant Responses to Infection. Physiol. Mol. Plant Pathol. 1991, 39, 71–93. [Google Scholar] [CrossRef]
- Lehman, M.C.; Pahls, D.R.; Meredith, J.M.; Sommer, R.D.; Heinekey, D.M.; Cundari, T.R.; Ison, E.A. Oxyfunctionalization with Cp∗IrIII(NHC)(Me)(Cl) with O2: Identification of a Rare Bimetallic IrIV μ-Oxo Intermediate. J. Am. Chem. Soc. 2015, 137, 3574–3584. [Google Scholar] [CrossRef] [PubMed]
- Marschner, H. The Soil-Root Interface (Rhizosphere) in Relation to Mineral Nutrition. In Mineral Nutrition of Higher Plants; Academic Press: London, UK, 1995; pp. 537–595. ISBN 978-0-12-473542-2. [Google Scholar]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-Type ATPase Heavy Metal Transporters with Roles in Essential Zinc Homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef]
- Broadley, M.R.; White, P.J.; Hammond, J.P.; Zelko, I.; Lux, A. Zinc in Plants. New Phytol. 2007, 173, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Cakmak, I. Tansley Review No. 111: Possible Roles of Zinc in Protecting Plant Cells from Damage by Reactive Oxygen Species. New Phytol. 2000, 146, 185–205. [Google Scholar] [CrossRef]
- Cabot, C.; Martos, S.; Llugany, M.; Gallego, B.; Tolrà, R.; Poschenrieder, C. A Role for Zinc in Plant Defense Against Pathogens and Herbivores. Front Plant Sci. 2019, 10, 1171. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Zhang, Y.; Wang, W.; Irish, V.F.; Huang, T. RABBIT EARS Regulates the Transcription of TCP4 during Petal Development in Arabidopsis. J. Exp. Bot. 2016, 67, 6473–6480. [Google Scholar] [CrossRef]
- Payne, T.; Johnson, S.D.; Koltunow, A.M. KNUCKLES (KNU) Encodes a C2H2 Zinc-Finger Protein That Regulates Development of Basal Pattern Elements of the Arabidopsis gynoecium. Development 2004, 131, 3737–3749. [Google Scholar] [CrossRef]
- Pedas, P.; Husted, S. Zinc Transport Mediated by Barley ZIP Proteins Are Induced by Low PH. Plant Signal Behav. 2009, 4, 842–845. [Google Scholar] [CrossRef]
- Guerinot, M.L. The ZIP Family of Metal Transporters. Biochim. Biophys. Acta (BBA) Biomembr. 2000, 1465, 190–198. [Google Scholar] [CrossRef]
- Palmer, C.; Guerinot, M.L. A Question of Balance: Facing the Challenges of Cu, Fe and Zn Homeostasis. Nat. Chem. Biol. 2009, 5, 333–340. [Google Scholar] [CrossRef]
- Ricachenevsky, F.K.; Menguer, P.K.; Sperotto, R.A.; Fett, J.P. Got to Hide Your Zn Away: Molecular Control of Zn Accumulation and Biotechnological Applications. Plant Sci. 2015, 236, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Grotz, N.; Guerinot, M.L. Molecular Aspects of Cu, Fe and Zn Homeostasis in Plants. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2006, 1763, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Astudillo, C.; Fernandez, A.; Blair, M.; Cichy, K. The Phaseolus Vulgaris ZIP Gene Family: Identification, Characterization, Mapping, and Gene Expression. Front. Plant Sci. 2013, 4, 286. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.-Z.; Zhou, X.; Xing, F.; Ling, L.-L.; Chun, C.-P.; Cao, L.; Aarts, M.G.M.; Peng, L.-Z. Genome-Wide Identification, Cloning and Functional Analysis of the Zinc/Iron-Regulated Transporter-Like Protein (ZIP) Gene Family in Trifoliate Orange (Poncirus trifoliata L. Raf.). Front. Plant Sci. 2017, 8, 588. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.R.; Feng, Y.; Chao, Y.E. Genomic Analysis and Expression Pattern of OsZIP1, OsZIP3, and OsZIP4 in Two Rice (Oryza sativa L.) Genotypes with Different Zinc Efficiency. Russ. J. Plant Physiol. 2008, 55, 400–409. [Google Scholar] [CrossRef]
- Evens, N.P.; Buchner, P.; Williams, L.E.; Hawkesford, M.J. The Role of ZIP Transporters and Group F BZIP Transcription Factors in the Zn-deficiency Response of Wheat (Triticum aestivum). Plant J. 2017, 92, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.K.; Ganie, S.A.; Rana, M.K.; Sharma, T.R. Genome-Wide Analysis of Zinc Transporter Genes of Maize (Zea mays). Plant. Mol. Biol. Rep. 2014, 32, 605–616. [Google Scholar] [CrossRef]
- Migeon, A.; Blaudez, D.; Wilkins, O.; Montanini, B.; Campbell, M.M.; Richaud, P.; Thomine, S.; Chalot, M. Genome-Wide Analysis of Plant Metal Transporters, with an Emphasis on Poplar. Cell Mol. Life Sci. 2010, 67, 3763–3784. [Google Scholar] [CrossRef]
- Gainza-Cortés, F.; Pérez-Dïaz, R.; Pérez-Castro, R.; Tapia, J.; Casaretto, J.A.; González, S.; Peña-Cortés, H.; Ruiz-Lara, S.; González, E. Characterization of a Putative Grapevine Zn Transporter, VvZIP3, Suggests Its Involvement in Early Reproductive Development in Vitis vinifera L. BMC Plant Biol. 2012, 12, 111. [Google Scholar] [CrossRef]
- Grimplet, J.; Adam-Blondon, A.-F.; Bert, P.-F.; Bitz, O.; Cantu, D.; Davies, C.; Delrot, S.; Pezzotti, M.; Rombauts, S.; Cramer, G.R. The Grapevine Gene Nomenclature System. BMC Genom. 2014, 15, 1077. [Google Scholar] [CrossRef]
- Mullins, M.G.; Bouquet, A.; Williams, L.E. Biology of the Grapevine (The Biology of Horticultural Crops); Cambridge University Press: Cambridge, UK, 1992; ISBN 978-0-521-03867-6. [Google Scholar]
- Boss, P.K.; Vivier, M.; Matsumoto, S.; Dry, I.B.; Thomas, M.R. A CDNA from Grapevine (Vitis vinifera L.), Which Shows Homology to AGAMOUS and SHATTERPROOF, Is Not Only Expressed in Flowers but Also throughout Berry Development. Plant Mol. Biol. 2001, 45, 541–553. [Google Scholar] [CrossRef]
- Boss, P.; Sensi, E.; Hua, C.; Davies, C.; Thomas, M. Cloning and Characterisation of Grapevine (Vitis vinifera L.) MADS-Box Genes Expressed during Inflorescence and Berry Development. Plant Sci. 2002, 162, 887–895. [Google Scholar] [CrossRef]
- Calonje, M.; Cubas, P.; Martínez-Zapater, J.M.; Carmona, M.J. Floral Meristem Identity Genes Are Expressed during Tendril Development in Grapevine. Plant Physiol. 2004, 135, 1491–1501. [Google Scholar] [CrossRef] [PubMed]
- Sandelin, A.; Alkema, W.; Engström, P.; Wasserman, W.W.; Lenhard, B. JASPAR: An Open-Access Database for Eukaryotic Transcription Factor Binding Profiles. Nucleic Acids Res. 2004, 32, D91–D94. [Google Scholar] [CrossRef]
- Sandelin, A.; Wasserman, W.W.; Lenhard, B. ConSite: Web-Based Prediction of Regulatory Elements Using Cross-Species Comparison. Nucleic Acids Res. 2004, 32, W249–W252. [Google Scholar] [CrossRef]
- Shore, P.; Sharrocks, A.D. The MADS-Box Family of Transcription Factors. Eur. J. Biochem. 1995, 229, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Riquelme, J.; Lijavetzky, D.; Martinez-Zapater, J.M.; Carmona, M.J. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiol. 2009, 149, 354–369. [Google Scholar] [CrossRef]
- Pope, S.D.; Medzhitov, R. Emerging Principles of Gene Expression Programs and Their Regulation. Mol. Cell 2018, 71, 389–397. [Google Scholar] [CrossRef]
- Li, X.; MacArthur, S.; Bourgon, R.; Nix, D.; Pollard, D.A.; Iyer, V.N.; Hechmer, A.; Simirenko, L.; Stapleton, M.; Hendriks, C.L.L.; et al. Transcription Factors Bind Thousands of Active and Inactive Regions in the Drosophila blastoderm. PLoS Biol. 2008, 6, e27. [Google Scholar] [CrossRef]
- Georges, A.B.; Benayoun, B.A.; Caburet, S.; Veitia, R.A. Generic Binding Sites, Generic DNA-Binding Domains: Where Does Specific Promoter Recognition Come From? The FASEB J. 2010, 24, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Tsai, C.-J.; Ma, B.; Nussinov, R. Mechanisms of Transcription Factor Selectivity. Trends Genet. 2010, 26, 75–83. [Google Scholar] [CrossRef]
- Merika, M.; Thanos, D. Enhanceosomes. Curr. Opin. Genet. Dev. 2001, 11, 205–208. [Google Scholar] [CrossRef]
- Fox, T.C.; Guerinot, M.L. Molecular Biology of Cation Transport in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 669–696. [Google Scholar] [CrossRef] [PubMed]
- Ricachenevsky, F.; Menguer, P.; Sperotto, R.; Williams, L.; Fett, J. Roles of Plant Metal Tolerance Proteins (MTP) in Metal Storage and Potential Use in Biofortification Strategies. Front. Plant Sci. 2013, 4, 144. [Google Scholar] [CrossRef]
- Williams, L.E.; Mills, R.F. P1B-ATPases—An Ancient Family of Transition Metal Pumps with Diverse Functions in Plants. Trends Plant Sci. 2005, 10, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Verret, F.; Gravot, A.; Auroy, P.; Leonhardt, N.; David, P.; Nussaume, L.; Vavasseur, A.; Richaud, P. Overexpression of AtHMA4 Enhances Root-to-Shoot Translocation of Zinc and Cadmium and Plant Metal Tolerance. FEBS Lett. 2004, 576, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Shigaki, T.; Rees, I.; Nakhleh, L.; Hirschi, K.D. Identification of Three Distinct Phylogenetic Groups of CAX Cation/Proton Antiporters. J. Mol. Evol. 2006, 63, 815–825. [Google Scholar] [CrossRef]
- Socha, A.; Guerinot, M.L. Mn-Euvering Manganese: The Role of Transporter Gene Family Members in Manganese Uptake and Mobilization in Plants. Front. Plant Sci. 2014, 5, 106. [Google Scholar] [CrossRef] [PubMed]
- Song, W.-Y.; Choi, K.S.; Kim, D.Y.; Geisler, M.; Park, J.; Vincenzetti, V.; Schellenberg, M.; Kim, S.H.; Lim, Y.P.; Noh, E.W.; et al. Arabidopsis PCR2 Is a Zinc Exporter Involved in Both Zinc Extrusion and Long-Distance Zinc Transport. Plant Cell 2010, 22, 2237–2252. [Google Scholar] [CrossRef] [PubMed]
- Takatsuji, H. Zinc-Finger Proteins: The Classical Zinc Finger Emerges in Contemporary Plant Science. Plant Mol. Biol. 1999, 39, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Guo, Y.; Wu, C.; Yang, G.; Li, Y.; Zheng, C. Genome-Wide Analysis of CCCH Zinc Finger Family in Arabidopsis and Rice. BMC Genom. 2008, 9, 44. [Google Scholar] [CrossRef]
- González, S. Caracterización Estructural y Funcional de Genes Que Codifican Para Proteinas Asociadas a La Red Regulatoria Del Desarrollo Floral y Frutal En Vitis vinifera Cv. Carménère; Universidad de Talca: Talca, Chile, 2013; Available online: http://dspace.utalca.cl/handle/1950/11903 (accessed on 15 August 2023).
- Agarwal, P.; Arora, R.; Ray, S.; Singh, A.K.; Singh, V.P.; Takatsuji, H.; Kapoor, S.; Tyagi, A.K. Genome-Wide Identification of C2H2 Zinc-Finger Gene Family in Rice and Their Phylogeny and Expression Analysis. Plant Mol. Biol. 2007, 65, 467–485. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Weber, M.; Schmidt, H.; Rensch, S.; Trampczynska, A.; Hansen, T.H.; Husted, S.; Schjoerring, J.K.; Talke, I.N.; Krämer, U.; et al. Elevated Nicotianamine Levels in Arabidopsis Halleri Roots Play a Key Role in Zinc Hyperaccumulation. Plant Cell 2012, 24, 708–723. [Google Scholar] [CrossRef] [PubMed]
- Haydon, M.J.; Kawachi, M.; Wirtz, M.; Hillmer, S.; Hell, R.; Krämer, U. Vacuolar Nicotianamine Has Critical and Distinct Roles under Iron Deficiency and for Zinc Sequestration in Arabidopsis. Plant Cell 2012, 24, 724–737. [Google Scholar] [CrossRef]
- Hanikenne, M.; Talke, I.N.; Haydon, M.J.; Lanz, C.; Nolte, A.; Motte, P.; Kroymann, J.; Weigel, D.; Krämer, U. Evolution of Metal Hyperaccumulation Required cis-Regulatory Changes and Triplication of HMA4. Nature 2008, 453, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Aerts, N.; de Bruijn, S.; van Mourik, H.; Angenent, G.C.; van Dijk, A.D.J. Comparative Analysis of Binding Patterns of MADS-Domain Proteins in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 131. [Google Scholar] [CrossRef] [PubMed]
- Sehra, B.; Franks, R.G. Redundant CArG Box cis-Motif Activity Mediates SHATTERPROOF2 Transcriptional Regulation during Arabidopsis thaliana Gynoecium Development. Front. Plant Sci. 2017, 8, 1712. [Google Scholar] [CrossRef]
- Smaczniak, C.; Muiño, J.M.; Chen, D.; Angenent, G.C.; Kaufmann, K. Differences in DNA Binding Specificity of Floral Homeotic Protein Complexes Predict Organ-Specific Target Genes. Plant Cell 2017, 29, 1822–1835. [Google Scholar] [CrossRef]
- Ó’Maoiléidigh, D.S.; Wuest, S.E.; Rae, L.; Raganelli, A.; Ryan, P.T.; Kwaśniewska, K.; Das, P.; Lohan, A.J.; Loftus, B.; Graciet, E.; et al. Control of Reproductive Floral Organ Identity Specification in Arabidopsis by the C Function Regulator AGAMOUS. Plant Cell 2013, 25, 2482–2503. [Google Scholar] [CrossRef]
- Gustafson-Brown, C.; Savidge, B.; Yanofsky, M.F. Regulation of the Arabidopsis Floral Homeotic Gene APETALA1. Cell 1994, 76, 131–143. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, Y.; Li, Z.; Song, C.; Wang, X. Advancements in Plant Regeneration and Genetic Transformation of Grapevine (Vitis Spp.). J. Integr. Agric. 2021, 20, 1407–1434. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral Dip: A Simplified Method for Agrobacterium-Mediated Transformation of Arabidopsis Thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Logemann, E.; Birkenbihl, R.P.; Ülker, B.; Somssich, I.E. An Improved Method for Preparing Agrobacterium Cells That Simplifies the Arabidopsis Transformation Protocol. Plant Methods 2006, 2, 16. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.-S.; Niu, Q.-W.; Chua, N.-H. Agrobacterium-Mediated Transformation of Arabidopsis Thaliana Using the Floral Dip Method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, K.; Muiño, J.M.; Jauregui, R.; Airoldi, C.A.; Smaczniak, C.; Krajewski, P.; Angenent, G.C. Target Genes of the MADS Transcription Factor SEPALLATA3: Integration of Developmental and Hormonal Pathways in the Arabidopsis Flower. PLoS Biology 2009, 7, e1000090. [Google Scholar] [CrossRef] [PubMed]
- Immink, R.G.H.; Kaufmann, K.; Angenent, G.C. The ‘ABC’ of MADS Domain Protein Behaviour and Interactions. Semin. Cell Dev. Biol. 2010, 21, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Mena, C.; de Folter, S.; Costa, M.M.R.; Angenent, G.C.; Sablowski, R. Transcriptional Program Controlled by the Floral Homeotic Gene AGAMOUS during Early Organogenesis. Development 2005, 132, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Gregis, V.; Sessa, A.; Colombo, L.; Kater, M.M. AGAMOUS-LIKE24 and Short Vegetative Phase Determine Floral Meristem Identity in Arabidopsis. Plant J. 2008, 56, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mara, C. Regulatory Mechanisms for Floral Homeotic Gene Expression. Semin. Cell Dev. Biol. 2010, 21, 80–86. [Google Scholar] [CrossRef]
- Weigel, D.; Alvarez, J.; Smyth, D.R.; Yanofsky, M.F.; Meyerowitz, E.M. LEAFY Controls Floral Meristem Identity in Arabidopsis. Cell 1992, 69, 843–859. [Google Scholar] [CrossRef]
- Liu, C.; Xi, W.; Shen, L.; Tan, C.; Yu, H. Regulation of Floral Patterning by Flowering Time Genes. Dev. Cell 2009, 16, 711–722. [Google Scholar] [CrossRef]
- Posé, D.; Yant, L.; Schmid, M. The End of Innocence: Flowering Networks Explode in Complexity. Curr. Opin. Plant Biol. 2012, 15, 45–50. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS Fusions: Beta-Glucuronidase as a Sensitive and Versatile Gene Fusion Marker in Higher Plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Schubert, D.; Lechtenberg, B.; Forsbach, A.; Gils, M.; Bahadur, S.; Schmidt, R. Silencing in Arabidopsis T-DNA Transformants: The Predominant Role of a Gene-Specific RNA Sensing Mechanism versus Position Effects. Plant Cell 2004, 16, 2561–2572. [Google Scholar] [CrossRef] [PubMed]
- Tilly, J.J.; Allen, D.W.; Jack, T. The CArG Boxes in the Promoter of the Arabidopsis Floral Organ Identity Gene APETALA3 Mediate Diverse Regulatory Effects. Development 1998, 125, 1647–1657. [Google Scholar] [CrossRef] [PubMed]
- Meneghetti, S.; Gardiman, M.; Calò, A. Flower Biology of Grapevine. A Review. Adv. Hortic. Sci. 2006, 20, 317–325. [Google Scholar] [CrossRef]
- Krizek, B.A.; Fletcher, J.C. Molecular Mechanisms of Flower Development: An Armchair Guide. Nat. Rev. Genet. 2005, 6, 688–698. [Google Scholar] [CrossRef]
- Vasconcelos, M.C.; Greven, M.; Winefield, C.; Trought, M.; Raw, V. The Flowering Process of Vitis Vinifera: A Review. Am. J. Enol. Vitic. 2009, 60, 411–434. [Google Scholar] [CrossRef]
- Kamal, N.; Ochßner, I.; Schwandner, A.; Viehöver, P.; Hausmann, L.; Töpfer, R.; Weisshaar, B.; Holtgräwe, D. Characterization of Genes and Alleles Involved in the Control of Flowering Time in Grapevine. PLoS ONE 2019, 14, e0214703. [Google Scholar] [CrossRef]
- Joly, D.; Perrin, M.; Gertz, C.; Kronenberger, J.; Demangeat, G.; Masson, J. Expression Analysis of Flowering Genes from Seedling-Stage to Vineyard Life of Grapevine Cv. Riesling*. Plant Sci. 2004, 166, 1427–1436. [Google Scholar] [CrossRef]
- Sreekantan, L.; Thomas, M.R. VvFT and VvMADS8, the Grapevine Homologues of the Floral Integrators FT and SOC1, Have Unique Expression Patterns in Grapevine and Hasten Flowering in Arabidopsis. Funct. Plant Biol. 2006, 33, 1129–1139. [Google Scholar] [CrossRef]
- Carmona, M.J.; Cubas, P.; Calonje, M.; Martínez-Zapater, J.M. Flowering Transition in Grapevine (Vitis vinifera L.). Can. J. Bot. 2007, 85, 701–711. [Google Scholar] [CrossRef]
- Sreekantan, L.; Torregrosa, L.; Fernandez, L.; Thomas, M.R. VvMADS9, a Class B MADS-Box Gene Involved in Grapevine Flowering, Shows Different Expression Patterns in Mutants with Abnormal Petal and Stamen Structures. Funct. Plant Biol. 2006, 33, 877–886. [Google Scholar] [CrossRef]
- Poupin, M.J.; Federici, F.; Medina, C.; Matus, J.T.; Timmermann, T.; Arce-Johnson, P. Isolation of the Three Grape Sub-Lineages of B-Class MADS-Box TM6, PISTILLATA and APETALA3 Genes Which Are Differentially Expressed during Flower and Fruit Development. Gene 2007, 404, 10–24. [Google Scholar] [CrossRef]
- Hanania, U.; Velcheva, M.; Sahar, N.; Perl, A. An Improved Method for Isolating High-Quality DNA From Vitis vinifera Nuclei. Plant Mol. Biol. Rep. 2004, 22, 173–177. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-Acting Regulatory DNA Elements (PLACE) Database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and Modifications of Primer Design Program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Qingshun, L.Q. Protocol: Streamline Cloning of Genes into Binary Vectors in Agrobacterium via the Gateway® TOPO Vector System. Plant Methods 2008, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Ee, S.-F.; Khairunnisa, M.B.; Mohamed-Hussein, Z.-A.; Shaharuddin, N.; Zainal, Z. Effective Hygromycin Concentration for Selection of Agrobacterium-Mediated Transgenic Arabidopsis thaliana. Malays. Appl. Biol. J. 2014, 43, 119–123. [Google Scholar]
- Tapia, G.; Verdugo, I.; Yañez, M.; Ahumada, I.; Theoduloz, C.; Cordero, C.; Poblete, F.; González, E.; Ruiz-Lara, S. Involvement of Ethylene in Stress-Induced Expression of the TLC1.1 Retrotransposon from Lycopersicon chilense Dun. Plant Physiol. 2005, 138, 2075–2086. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, J.; Jiang, J.; Chen, S.; Guan, Z.; Liao, Y.; Chen, F. Reference Genes for Normalizing Transcription in Diploid and Tetraploid Arabidopsis. Sci. Rep. 2014, 4, 6781. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]


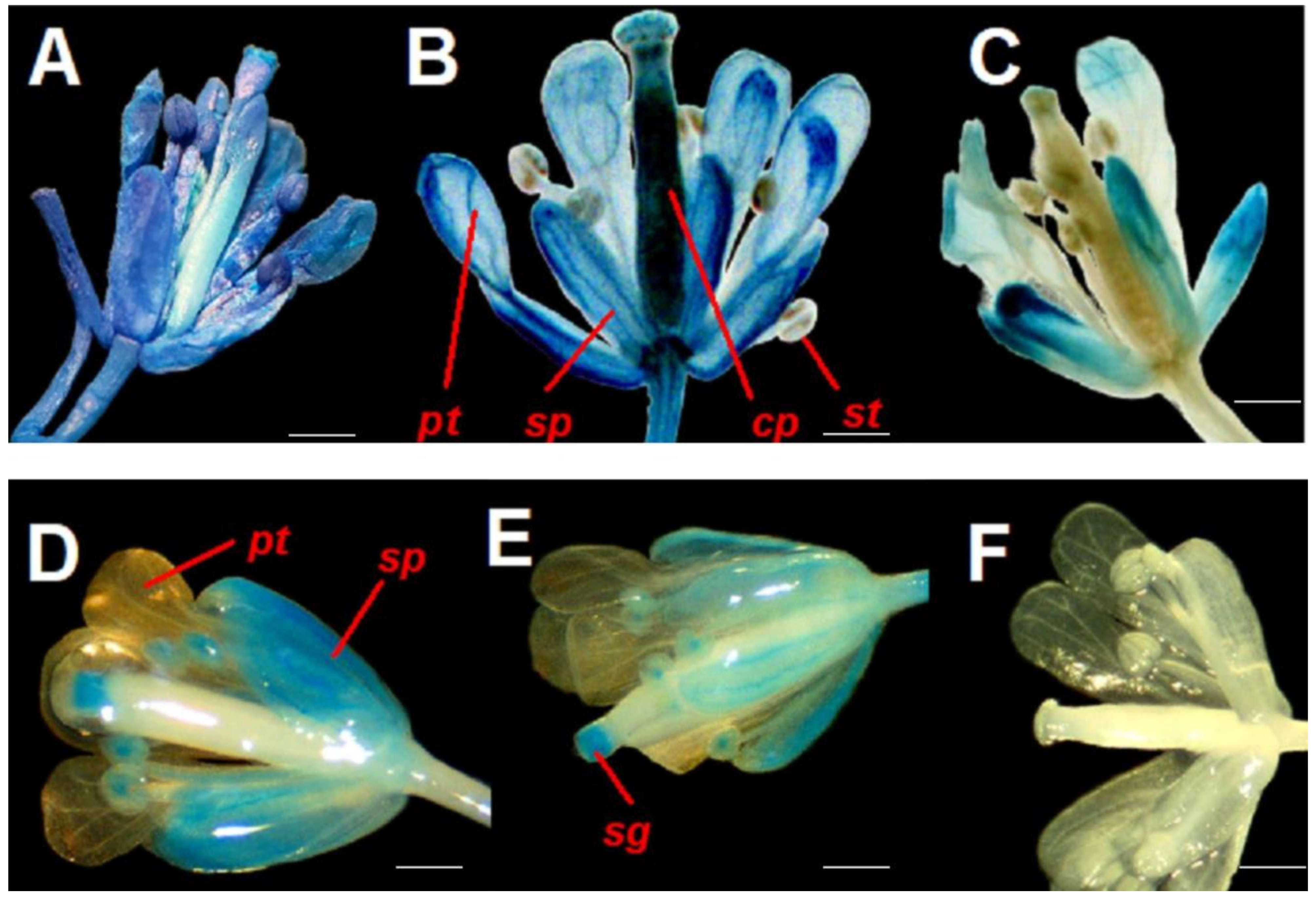

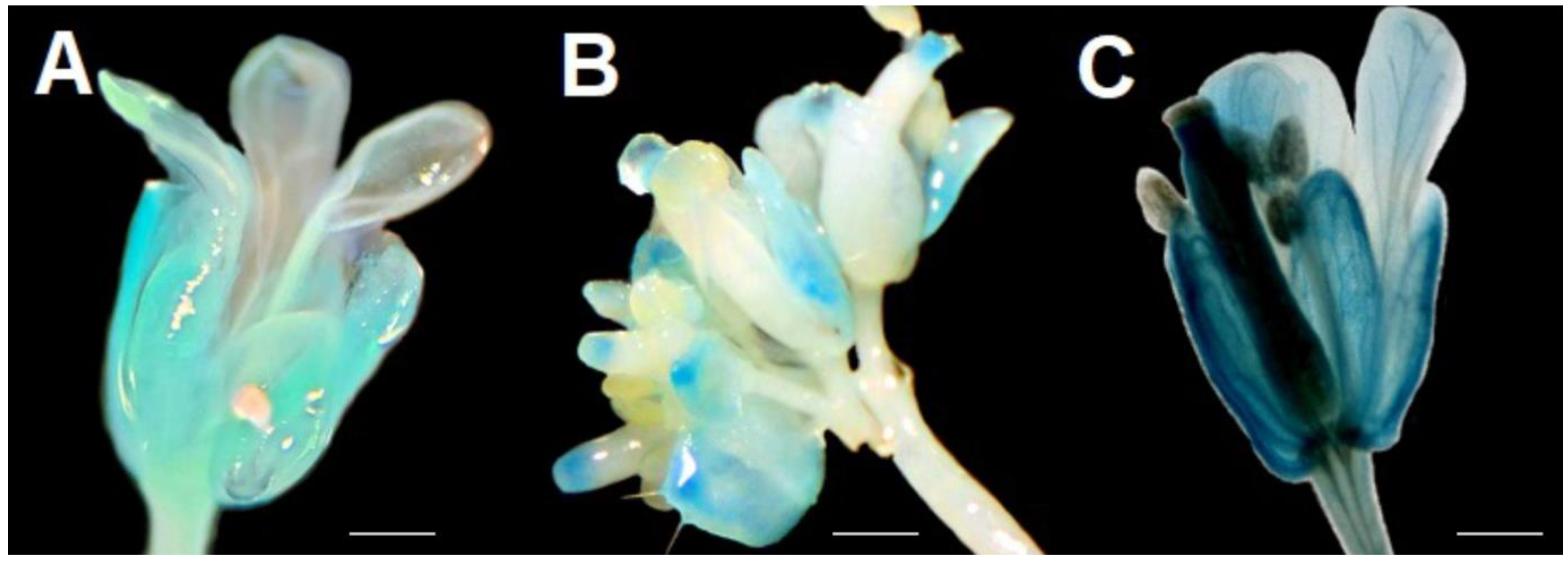
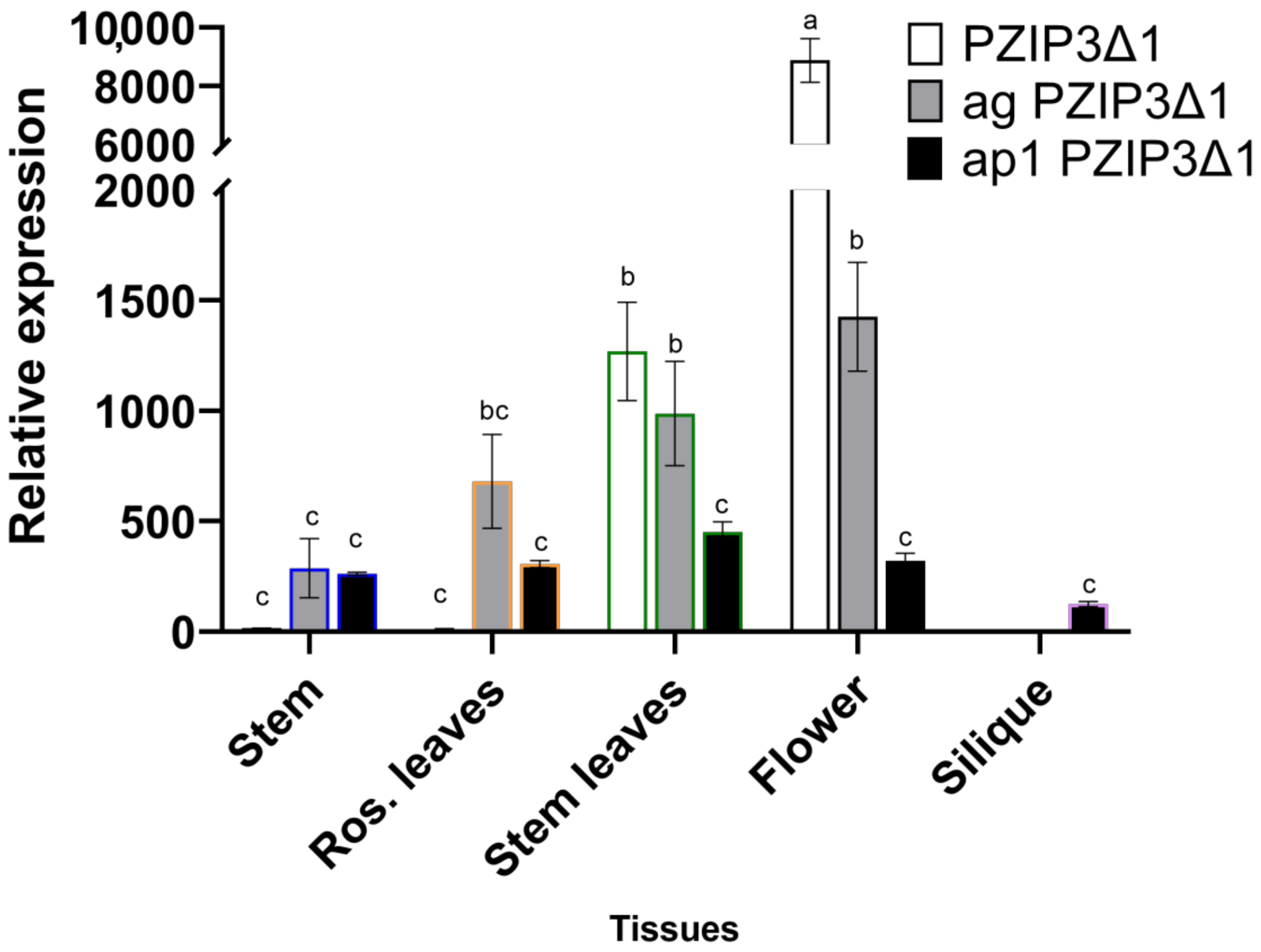

| Response Element | Transcription Factor | Predicted Sequence | Distance to Transcription of Start Site (TSS) |
|---|---|---|---|
| AG | MADS | GATTCCTCATTTGGGTT | 1115 |
| AP1 | MADS | TAAACAAAAATGGACAGCAC | 181 |
| WRKY71OS (GARE) | Repressor of the GA signaling pathway | GTAC | 346 |
| WRKY71OS (GARE) | Repressor of the GA signaling pathway | GTAC | 1521 |
| ABRELATERD1 (ABA) | ABRE (ABA-responsive element) | CACGTGC | 832 |
| ABRELATERD1 (ABA) | ABRE (ABA-responsive element) | CAAGT | 77 |
| Athb-1 | Homeodomain–Leucine Zipper I (HD-Zip I) | AATAATTG | 330 |
| Athb-1 | Homeodomain–Leucine Zipper I (HD-Zip I) | CAATTATT | 111 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saavedra Núñez, G.; González-Villanueva, E.; Ramos, P. Floral Development on Vitis vinifera Is Associated with MADS-Box Transcription Factors through the Transcriptional Regulation of VviZIP3. Plants 2023, 12, 3322. https://doi.org/10.3390/plants12183322
Saavedra Núñez G, González-Villanueva E, Ramos P. Floral Development on Vitis vinifera Is Associated with MADS-Box Transcription Factors through the Transcriptional Regulation of VviZIP3. Plants. 2023; 12(18):3322. https://doi.org/10.3390/plants12183322
Chicago/Turabian StyleSaavedra Núñez, Germán, Enrique González-Villanueva, and Patricio Ramos. 2023. "Floral Development on Vitis vinifera Is Associated with MADS-Box Transcription Factors through the Transcriptional Regulation of VviZIP3" Plants 12, no. 18: 3322. https://doi.org/10.3390/plants12183322
APA StyleSaavedra Núñez, G., González-Villanueva, E., & Ramos, P. (2023). Floral Development on Vitis vinifera Is Associated with MADS-Box Transcription Factors through the Transcriptional Regulation of VviZIP3. Plants, 12(18), 3322. https://doi.org/10.3390/plants12183322







