Antioxidant, Anti-Inflammatory and Antiproliferative Effects of Osmanthus fragrans (Thunb.) Lour. Flower Extracts
Abstract
:1. Introduction
2. Results
2.1. Total Phytochemical Analysis
2.2. Antioxidant Activity
2.3. Anti-Inflammatory Activities
2.4. Anti-Prostate Cancer Activity
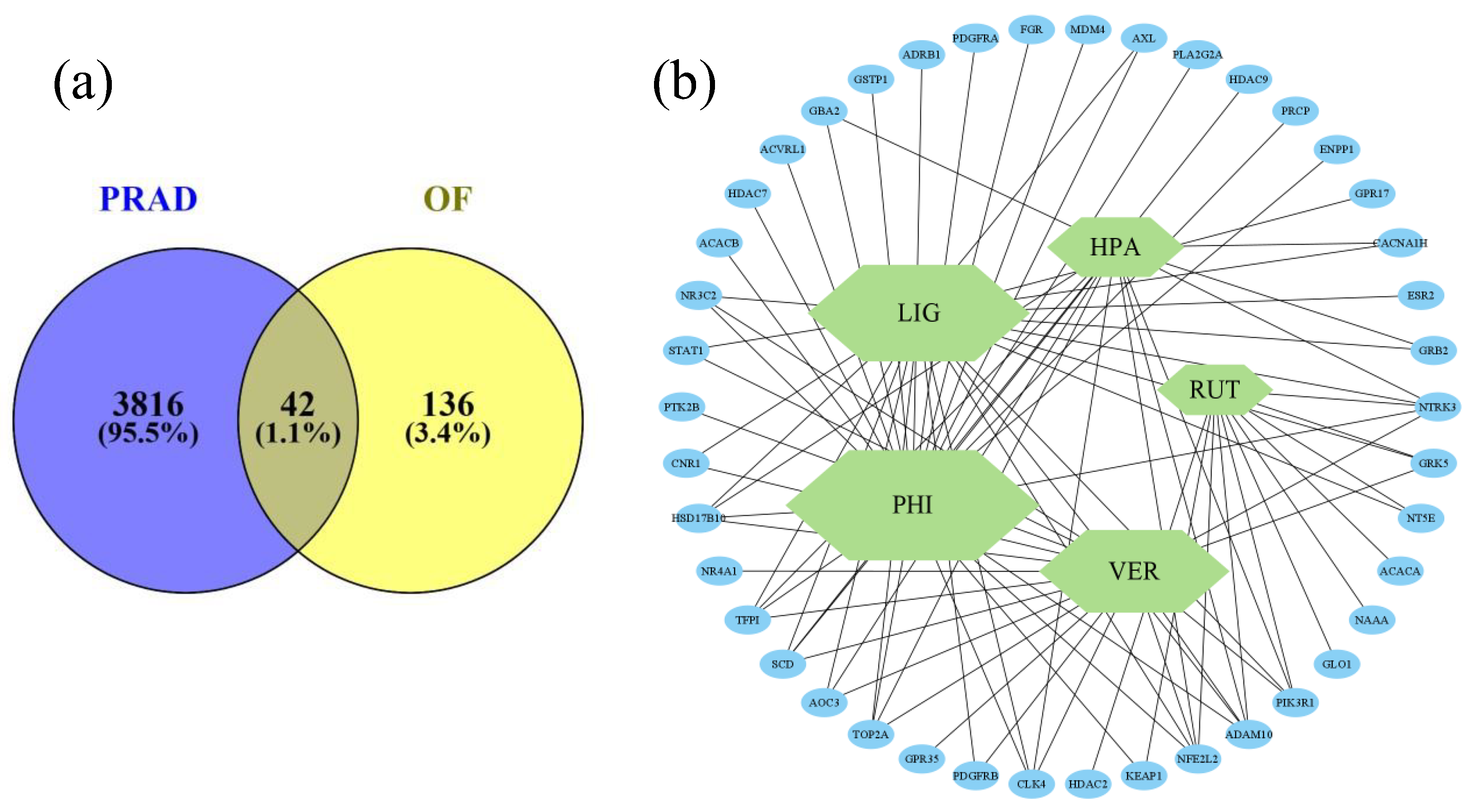
2.5. Network Pharmacology
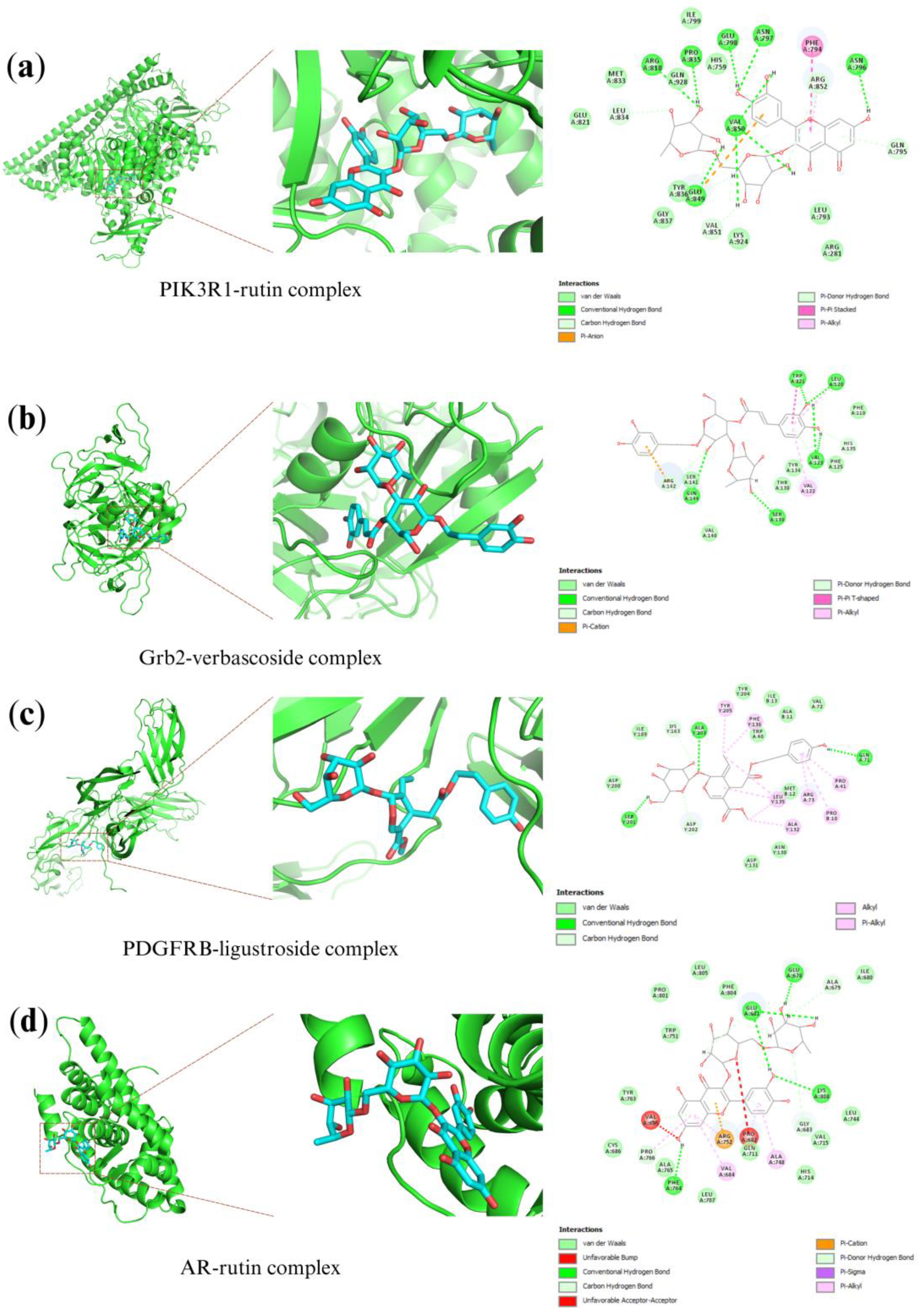
2.6. Molecular Docking Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Collection and Preparation of Plant Extracts
4.2. Total Phytochemical Analysis
4.2.1. Total Phenolic Content (TPC) Assay [16]
4.2.2. Total Flavonoids Content (TFC) Assay [16]
4.2.3. Total Tannin Content (TTC) Assay [16]
4.3. Antioxidant Activity
4.3.1. DPPH Free Radical Scavenging Assay [16]
4.3.2. Ferric Reducing Antioxidant Power (FRAP) Assay [16]
4.4. Anti-Inflammatory Activity
4.4.1. Murine Macrophage Cell Line RAW 264.7 Culture
4.4.2. Measurement of NO Production by Griess Reaction and Cell Viability [84,85]
4.5. Anti-Prostate Cancer Activity
4.5.1. Human Prostate Cancer Cell Line DU-145
4.5.2. Cell Treatment and Cell Viability with WST-1 Assay [86]
4.6. Data Treatment and Statistical Analysis
4.7. Network Pharmacology
4.7.1. Target Prediction and Identification
4.7.2. Protein-Protein Interaction (PPI) Network Construction
4.7.3. Gene Ontology Term and KEGG Pathway Enrichment Analysis
4.7.4. Molecular Docking Validation
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Komakech, R.; Yim, N.-H.; Shim, K.-S.; Jung, H.; Byun, J.-E.; Lee, J.; Okello, D.; Matsabisa, M.G.; Erhabor, J.O.; Oyenihi, O.; et al. Root Extract of a Micropropagated Prunus africana Medicinal Plant Induced Apoptosis in Human Prostate Cancer Cells (PC-3) via Caspase-3 Activation. Evid.-Based Complement. Altern. Med. 2022, 2022, 8232851. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lu, B.; He, M.; Wang, Y.; Wang, Z.; Du, L. Prostate Cancer Incidence and Mortality: Global Status and Temporal Trends in 89 Countries from 2000 to 2019. Front. Public Health 2022, 10, 811044. [Google Scholar] [CrossRef] [PubMed]
- Balea, Ş.S.; Pârvu, A.E.; Pârvu, M.; Vlase, L.; Dehelean, C.A.; Pop, T.I. Antioxidant, Anti-Inflammatory and Antiproliferative Effects of the Vitis vinifera L. Var. Fetească Neagră and Pinot Noir Pomace Extracts. Front. Pharmacol. 2020, 11, 990. [Google Scholar] [CrossRef] [PubMed]
- Thomas-Charles, C.; Fennell, H. Anti-Prostate Cancer Activity of Plant-Derived Bioactive Compounds: A Review. Curr. Mol. Biol. Rep. 2019, 5, 140–151. [Google Scholar] [CrossRef]
- Livermore, K.E.; Munkley, J.; Elliott, D.J. Androgen Receptor and Prostate Cancer. AIMS Mol. Sci. 2016, 3, 280–299. [Google Scholar] [CrossRef]
- Rizzo, M. Mechanisms of Docetaxel Resistance in Prostate Cancer: The Key Role Played by MiRNAs. Biochim. Biophys. Acta BBA-Rev. Cancer 2021, 1875, 188481. [Google Scholar] [CrossRef]
- Fu, C.-C.; Xu, F.-Y.; Qian, Y.-C.; Koo, H.-L.; Duan, Y.-F.; Weng, G.-M.; Fan, T.-P.; Chen, M.-X.; Zhu, F.-Y. Secondary Metabolites of Osmanthus fragrans: Metabolism and Medicinal Value. Front. Pharmacol. 2022, 13, 922204. [Google Scholar] [CrossRef]
- Wang, B.; Luan, F.; Bao, Y.; Peng, X.; Rao, Z.; Tang, Q.; Zeng, N. Traditional Uses, Phytochemical Constituents and Pharmacological Properties of Osmanthus fragrans: A Review. J. Ethnopharmacol. 2022, 293, 115273. [Google Scholar] [CrossRef]
- Wu, C.; Chen, C.; Hsieh, P.; Lee, Y.; Kuo, W.W.; Wu, R.C.; Hung, C.; Yang, Y.; Lin, V.C. Verbascoside Inhibits the Epithelial-mesenchymal Transition of Prostate Cancer Cells through High-mobility Group Box 1/Receptor for Advanced Glycation End-products/TGF-β Pathway. Environ. Toxicol. 2021, 36, 1080–1089. [Google Scholar] [CrossRef]
- Seca, A.; Pinto, D. Plant Secondary Metabolites as Anticancer Agents: Successes in Clinical Trials and Therapeutic Application. Int. J. Mol. Sci. 2018, 19, 263. [Google Scholar] [CrossRef]
- Keita, K.; Darkoh, C.; Okafor, F. Secondary Plant Metabolites as Potent Drug Candidates against Antimicrobial-Resistant Pathogens. SN Appl. Sci. 2022, 4, 209. [Google Scholar] [CrossRef] [PubMed]
- Bachheti, R.K.; Worku, L.A.; Gonfa, Y.H.; Zebeaman, M.; Deepti; Pandey, D.P.; Bachheti, A. Prevention and Treatment of Cardiovascular Diseases with Plant Phytochemicals: A Review. Evid.-Based Complement. Altern. Med. 2022, 2022, 5741198. [Google Scholar] [CrossRef] [PubMed]
- Shehadeh, M.B.; Suaifan, G.A.R.Y.; Abu-Odeh, A.M. Plants Secondary Metabolites as Blood Glucose-Lowering Molecules. Molecules 2021, 26, 4333. [Google Scholar] [CrossRef] [PubMed]
- Velu, G.; Palanichamy, V.; Rajan, A.P. Phytochemical and Pharmacological Importance of Plant Secondary Metabolites in Modern Medicine. In Bioorganic Phase in Natural Food: An Overview; Springer International Publishing: Cham, Switzerland, 2018; pp. 135–156. [Google Scholar]
- Luna-Guevara, M.L.; Luna-Guevara, J.J.; Hernández-Carranza, P.; Ruíz-Espinosa, H.; Ochoa-Velasco, C.E. Phenolic Compounds: A Good Choice against Chronic Degenerative Diseases. Stud. Nat. Prod. Chem. 2018, 59, 79–108. [Google Scholar]
- Tsai, P.-W.; Hsueh, C.-C.; Yang, H.-C.; Tsai, H.-Y.; Chen, B.-Y. Interactive Deciphering Electron-Shuttling Characteristics of Agricultural Wastes with Potential Bioenergy-Steered Anti-COVID-19 Activity via Microbial Fuel Cells. J. Taiwan Inst. Chem. Eng. 2022, 136, 104426. [Google Scholar] [CrossRef]
- Rahman, M.M.; Rahaman, M.S.; Islam, M.R.; Rahman, F.; Mithi, F.M.; Alqahtani, T.; Almikhlafi, M.A.; Alghamdi, S.Q.; Alruwaili, A.S.; Hossain, M.S.; et al. Role of Phenolic Compounds in Human Disease: Current Knowledge and Future Prospects. Molecules 2021, 27, 233. [Google Scholar] [CrossRef]
- Maleki, S.J.; Crespo, J.F.; Cabanillas, B. Anti-Inflammatory Effects of Flavonoids. Food Chem. 2019, 299, 125124. [Google Scholar] [CrossRef]
- Biharee, A.; Sharma, A.; Kumar, A.; Jaitak, V. Antimicrobial Flavonoids as a Potential Substitute for Overcoming Antimicrobial Resistance. Fitoterapia 2020, 146, 104720. [Google Scholar] [CrossRef]
- Badshah, S.L.; Faisal, S.; Muhammad, A.; Poulson, B.G.; Emwas, A.H.; Jaremko, M. Antiviral Activities of Flavonoids. Biomed. Pharmacother. 2021, 140, 111596. [Google Scholar] [CrossRef]
- Hosseinzade, A.; Sadeghi, O.; Naghdipour Biregani, A.; Soukhtehzari, S.; Brandt, G.S.; Esmaillzadeh, A. Immunomodulatory Effects of Flavonoids: Possible Induction of T CD4+ Regulatory Cells through Suppression of MTOR Pathway Signaling Activity. Front. Immunol. 2019, 10, 51. [Google Scholar] [CrossRef]
- Peng, K.; Wang, G.; Wang, Y.; Chen, B.; Sun, Y.; Mo, W.; Li, G.; Huang, Y. Condensed Tannins Enhanced Antioxidant Capacity and Hypoxic Stress Survivability but Not Growth Performance and Fatty Acid Profile of Juvenile Japanese Seabass (Lateolabrax japonicus). Anim. Feed. Sci. Technol. 2020, 269, 114671. [Google Scholar] [CrossRef]
- Sharifi-Rad, J.; Quispe, C.; Castillo, C.M.S.; Caroca, R.; Lazo-Vélez, M.A.; Antonyak, H.; Polishchuk, A.; Lysiuk, R.; Oliinyk, P.; De Masi, L.; et al. Ellagic Acid: A Review on Its Natural Sources, Chemical Stability, and Therapeutic Potential. Oxid. Med. Cell Longev. 2022, 2022, 3848084. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Goel, N. Phenolic Acids: Natural Versatile Molecules with Promising Therapeutic Applications. Biotechnol. Rep. 2019, 24, e00370. [Google Scholar] [CrossRef]
- Riveiro, M.; De Kimpe, N.; Moglioni, A.; Vazquez, R.; Monczor, F.; Shayo, C.; Davio, C. Coumarins: Old Compounds with Novel Promising Therapeutic Perspectives. Curr. Med. Chem. 2010, 17, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Kim, H.J.; Ali, M.S.; Lee, Y.S. An Update on Bioactive Plant Lignans. Nat. Prod. Rep. 2005, 22, 696. [Google Scholar] [CrossRef]
- Tsao, R. Chemistry and Biochemistry of Dietary Polyphenols. Nutrients 2010, 2, 1231–1246. [Google Scholar] [CrossRef] [PubMed]
- Afshari, A.; Sayyed-Alangi, S.Z. Antioxidant Effect of Leaf Extracts from Cressa cretica against Oxidation Process in Soybean Oil. Food Sci. Nutr. 2017, 5, 324–333. [Google Scholar] [CrossRef]
- Galgano, F.; Tolve, R.; Scarpa, T.; Caruso, M.C.; Lucini, L.; Senizza, B.; Condelli, N. Extraction Kinetics of Total Polyphenols, Flavonoids, and Condensed Tannins of Lentil Seed Coat: Comparison of Solvent and Extraction Methods. Foods 2021, 10, 1810. [Google Scholar] [CrossRef]
- Hussain, Z.T.E.; Yagi, S.; Mahomoodally, M.F.; Mohammed, I.; Zengin, G. A Comparative Study of Different Solvents and Extraction Techniques on the Anti-Oxidant and Enzyme Inhibitory Activities of Adansonia digitata L. (Baobab) Fruit Pulp. S. Afr. J. Bot. 2019, 126, 207–213. [Google Scholar] [CrossRef]
- Ansori, M.; Wahyuningsih; Fathonah, S.; Rosidah; Yulianti, N.A.H. The Difference in Antioxidant Capacity and Tannin Level in the Production of Parijoto Fruit Extract Based Dodol (Sweet Toffeelike Sugar Palm-Based Confection) Using 4 Different Types of Solvent. IOP Conf. Ser. Earth Environ. Sci. 2021, 700, 012067. [Google Scholar] [CrossRef]
- Kulbat-Warycha, K.; Oracz, J.; Żyżelewicz, D. Bioactive Properties of Extracts from Plectranthus barbatus (Coleus forskohlii) Roots Received Using Various Extraction Methods. Molecules 2022, 27, 8986. [Google Scholar] [CrossRef]
- Liu, Z.; Ren, Z.; Zhang, J.; Chuang, C.-C.; Kandaswamy, E.; Zhou, T.; Zuo, L. Role of ROS and Nutritional Antioxidants in Human Diseases. Front. Physiol. 2018, 9, 477. [Google Scholar] [CrossRef] [PubMed]
- Ponnampalam, E.N.; Kiani, A.; Santhiravel, S.; Holman, B.W.B.; Lauridsen, C.; Dunshea, F.R. The Importance of Dietary Antioxidants on Oxidative Stress, Meat and Milk Production, and Their Preservative Aspects in Farm Animals: Antioxidant Action, Animal Health, and Product Quality—Invited Review. Animals 2022, 12, 3279. [Google Scholar] [CrossRef] [PubMed]
- Thyagarajan, A.; Sahu, R.P. Potential Contributions of Antioxidants to Cancer Therapy: Immunomodulation and Radiosensitization. Integr. Cancer Ther. 2018, 17, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zhou, L.; Huang, Z.; Li, B.; Nice, E.C.; Xu, J.; Huang, C. Antioxidant Therapy in Cancer: Rationale and Progress. Antioxidants 2022, 11, 1128. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-Y.; Huang, F.-L.; Shi, L.-S.; Ka, S.-M.; Wang, J.-Y.; Tsai, Y.-C.; Hung, T.-J.; Ye, Y.-L. The Ethanol Extract of Osmanthus fragrans Flowers Reduces Oxidative Stress and Allergic Airway Inflammation in an Animal Model. Evid.-Based Complement. Altern. Med. 2013, 2013, 304290. [Google Scholar] [CrossRef]
- Tsai, P.-W.; Rogio, K.G.G.; Hsieh, C.-Y.; Cruz, K.A.D.C.; Lee, C.-J.; Hsueh, C.-C.; Huang, T.-N.; Lu, W.-Z.; Xie, Z.-L.; Jheng, Y.-N.; et al. Optimal Stimulation of Citrus Reticulate for Bioenergy Extraction in MFCs and Antioxidant Activity via Traditional Chinese Medicine Processing Methods. J. Taiwan Inst. Chem. Eng. 2023, 143, 104690. [Google Scholar] [CrossRef]
- Lin, W.-T.; He, Y.-H.; Lo, Y.-H.; Chiang, Y.-T.; Wang, S.-Y.; Bezirganoglu, I.; Kumar, K.J.S. Essential Oil from Glossogyne tenuifolia Inhibits Lipopolysaccharide-Induced Inflammation-Associated Genes in Macro-Phage Cells via Suppression of NF-ΚB Signaling Pathway. Plants 2023, 12, 1241. [Google Scholar] [CrossRef]
- Lee, D.-G.; Lee, S.-M.; Bang, M.-H.; Park, H.-J.; Lee, T.-H.; Kim, Y.-H.; Kim, J.-Y.; Baek, N.-I. Lignans from the Flowers of Osmanthus fragrans Var. Aurantiacus and Their Inhibition Effect on NO Production. Arch. Pharm. Res. 2011, 34, 2029–2035. [Google Scholar] [CrossRef]
- Deep, G.; Panigrahi, G.K. Hypoxia-Induced Signaling Promotes Prostate Cancer Progression: Exosomes Role as Messenger of Hypoxic Response in Tumor Microenvironment. Crit. Rev. Oncog. 2015, 20, 419–434. [Google Scholar] [CrossRef]
- Ardi, V.C.; Kupriyanova, T.A.; Deryugina, E.I.; Quigley, J.P. Human Neutrophils Uniquely Release TIMP-Free MMP-9 to Provide a Potent Catalytic Stimulator of Angiogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 20262–20267. [Google Scholar] [CrossRef] [PubMed]
- Andrisic, L.; Dudzik, D.; Barbas, C.; Milkovic, L.; Grune, T.; Zarkovic, N. Short Overview on Metabolomics Approach to Study Pathophysiology of Oxidative Stress in Cancer. Redox Biol. 2018, 14, 47–58. [Google Scholar] [CrossRef] [PubMed]
- de Souza, T.A.; de Campos, C.B.; De Biasi Bassani Gonçalves, A.; Nunes, F.C.; Monteiro, L.N.; de Oliveira Vasconcelos, R.; Cassali, G.D. Relationship between the Inflammatory Tumor Microenvironment and Different Histologic Types of Canine Mammary Tumors. Res. Vet. Sci. 2018, 119, 209–214. [Google Scholar] [CrossRef]
- Lee, D.-G. 24-Ethylcholesta-4,24(28)-Dien-3,6-Dione from Osmanthus fragrans var. aurantiacus Flowers Inhibits the Growth of Human Colon Cancer Cell Line, HCT-116. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 206–210. [Google Scholar] [CrossRef]
- Grizzi, F.; Chiriva-Internati, M. Cancer: Looking for Simplicity and Finding Complexity. Cancer Cell Int. 2006, 6, 4. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative Stress, Inflammation, and Cancer: How Are They Linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Mäkelä, S.; Aittokallio, T. Network Pharmacology Applications to Map the Unexplored Target Space and Therapeutic Potential of Natural Products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef]
- Li, S.; Fan, T.-P.; Jia, W.; Lu, A.; Zhang, W. Network Pharmacology in Traditional Chinese Medicine. Evid.-Based Complement. Altern. Med. 2014, 2014, 138460. [Google Scholar] [CrossRef]
- Poornima, P.; Kumar, J.D.; Zhao, Q.; Blunder, M.; Efferth, T. Network Pharmacology of Cancer: From Understanding of Complex Interactomes to the Design of Multi-Target Specific Therapeutics from Nature. Pharmacol. Res. 2016, 111, 290–302. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network Pharmacology: The next Paradigm in Drug Discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Guo, Y.; Nie, Q.; MacLean, A.L.; Li, Y.; Lei, J.; Li, S. Multiscale Modeling of Inflammation-Induced Tumorigenesis Reveals Competing Oncogenic and Oncoprotective Roles for Inflammation. Cancer Res. 2017, 77, 6429–6441. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wang, M.; Olounfeh, K.M.; Zhao, N.; Wang, S.; Meng, F. Network Pharmacology-Based Identifcation of Potential Targets of the Flower of Trollius Chinensis Bunge Acting on Anti-Inflammatory Effectss. Sci. Rep. 2019, 9, 8109. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-Y.; Tsai, Y.-C.; Li, K.-Y. Phenolic Antioxidants Isolated from the Flowers of Osmanthus fragrans. Molecules 2012, 17, 10724–10737. [Google Scholar] [CrossRef] [PubMed]
- Bitting, R.L.; Armstrong, A.J. Targeting the PI3K/Akt/MTOR Pathway in Castration-Resistant Prostate Cancer. Endocr. Relat. Cancer 2013, 20, R83–R99. [Google Scholar] [CrossRef]
- Mellor, P.; Furber, L.A.; Nyarko, J.N.K.; Anderson, D.H. Multiple Roles for the P85α Isoform in the Regulation and Function of PI3K Signalling and Receptor Trafficking. Biochem. J. 2012, 441, 23–37. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, Y.; McIlroy, J.; Rordorf-Nikolic, T.; Orr, G.A.; Backer, J.M. Regulation of the P85/P110 Phosphatidylinositol 3′-Kinase: Stabilization and Inhibition of the P110α Catalytic Subunit by the P85 Regulatory Subunit. Mol. Cell Biol. 1998, 18, 1379–1387. [Google Scholar] [CrossRef]
- Chakraborty, G.; Nandakumar, S.; Hirani, R.; Nguyen, B.; Stopsack, K.H.; Kreitzer, C.; Rajanala, S.H.; Ghale, R.; Mazzu, Y.Z.; Pillarsetty, N.V.K.; et al. The Impact of PIK3R1 Mutations and Insulin–PI3K–Glycolytic Pathway Regulation in Prostate Cancer. Clin. Cancer Res. 2022, 28, 3603–3617. [Google Scholar] [CrossRef]
- Giubellino, A.; Burke, T.R.; Bottaro, D.P. Grb2 Signaling in Cell Motility and Cancer. Expert. Opin. Ther. Targets 2008, 12, 1021–1033. [Google Scholar] [CrossRef]
- Olivier, J. A Drosophila SH2-SH3 Adaptor Protein Implicated in Coupling the Sevenless Tyrosine Kinase to an Activator of Ras Guanine Nucleotide Exchange, Sos. Cell 1993, 73, 179–191. [Google Scholar] [CrossRef]
- Pawson, T.; Scott, J.D. Signaling Through Scaffold, Anchoring, and Adaptor Proteins. Science 1997, 278, 2075–2080. [Google Scholar] [CrossRef]
- Lowenstein, E.J.; Daly, R.J.; Batzer, A.G.; Li, W.; Margolis, B.; Lammers, R.; Ullrich, A.; Skolnik, E.Y.; Bar-Sagi, D.; Schlessinger, J. The SH2 and SH3 Domain-Containing Protein GRB2 Links Receptor Tyrosine Kinases to Ras Signaling. Cell 1992, 70, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Iwata, T.; Sedukhina, A.S.; Kubota, M.; Oonuma, S.; Maeda, I.; Yoshiike, M.; Usuba, W.; Minagawa, K.; Hames, E.; Meguro, R.; et al. A New Bioinformatics Approach Identifies Overexpression of GRB2 as a Poor Prognostic Biomarker for Prostate Cancer. Sci. Rep. 2021, 11, 5696. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-Y.; Li, E.-M.; Wu, Z.-Y.; Cao, H.-H.; Shen, J.-H.; Xu, X.-E.; Chen, B.; Wu, J.-Y.; Xu, L.-Y. Overexpression of GRB2 Is Correlated with Lymph Node Metastasis and Poor Prognosis in Esophageal Squamous Cell Carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 3132–3140. [Google Scholar] [PubMed]
- Yu, G.Z.; Chen, Y.; Wang, J.J. Overexpression of Grb2/HER2 Signaling in Chinese Gastric Cancer: Their Relationship with Clinicopathological Parameters and Prognostic Significance. J. Cancer Res. Clin. Oncol. 2009, 135, 1331–1339. [Google Scholar] [CrossRef]
- Appiah-Kubi, K.; Lan, T.; Wang, Y.; Qian, H.; Wu, M.; Yao, X.; Wu, Y.; Chen, Y. Platelet-Derived Growth Factor Receptors (PDGFRs) Fusion Genes Involvement in Hematological Malignancies. Crit. Rev. Oncol. Hematol. 2017, 109, 20–34. [Google Scholar] [CrossRef]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of Platelet-Derived Growth Factors in Physiology and Medicine. Genes. Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef]
- Nordby, Y.; Richardsen, E.; Rakaee, M.; Ness, N.; Donnem, T.; Patel, H.R.H.; Busund, L.-T.; Bremnes, R.M.; Andersen, S. High Expression of PDGFR-β in Prostate Cancer Stroma Is Independently Associated with Clinical and Biochemical Prostate Cancer Recurrence. Sci. Rep. 2017, 7, 43378. [Google Scholar] [CrossRef]
- Hofer, M.D.; Fecko, A.; Shen, R.; Setlur, S.R.; Pienta, K.G.; Tomlins, S.A.; Chinnaiyan, A.M.; Rubin, M.A. Expression of the Platelet-Derived Growth Factor Receptor in Prostate Cancer and Treatment Implications with Tyrosine Kinase Inhibitors. Neoplasia 2004, 6, 503–512. [Google Scholar] [CrossRef]
- Arvidsson, A.K.; Rupp, E.; Nånberg, E.; Downward, J.; Rönnstrand, L.; Wennström, S.; Schlessinger, J.; Heldin, C.H.; Claesson-Welsh, L. Tyr-716 in the Platelet-Derived Growth Factor Beta-Receptor Kinase Insert Is Involved in GRB2 Binding and Ras Activation. Mol. Cell Biol. 1994, 14, 6715–6726. [Google Scholar] [CrossRef]
- Rodriguez-Viciana, P.; Warne, P.H.; Dhand, R.; Vanhaesebroeck, B.; Gout, I.; Fry, M.J.; Waterfield, M.D.; Downward, J. Phosphatidylinositol-3-OH Kinase Direct Target of Ras. Nature 1994, 370, 527–532. [Google Scholar] [CrossRef]
- Singh, D.; Febbo, P.G.; Ross, K.; Jackson, D.G.; Manola, J.; Ladd, C.; Tamayo, P.; Renshaw, A.A.; D’Amico, A.V.; Richie, J.P.; et al. Gene Expression Correlates of Clinical Prostate Cancer Behavior. Cancer Cell 2002, 1, 203–209. [Google Scholar] [CrossRef]
- Obaidullah, A.J.; Alanazi, M.M.; Alsaif, N.A.; Alanazi, A.S.; Albassam, H.; AZ, A.; Alwassil, O.I.; Alqahtani, A.M.; Tareq, A.M. Network Pharmacology- and Molecular Docking-Based Identification of Potential Phytocompounds from Argyreia capitiformis in the Treatment of Inflammation. Evid.-Based Complement. Altern. Med. 2022, 2022, 8037488. [Google Scholar] [CrossRef]
- Pandey, P.; Khan, F.; Qari, H.A.; Oves, M. Rutin (Bioflavonoid) as Cell Signaling Pathway Modulator: Prospects in Treatment and Chemoprevention. Pharmaceuticals 2021, 14, 1069. [Google Scholar] [CrossRef]
- Hoai, T.T.; Yen, P.T.; Dao, T.T.B.; Long, L.H.; Anh, D.X.; Minh, L.H.; Anh, B.Q.; Thuong, N.T. Evaluation of the Cytotoxic Effect of Rutin Prenanoemulsion in Lung and Colon Cancer Cell Lines. J. Nanomater. 2020, 2020, 8867669. [Google Scholar] [CrossRef]
- Paudel, K.R.; Wadhwa, R.; Tew, X.N.; Lau, N.J.X.; Madheswaran, T.; Panneerselvam, J.; Zeeshan, F.; Kumar, P.; Gupta, G.; Anand, K.; et al. Rutin Loaded Liquid Crystalline Nanoparticles Inhibit Non-Small Cell Lung Cancer Proliferation and Migration In Vitro. Life Sci. 2021, 276, 119436. [Google Scholar] [CrossRef]
- Perk, A.A.; Shatynska-Mytsyk, I.; Gerçek, Y.C.; Boztaş, K.; Yazgan, M.; Fayyaz, S.; Farooqi, A.A. Rutin Mediated Targeting of Signaling Machinery in Cancer Cells. Cancer Cell Int. 2014, 14, 124. [Google Scholar] [CrossRef]
- Messner, E.A.; Steele, T.M.; Tsamouri, M.M.; Hejazi, N.; Gao, A.C.; Mudryj, M.; Ghosh, P.M. The Androgen Receptor in Prostate Cancer: Effect of Structure, Ligands and Spliced Variants on Therapy. Biomedicines 2020, 8, 422. [Google Scholar] [CrossRef]
- Tan, M.E.; Li, J.; Xu, H.E.; Melcher, K.; Yong, E. Androgen Receptor: Structure, Role in Prostate Cancer and Drug Discovery. Acta Pharmacol. Sin. 2015, 36, 3–23. [Google Scholar] [CrossRef]
- Fei, Q.; Li, X.; Zhu, Q.; Wang, Y.; Ge, R.; Jin, X. Rutin Inhibits Androgen Synthesis and Metabolism in Rat Immature Leydig Cells In Vitro. Andrologia 2021, 53, e14221. [Google Scholar] [CrossRef]
- Mulani, S.K.; Guh, J.-H.; Mong, K.-K.T. A General Synthetic Strategy and the Anti-Proliferation Properties on Prostate Cancer Cell Lines for Natural Phenylethanoid Glycosides. Org. Biomol. Chem. 2014, 12, 2926. [Google Scholar] [CrossRef]
- Lee, K.-W.; Kim, H.J.; Lee, Y.S.; Park, H.-J.; Choi, J.-W.; Ha, J.; Lee, K.-T. Acteoside Inhibits Human Promyelocytic HL-60 Leukemia Cell Proliferation via Inducing Cell Cycle Arrest at G0/G1 Phase and Differentiation into Monocyte. Carcinogenesis 2007, 28, 1928–1936. [Google Scholar] [CrossRef]
- García-Caballero, M.; Torres-Vargas, J.A.; Marrero, A.D.; Martínez-Poveda, B.; Medina, M.Á.; Quesada, A.R. Angioprevention of Urologic Cancers by Plant-Derived Foods. Pharmaceutics 2022, 14, 256. [Google Scholar] [CrossRef]
- Soonthornsit, N.; Pitaksutheepong, C.; Hemstapat, W.; Utaisincharoen, P.; Pitaksuteepong, T. In Vitro Anti-Inflammatory Activity of Morus alba L. Stem Extract in LPS-Stimulated RAW 264.7 Cells. Evid.-Based Complement. Altern. Med. 2017, 2017, 3928956. [Google Scholar] [CrossRef]
- Kang, J.-K.; Chung, Y.-C.; Hyun, C.-G. Anti-Inflammatory Effects of 6-Methylcoumarin in LPS-Stimulated RAW 264.7 Macrophages via Regulation of MAPK and NF-ΚB Signaling Pathways. Molecules 2021, 26, 5351. [Google Scholar] [CrossRef]
- Park, J.S.; Kwon, J.K.; Kim, H.R.; Kim, H.J.; Kim, B.S.; Jung, J.Y. Farnesol Induces Apoptosis of DU145 Prostate Cancer Cells through the PI3K/Akt and MAPK Pathways. Int. J. Mol. Med. 2014, 33, 1169–1176. [Google Scholar] [CrossRef]
- Nickel, J.; Gohlke, B.-O.; Erehman, J.; Banerjee, P.; Rong, W.W.; Goede, A.; Dunkel, M.; Preissner, R. SuperPred: Update on Drug Classification and Target Prediction. Nucleic Acids Res. 2014, 42, W26–W31. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An Enhanced Web Server for Large-Scale Expression Profiling and Interactive Analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Koopmans, F.; van Nierop, P.; Andres-Alonso, M.; Byrnes, A.; Cijsouw, T.; Coba, M.P.; Cornelisse, L.N.; Farrell, R.J.; Goldschmidt, H.L.; Howrigan, D.P.; et al. SynGO: An Evidence-Based, Expert-Curated Knowledge Base for the Synapse. Neuron 2019, 103, 217–234.e4. [Google Scholar] [CrossRef]
- Khan, A.; Mathelier, A. Intervene: A Tool for Intersection and Visualization of Multiple Gene or Genomic Region Sets. BMC Bioinform. 2017, 18, 287. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. CytoHubba: Identifying Hub Objects and Sub-Networks from Complex Interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Sanches, K.; Dias, R.V.R.; da Silva, P.H.; Fossey, M.A.; Caruso, Í.P.; de Souza, F.P.; de Oliveira, L.C.; de Melo, F.A. Grb2 Dimer Interacts with Coumarin through SH2 Domains: A Combined Experimental and Molecular Modeling Study. Heliyon 2019, 5, e02869. [Google Scholar] [CrossRef]
- Mao, X.; Ren, Z.; Parker, G.N.; Sondermann, H.; Pastorello, M.A.; Wang, W.; McMurray, J.S.; Demeler, B.; Darnell, J.E.; Chen, X. Structural Bases of Unphosphorylated STAT1 Association and Receptor Binding. Mol. Cell 2005, 17, 761–771. [Google Scholar] [CrossRef]
- Halder, D.; Das, S.; Aiswarya, R.; Jeyaprakash, R.S. Molecular Docking and Dynamics Based Approach for the Identification of Kinase Inhibitors Targeting PI3Kα against Non-Small Cell Lung Cancer: A Computational Study. RSC Adv. 2022, 12, 21452–21467. [Google Scholar] [CrossRef]
- Ward, R.A.; Anderton, M.J.; Bethel, P.; Breed, J.; Cook, C.; Davies, E.J.; Dobson, A.; Dong, Z.; Fairley, G.; Farrington, P.; et al. Discovery of a Potent and Selective Oral Inhibitor of ERK1/2 (AZD0364) That Is Efficacious in Both Monotherapy and Combination Therapy in Models of Nonsmall Cell Lung Cancer (NSCLC). J. Med. Chem. 2019, 62, 11004–11018. [Google Scholar] [CrossRef]



| Sample | Total Phenolic Content (g GAE per kg Sample) | Total Flavonoid Content (g RE per kg Sample) | Total Tannin Content (g CE per kg Sample) |
|---|---|---|---|
| OF-F-W | 170.860 ± 2.868 | 17.820 ± 0.544 | 38.590 ± 0.854 |
| OF-F-E | 233.360 ± 3.613 | 17.200 ± 0.771 | 93.350 ± 1.003 |
| Equation of the line: | y = 0.0038x − 0.0132 (r2 = 0.9998) | y = 0.0049x + 0.0287 (r2 = 0.9994) | y = 0.0078x − 0.0242 (r2 = 0.9974) |
| Extract | DPPH IC50 (kg/L) | FRAP (Trolox g/kg) |
|---|---|---|
| OF-F-E | 0.337 ± 0.008 | 467.300 ± 2.784 |
| OF-F-W | 0.173 ± 0.004 | 830.600 ± 6.843 |
| Ascorbic acid | 0.111 ± 0.003 | - |
| Trolox calibration curve: y = 0.0038x + 0.0074 R2 = 0.998 |
| Extract | IC30 (mg/mL) | Cell Viability |
|---|---|---|
| OF-F-E | N.D. | Non toxic |
| OF-F-W | 0.3218 | Non toxic |
| Protein Target | Metabolite | Number of Interactions | LibDock Score | |||
|---|---|---|---|---|---|---|
| vdW | H-Bond | Other | Unfavorable | |||
| PIK3R1 | PHY | 14 | 3 | 7 | - | 121.49 |
| LIG | 17 | 3 | 9 | - | 136.26 | |
| VER | 14 | 7 | 4 | 2 | 162.70 | |
| HPA | 9 | 1 | 2 | - | 77.54 | |
| RUT | 10 | 15 | 3 | - | 163.22 | |
| 5–FU | 6 | 3 | 1 | 2 | 63.15 | |
| Grb2 | PHY | 5 | 9 | 3 | 3 | 84.84 |
| LIG | 8 | 3 | 6 | - | 98.15 | |
| VER | 6 | 10 | 4 | - | 115.42 | |
| HPA | 5 | 4 | 2 | 1 | 70.23 | |
| RUT | 6 | 7 | 8 | - | 104.35 | |
| 5–FU | - | - | - | - | None | |
| PDGFRB | PHY | 13 | 9 | 5 | - | 81.56 |
| LIG | 9 | 5 | 9 | - | 136.46 | |
| VER | 12 | 4 | 5 | - | 129.82 | |
| HPA | 3 | 4 | 3 | - | 79.33 | |
| RUT | 9 | 5 | 8 | 1 | 115.99 | |
| 5–FU | 4 | 4 | 4 | - | 60.04 | |
| AR | PHY | 12 | 2 | 9 | - | 127.89 |
| LIG | 10 | 7 | 11 | 1 | 165.10 | |
| VER | 7 | 11 | 7 | - | 169.33 | |
| HPA | 7 | 3 | 3 | - | 73.25 | |
| RUT | 13 | 12 | 6 | 2 | 173.17 | |
| 5–FU | 5 | 4 | 2 | - | 66.76 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.K.-H.; Bueno, P.R.P.; Garcia, P.J.B.; Lee, M.-J.; De Castro-Cruz, K.A.; Leron, R.B.; Tsai, P.-W. Antioxidant, Anti-Inflammatory and Antiproliferative Effects of Osmanthus fragrans (Thunb.) Lour. Flower Extracts. Plants 2023, 12, 3168. https://doi.org/10.3390/plants12173168
Huang SK-H, Bueno PRP, Garcia PJB, Lee M-J, De Castro-Cruz KA, Leron RB, Tsai P-W. Antioxidant, Anti-Inflammatory and Antiproliferative Effects of Osmanthus fragrans (Thunb.) Lour. Flower Extracts. Plants. 2023; 12(17):3168. https://doi.org/10.3390/plants12173168
Chicago/Turabian StyleHuang, Steven Kuan-Hua, Paolo Robert P. Bueno, Patrick Jay B. Garcia, Mon-Juan Lee, Kathlia A. De Castro-Cruz, Rhoda B. Leron, and Po-Wei Tsai. 2023. "Antioxidant, Anti-Inflammatory and Antiproliferative Effects of Osmanthus fragrans (Thunb.) Lour. Flower Extracts" Plants 12, no. 17: 3168. https://doi.org/10.3390/plants12173168
APA StyleHuang, S. K.-H., Bueno, P. R. P., Garcia, P. J. B., Lee, M.-J., De Castro-Cruz, K. A., Leron, R. B., & Tsai, P.-W. (2023). Antioxidant, Anti-Inflammatory and Antiproliferative Effects of Osmanthus fragrans (Thunb.) Lour. Flower Extracts. Plants, 12(17), 3168. https://doi.org/10.3390/plants12173168









