CRISPR/Cas Technology Revolutionizes Crop Breeding
Abstract
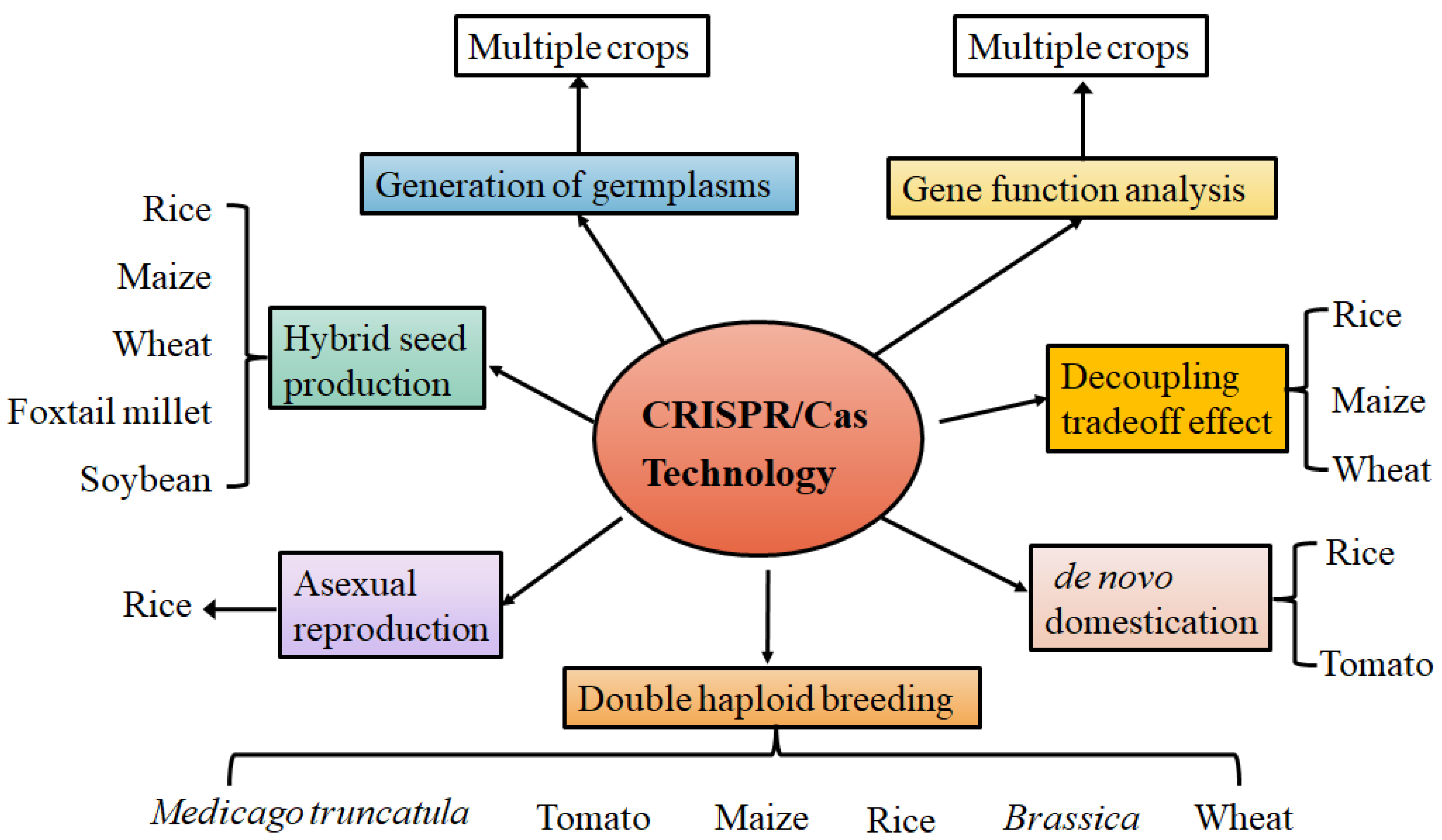
:1. Introduction
2. Exploring Gene Functions and Creating Desired Germplasms
3. Ushering in a New Era of Crop De Novo Domestication
4. Breaking Breeding Bottlenecks of Tradeoff Effects
5. Accelerating Conventional Production of Crop Hybrid Seed
6. Promoting Hybrid Rice Asexual Reproduction
7. Facilitating Double Haploid Breeding Technology
8. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chakraborty, S.; Pattanayak, A.; Mandal, S.; Das, M.; Roychowdhury, R. An Overview of Climate Change: Causes, Trends and Implications. In Crop Improvement in the Era of Climate Change; IK International Publishing House: New Delhi, India, 2014; pp. 1–29. [Google Scholar]
- Razzaq, A.; Wani, S.H.; Saleem, F.; Yu, M.; Zhou, M.; Shabala, S. Rewilding crops for climate resilience: Economic analysis and de novo domestication strategies. J. Exp. Bot. 2021, 72, 6123–6139. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.X. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Xiong, J.; Hu, F.; Ren, J.; Huang, Y.; Liu, C.; Wang, K. Synthetic apomixis: The beginning of a new era. Curr. Opin. Biotechnol. 2023, 79, 102877. [Google Scholar] [CrossRef] [PubMed]
- Jacquier, N.M.A.; Gilles, L.M.; Pyott, D.E.; Martinant, J.P.; Rogowsky, P.M.; Widiez, T. Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat. Plants 2020, 6, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Awan, M.J.A.; Pervaiz, K.; Rasheed, A.; Amin, I.; Saeed, N.A.; Dhugga, K.S.; Mansoor, S. Genome edited wheat-current advances for the second green revolution. Biotechnol. Adv. 2022, 60, 108006. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, M.F.; Mohd Noor, S.N.; Abd-Aziz, N.; Pua, T.L.; Tan, B.C. Green Revolution to Gene Revolution: Technological Advances in Agriculture to Feed the World. Plants 2022, 11, 1297. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Q.; Liu, H.Y.; Wang, K.; Zhang, S.X.; Ye, X.G. Development and innovation of haploid induction technologies in plants. Yi Chuan 2020, 42, 466–482. [Google Scholar] [CrossRef]
- Lv, J.; Kelliher, T. Recent Advances in Engineering of In Vivo Haploid Induction Systems. Methods Mol. Biol. 2023, 2653, 365–383. [Google Scholar] [CrossRef]
- Wang, J.Y.; Doudna, J.A. CRISPR technology: A decade of genome editing is only the beginning. Science 2023, 379, eadd8643. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Kim, M.J.; Kim, J.Y. CRISPR-Mediated Engineering across the Central Dogma in Plant Biology for Basic Research and Crop Improvement. Mol. Plant 2021, 14, 127–150. [Google Scholar] [CrossRef]
- Shelake, R.M.; Kadam, U.S.; Kumar, R.; Pramanik, D.; Singh, A.K.; Kim, J.Y. Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives. Plant Commun. 2022, 3, 100417. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, R.; Meng, X.; Chen, S.; Zong, Y.; Lu, C.; Qiu, J.L.; Chen, Y.H.; Li, J.; Gao, C. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 2020, 38, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Gootenberg, J.S.; Abudayyeh, O.O. Discovery of Diverse CRISPR-Cas Systems and Expansion of the Genome Engineering Toolbox. Biochemistry 2023. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Yan, J. De Novo Domestication: An Alternative Route toward New Crops for the Future. Mol. Plant 2019, 12, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Adeyinka, O.S.; Tabassum, B.; Koloko, B.L.; Ogungbe, I.V. Enhancing the quality of staple food crops through CRISPR/Cas-mediated site-directed mutagenesis. Planta 2023, 257, 78. [Google Scholar] [CrossRef]
- Nerkar, G.; Devarumath, S.; Purankar, M.; Kumar, A.; Valarmathi, R.; Devarumath, R.; Appunu, C. Advances in Crop Breeding Through Precision Genome Editing. Front. Genet. 2022, 13, 880195. [Google Scholar] [CrossRef]
- Ahmad, M. Plant breeding advancements with “CRISPR-Cas” genome editing technologies will assist future food security. Front. Plant Sci. 2023, 14, 1133036. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Z.; Unver, T.; Zhang, B. CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 2021, 29, 207–221. [Google Scholar] [CrossRef]
- Chen, W.K.; Chen, L.; Zhang, X.; Yang, N.; Guo, J.H.; Wang, M.; Ji, S.H.; Zhao, X.Y.; Yin, P.F.; Cai, L.C.; et al. Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 2022, 375, eabg7985. [Google Scholar] [CrossRef]
- Kong, D.; Wang, B.; Wang, H. UPA2 and ZmRAVL1: Promising targets of genetic improvement of maize plant architecture. J. Integr. Plant Biol. 2020, 62, 394–397. [Google Scholar] [CrossRef]
- Tian, J.; Wang, C.; Xia, J.; Wu, L.; Xu, G.; Wu, W.; Li, D.; Qin, W.; Han, X.; Chen, Q.; et al. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 2019, 365, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, J.; Lu, J.; Xiong, Y.; Zhao, Z.; Yu, X.; Zheng, X.; Li, J.; Lin, Q.; Ren, Y.; et al. A natural gene drive system confers reproductive isolation in rice. Cell 2023, 186, 3577–3592. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Zheng, X.; Wang, W.; Yin, X.; Liu, H.; Ma, C.; Niu, X.; Zhu, J.K.; Wang, F. Creation of aromatic maize by CRISPR/Cas. J. Integr. Plant Biol. 2021, 63, 1664–1670. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Qi, X.; Zhu, J.; Liu, C.; Zhang, X.; Cheng, B.; Mao, L.; Xie, C. Supersweet and waxy: Meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol. J. 2019, 17, 1853–1855. [Google Scholar] [CrossRef]
- Gao, H.; Gadlage, M.J.; Lafitte, H.R.; Lenderts, B.; Yang, M.; Schroder, M.; Farrell, J.; Snopek, K.; Peterson, D.; Feigenbutz, L.; et al. Superior field performance of waxy corn engineered using CRISPR-Cas9. Nat. Biotechnol. 2020, 38, 579–581. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, Y.; Tabor, G.; Nian, H.; Phillips, J.; Wolters, P.; Yang, Q.; Balint-Kurti, P. A leucine-rich repeat receptor kinase gene confers quantitative susceptibility to maize southern leaf blight. New Phytol. 2023, 238, 1182–1197. [Google Scholar] [CrossRef]
- Lorenzo, C.D.; Debray, K.; Herwegh, D.; Develtere, W.; Impens, L.; Schaumont, D.; Vandeputte, W.; Aesaert, S.; Coussens, G.; De Boe, Y.; et al. BREEDIT: A multiplex genome editing strategy to improve complex quantitative traits in maize. Plant Cell 2023, 35, 218–238. [Google Scholar] [CrossRef]
- Hui, S.; Li, H.; Mawia, A.M.; Zhou, L.; Cai, J.; Ahmad, S.; Lai, C.; Wang, J.; Jiao, G.; Xie, L.; et al. Production of aromatic three-line hybrid rice using novel alleles of BADH2. Plant Biotechnol. J. 2022, 20, 59–74. [Google Scholar] [CrossRef]
- Song, X.; Chen, Z.; Du, X.; Li, B.; Fei, Y.; Tao, Y.; Wang, F.; Xu, Y.; Li, W.; Wang, J.; et al. Generation of new rice germplasms with low amylose content by CRISPR/CAS9-targeted mutagenesis of the FLOURY ENDOSPERM 2 gene. Front. Plant Sci. 2023, 14, 1138523. [Google Scholar] [CrossRef]
- Tian, Y.; Zhou, Y.; Gao, G.; Zhang, Q.; Li, Y.; Lou, G.; He, Y. Creation of Two-Line Fragrant Glutinous Hybrid Rice by Editing the Wx and OsBADH2 Genes via the CRISPR/Cas9 System. Int. J. Mol. Sci. 2023, 24, 849. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, Z.; Li, Y.; Xu, C.; Xia, H.; Zhuang, H.; Sun, S.; Guo, M.; Yan, C. Rapid improvement of rice eating and cooking quality through gene editing toward glutelin as target. J. Integr. Plant Biol. 2022, 64, 1860–1865. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Gu, Z.; Chen, Z.; Yu, J.; Chu, R.; Tan, H.; Zhao, D.; Fan, X.; Zhang, C.; Li, Q.; et al. Improving rice eating and cooking quality by coordinated expression of the major starch synthesis-related genes, SSII and Wx, in endosperm. Plant Mol. Biol. 2021, 106, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.S.; Basnet, R.; Ahmed, S.; Bao, J.; Shu, Q. Mutations of OsPLDa1 Increase Lysophospholipid Content and Enhance Cooking and Eating Quality in Rice. Plants 2020, 9, 390. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Wu, B.; Wang, J.; Zhu, W.; Nie, H.; Qian, J.; Huang, W.; Fang, Z. Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 2018, 16, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Wu, B.; Ji, Y. The Amino Acid Transporter OsAAP4 Contributes to Rice Tillering and Grain Yield by Regulating Neutral Amino Acid Allocation through Two Splicing Variants. Rice 2021, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Wang, J.; Miao, J.; Chen, J.; Wu, S.; Zhu, J.; Zhang, D.; Gu, H.; Cui, H.; Shi, S.; et al. The Spermine Synthase OsSPMS1 Regulates Seed Germination, Grain Size, and Yield. Plant Physiol. 2018, 178, 1522–1536. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gong, C.; Wu, J.; Yang, L.; Zhou, L.; Wu, B.; Gao, L.; Ling, F.; You, A.; Li, C.; et al. Improvement of Rice Agronomic Traits by Editing Type-B Response Regulators. Int. J. Mol. Sci. 2022, 23, 14165. [Google Scholar] [CrossRef]
- Li, C.; Zhou, L.; Wu, B.; Li, S.; Zha, W.; Li, W.; Zhou, Z.; Yang, L.; Shi, L.; Lin, Y.; et al. Improvement of Bacterial Blight Resistance in Two Conventionally Cultivated Rice Varieties by Editing the Noncoding Region. Cells 2022, 11, 2535. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, S.; Jiang, N.; Zhao, X.; Bai, Z.; Liu, J.; Yao, W.; Tang, Q.; Xiao, G.; Lv, C.; et al. Engineering of rice varieties with enhanced resistances to both blast and bacterial blight diseases via CRISPR/Cas9. Plant Biotechnol. J. 2022, 20, 876–885. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, R.; Huang, K.; Huang, S.; Wang, H.; Wei, Z.; Li, Z.; Bian, M.; Jiang, W.; Wu, T.; et al. The OsWRKY63-OsWRKY76-OsDREB1B module regulates chilling tolerance in rice. Plant J. 2022, 112, 383–398. [Google Scholar] [CrossRef]
- Zhao, W.; Wang, X.; Zhang, Q.; Zheng, Q.; Yao, H.; Gu, X.; Liu, D.; Tian, X.; Wang, X.; Li, Y.; et al. H3K36 demethylase JMJ710 negatively regulates drought tolerance by suppressing MYB48-1 expression in rice. Plant Physiol. 2022, 189, 1050–1064. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, R.; Gao, J.; Song, G.; Li, J.; Li, W.; Qi, Y.; Li, Y.; Li, G. CRISPR/Cas9-mediated genome editing for wheat grain quality improvement. Plant Biotechnol. J. 2021, 19, 1684–1686. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ke, W.; He, F.; Chai, L.; Cheng, X.; Xu, H.; Wang, X.; Du, D.; Zhao, Y.; Chen, X.; et al. A single nucleotide deletion in the third exon of FT-D1 increases the spikelet number and delays heading date in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2022, 20, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yang, H.; Zhang, Z.; Chen, Q.; Guo, W.; Rossi, V.; Xin, M.; Du, J.; Hu, Z.; Liu, J.; et al. An elite gamma-gliadin allele improves end-use quality in wheat. New Phytol. 2023, 239, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, R.; Liu, P.; Yang, M.; Xin, D.; Liu, C.; Zhang, Z.; Wu, X.; Chen, Q.; Zhao, Y. Design of high-monounsaturated fatty acid soybean seed oil using GmPDCTs knockout via a CRISPR-Cas9 system. Plant Biotechnol. J. 2023, 21, 1317. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ji, J.; Cheng, Y.; Zhang, S.; Wang, Z.; Duan, K.; Wang, Y. CRISPR/Cas9-mediated editing of GmTAP1 confers enhanced resistance to Phytophthora sojae in soybean. J. Integr. Plant Biol. 2023, 65, 1609–1612. [Google Scholar] [CrossRef]
- Zhu, X.; Zheng, K.; Lu, L.; Yu, H.; Wang, F.; Yang, X.; Bhat, J.A.; Zhao, B.; Wang, Y.; Li, H.; et al. Disruption of CHORISMATE SYNTHASE1 leads to yellow-green variegation in soybean leaves. J. Exp. Bot. 2023, 74, 4014–4030. [Google Scholar] [CrossRef]
- Kong, K.; Xu, M.; Xu, Z.; Lv, W.; Lv, P.; Begum, N.; Liu, B.; Liu, B.; Zhao, T. Dysfunction of GmVPS8a causes compact plant architecture in soybean. Plant Sci. 2023, 331, 111677. [Google Scholar] [CrossRef]
- Zhou, J.; Li, Z.; Li, Y.; Zhao, Q.; Luan, X.; Wang, L.; Liu, Y.; Liu, H.; Zhang, J.; Yao, D. Effects of Different Gene Editing Modes of CRISPR/Cas9 on Soybean Fatty Acid Anabolic Metabolism Based on GmFAD2 Family. Int. J. Mol. Sci. 2023, 24, 4769. [Google Scholar] [CrossRef]
- Zhu, X.G.; Zhu, J.K. Precision genome editing heralds rapid de novo domestication for new crops. Cell 2021, 184, 1133–1134. [Google Scholar] [CrossRef]
- Zsogon, A.; Cermak, T.; Voytas, D.; Peres, L.E. Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: Case study in tomato. Plant Sci. 2017, 256, 120–130. [Google Scholar] [CrossRef]
- Kumar, K.; Mandal, S.N.; Pradhan, B.; Kaur, P.; Kaur, K.; Neelam, K. From Evolution to Revolution: Accelerating Crop Domestication through Genome Editing. Plant Cell Physiol. 2022, 63, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Jian, L.M.; Yan, J.B.; Liu, J. De Novo Domestication in the Multi-Omics Era. Plant Cell Physiol. 2022, 63, 1592–1606. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.H.; Huang, S.W.; Han, B.; Li, J.Y. The integrated genomics of crop domestication and breeding. Cell 2022, 185, 2828–2839. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, K.; Moreira, J.D.; Peres, L.E.P.; Zsogon, A. De novo domestication of wild species to create crops with increased resilience and nutritional value. Curr. Opin. Plant Biol. 2021, 60, 102006. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.; Zaidi, S.S.E.A.; Amin, I.; Mansoor, S. A CRISPR Way for Fast-Forward Crop Domestication. Trends Plant Sci. 2019, 24, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Zsogon, A.; Cermak, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef]
- Li, T.D.; Yang, X.P.; Yu, Y.; Si, X.M.; Zhai, X.W.; Zhang, H.W.; Dong, W.X.; Gao, C.X.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, T.H.; Huang, X.Z.; Xu, C. A two-in-one breeding strategy boosts rapid utilization of wild species and elite cultivars. Plant Biotechnol. J. 2022, 20, 800–802. [Google Scholar] [CrossRef]
- Yu, H.; Lin, T.; Meng, X.B.; Du, H.L.; Zhang, J.K.; Liu, G.F.; Chen, M.J.; Jing, Y.H.; Kou, L.Q.; Li, X.X.; et al. A route to de novo domestication of wild allotetraploid rice. Cell 2021, 184, 1156–1170. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018, 19, 21–33. [Google Scholar] [CrossRef]
- Takatsuji, H. Regulating Tradeoffs to Improve Rice Production. Front. Plant Sci. 2017, 8, 171. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Long, X.Y.; Chern, M.; Chen, X.W. Understanding the molecular mechanisms of trade-offs between plant growth and immunity. Sci. China Life Sci. 2021, 64, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Meng, X.; Guo, H.; Cheng, Q.; Jing, Y.; Chen, M.; Liu, G.; Wang, B.; Wang, Y.; Li, J.; et al. Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol. 2022, 40, 1403–1411. [Google Scholar] [CrossRef]
- Saeed, S.; Usman, B.; Shim, S.H.; Khan, S.U.; Nizamuddin, S.; Saeed, S.; Shoaib, Y.; Jeon, J.S.; Jung, K.H. CRISPR/Cas-mediated editing of cis-regulatory elements for crop improvement. Plant Sci. 2022, 324, 111435. [Google Scholar] [CrossRef]
- Okita, T.W.; Delseny, M. Genome editing in plants: New advances and applications in plant biology and agriculture. Plant Sci. 2023, 328, 111577. [Google Scholar] [CrossRef]
- Zafar, S.A.; Zaidi, S.S.; Gaba, Y.; Singla-Pareek, S.L.; Dhankher, O.P.; Li, X.; Mansoor, S.; Pareek, A. Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J. Exp. Bot. 2020, 71, 470–479. [Google Scholar] [CrossRef]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef]
- Antony, G.; Zhou, J.H.; Huang, S.; Li, T.; Liu, B.; White, F.; Yang, B. Rice xa13 Recessive Resistance to Bacterial Blight Is Defeated by Induction of the Disease Susceptibility Gene Os-11N3. Plant Cell 2010, 22, 3864–3876. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Xu, X.M.; Gong, Q.; Li, Z.Y.; Li, Y.; Wang, S.; Yang, Y.Y.; Ma, W.X.; Liu, L.Y.; Zhu, B.; et al. Engineering Broad-Spectrum Bacterial Blight Resistance by Simultaneously Disrupting Variable TALE-Binding Elements of Multiple Susceptibility Genes in Rice. Mol. Plant 2019, 12, 1434–1446. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.R.; Meng, X.B.; Song, X.G.; Zhang, D.H.; Kou, L.Q.; Zhang, J.H.; Jing, Y.H.; Liu, G.F.; Liu, H.H.; Huang, X.H.; et al. Chilling-induced phosphorylation of IPA1 by OsSAPK6 activates chilling tolerance responses in rice. Cell Discov. 2022, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Luo, N.; Meng, X.; Song, X.; Jing, Y.; Kou, L.; Liu, G.; Huang, X.; Wang, Y.; Li, J.; et al. OsMPK4 promotes phosphorylation and degradation of IPA1 in response to salt stress to confer salt tolerance in rice. J. Genet. Genom. 2022, 49, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.M.; Shi, Z.Y.; Zhang, X.H.; Wang, M.X.; Zhang, L.; Zheng, K.Z.; Liu, J.Y.; Hu, X.M.; Di, C.R.; Qian, Q.; et al. Inducible overexpression of Ideal Plant Architecture1 improves both yield and disease resistance in rice. Nat. Plants 2019, 5, 389–400. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.J.; Zhu, X.B.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef]
- Song, X.G.; Lu, Z.F.; Yu, H.; Shao, G.N.; Xiong, J.S.; Meng, X.B.; Jing, Y.H.; Liu, G.F.; Xiong, G.S.; Duan, J.B.; et al. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 2017, 27, 1128–1141. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, H.; Ma, B.; Liu, G.F.; Wang, J.J.; Wang, J.M.; Gao, R.C.; Li, J.J.; Liu, J.Y.; Xu, J.; et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 2017, 8, 14789. [Google Scholar] [CrossRef]
- Wang, J.; Yu, H.; Xiong, G.S.; Lu, Z.F.; Jiao, Y.Q.; Meng, X.B.; Liu, G.F.; Chen, X.W.; Wang, Y.H.; Li, J.Y. Tissue-Specific Ubiquitination by IPA1 INTERACTING PROTEIN1 Modulates IPA1 Protein Levels to Regulate Plant Architecture in Rice. Plant Cell 2017, 29, 697–707. [Google Scholar] [CrossRef]
- Lu, Z.F.; Yu, H.; Xiong, G.S.; Wang, J.; Jiao, Y.Q.; Liu, G.F.; Jing, Y.H.; Meng, X.B.; Hu, X.M.; Qian, Q.; et al. Genome-Wide Binding Analysis of the Transcription Activator IDEAL PLANT ARCHITECTURE1 Reveals a Complex Network Regulating Rice Plant Architecture. Plant Cell 2013, 25, 3743–3759. [Google Scholar] [CrossRef]
- Jiao, Y.Q.; Wang, Y.H.; Xue, D.W.; Wang, J.; Yan, M.X.; Liu, G.F.; Dong, G.J.; Zeng, D.L.; Lu, Z.F.; Zhu, X.D.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Reynolds, M.P.; Ortiz, R. Mitigating tradeoffs in plant breeding. iScience 2021, 24, 102965. [Google Scholar] [CrossRef]
- Tan, W.; Miao, J.; Xu, B.; Zhou, C.; Wang, Y.; Gu, X.; Liang, S.; Wang, B.; Chen, C.; Zhu, J.; et al. Rapid production of novel beneficial alleles for improving rice appearance quality by targeting a regulatory element of SLG7. Plant Biotechnol. J. 2023, 21, 1305–1307. [Google Scholar] [CrossRef]
- Wei, X.; Liu, Q.; Sun, T.; Jiao, X.; Liu, C.; Hua, Y.; Chen, X.; Wang, K. Manipulation of genetic recombination by editing the transcriptional regulatory regions of a meiotic gene in hybrid rice. Plant Commun. 2023, 4, 100474. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, G.; Zhao, Y.; Zhang, R.; Tang, X.; Li, L.; Jia, X.; Guo, Y.; Wu, Y.; Han, Y.; et al. An efficient CRISPR-Cas12a promoter editing system for crop improvement. Nat. Plants 2023, 9, 588–604. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Liu, B.; Chen, Q.J.; Yang, B. High-efficiency prime editing enables new strategies for broad-spectrum resistance to bacterial blight of rice. Plant Biotechnol. J. 2023, 21, 1454–1464. [Google Scholar] [CrossRef] [PubMed]
- Bommert, P.; Lunde, C.; Nardmann, J.; Vollbrecht, E.; Running, M.; Jackson, D.; Hake, S.; Werr, W. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development 2005, 132, 1235–1245. [Google Scholar] [CrossRef]
- Bommert, P.; Nagasawa, N.S.; Jackson, D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 2013, 45, 334–337. [Google Scholar] [CrossRef]
- Taguchi-Shiobara, F.; Yuan, Z.; Hake, S.; Jackson, D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Gene Dev. 2001, 15, 2755–2766. [Google Scholar] [CrossRef]
- Rodriguez-Leal, D.; Xu, C.; Kwon, C.T.; Soyars, C.; Demesa-Arevalo, E.; Man, J.; Liu, L.; Lemmon, Z.H.; Jones, D.S.; Van Eck, J.; et al. Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 2019, 51, 786–792. [Google Scholar] [CrossRef]
- Il Je, B.; Gruel, J.; Lee, Y.K.; Bommert, P.; Arevalo, E.D.; Eveland, A.L.; Wu, Q.Y.; Goldshmidt, A.; Meeley, R.; Bartlett, M.; et al. Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat. Genet. 2016, 48, 785–791. [Google Scholar] [CrossRef]
- Basu, U.; Parida, S.K. Restructuring plant types for developing tailor-made crops. Plant Biotechnol. J. 2021, 21, 1106–1122. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat. Plants 2021, 7, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Cheng, X.; Shan, Q.W.; Zhang, Y.; Liu, J.X.; Gao, C.X.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstadler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 2017, 15, 367–378. [Google Scholar] [CrossRef]
- Wang, K. Fixation of hybrid vigor in rice: Synthetic apomixis generated by genome editing. aBIOTECH 2020, 1, 15–20. [Google Scholar] [CrossRef]
- Yu, H.; Li, J. Producing hybrid seeds like conventional rice. Cell Res. 2022, 32, 959–960. [Google Scholar] [CrossRef]
- Barman, H.N.; Sheng, Z.; Fiaz, S.; Zhong, M.; Wu, Y.; Cai, Y.; Wang, W.; Jiao, G.; Tang, S.; Wei, X.; et al. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019, 19, 109. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, Q.; Pu, L.; Zhang, H.; Li, X. CRISPR-Cas technology opens a new era for the creation of novel maize germplasms. Front. Plant Sci. 2022, 13, 1049803. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Y.; Wei, G.; Lei, T.; Wu, J.; Zheng, L.; Ma, H.; He, G.; Wang, N. POLLEN WALL ABORTION 1 is essential for pollen wall development in rice. Plant Physiol. 2022, 190, 2229–2245. [Google Scholar] [CrossRef]
- Ni, E.; Deng, L.; Chen, H.; Lin, J.; Ruan, J.; Liu, Z.; Zhuang, C.; Zhou, H. OsCER1 regulates humidity-sensitive genic male sterility through very-long-chain (VLC) alkane metabolism of tryphine in rice. Funct. Plant Biol. 2021, 48, 461–468. [Google Scholar] [CrossRef]
- Han, Y.; Zhou, S.D.; Fan, J.J.; Zhou, L.; Shi, Q.S.; Zhang, Y.F.; Liu, X.L.; Chen, X.; Zhu, J.; Yang, Z.N. OsMS188 Is a Key Regulator of Tapetum Development and Sporopollenin Synthesis in Rice. Rice 2021, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.J.; Sun, L.P.; Yu, P.; Yang, Z.F.; Zhang, P.P.; Zhang, Y.X.; Wu, W.X.; Chen, D.B.; Zhan, X.D.; Khan, R.M.; et al. The MYB transcription factor Baymax1 plays a critical role in rice male fertility. Theor. Appl. Genet. 2021, 134, 453–471. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Fang, R.; Chen, F.; Han, J.; Liu, Y.G.; Chen, L.; Zhu, Q. A novel CCCH-type zinc finger protein SAW1 activates OsGA20ox3 to regulate gibberellin homeostasis and anther development in rice. J. Integr. Plant Biol. 2020, 62, 1594–1606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, S.; Li, J.; Gao, J.; Song, G.; Li, W.; Geng, S.; Liu, C.; Lin, Y.; Li, Y.; et al. CRISPR/Cas9-targeted mutagenesis of TaDCL4, TaDCL5 and TaRDR6 induces male sterility in common wheat. Plant Biotechnol. J. 2023, 21, 839–853. [Google Scholar] [CrossRef]
- Fang, X.; Sun, X.; Yang, X.; Li, Q.; Lin, C.; Xu, J.; Gong, W.; Wang, Y.; Liu, L.; Zhao, L.; et al. MS1 is essential for male fertility by regulating the microsporocyte cell plate expansion in soybean. Sci. China Life Sci. 2021, 64, 1533–1545. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, C.; Zhu, J.; Liu, C.; Huang, C.; Li, X.; Xie, C. Genome Editing Enables Next-Generation Hybrid Seed Production Technology. Mol. Plant 2020, 13, 1262–1269. [Google Scholar] [CrossRef]
- Chen, R.; Xu, Q.; Liu, Y.; Zhang, J.; Ren, D.; Wang, G.; Liu, Y. Generation of Transgene-Free Maize Male Sterile Lines Using the CRISPR/Cas9 System. Front. Plant Sci. 2018, 9, 1180. [Google Scholar] [CrossRef]
- Fang, Y.; Yang, J.; Guo, X.; Qin, Y.; Zhou, H.; Liao, S.; Liu, F.; Qin, B.; Zhuang, C.; Li, R. CRISPR/Cas9-Induced Mutagenesis of TMS5 Confers Thermosensitive Genic Male Sterility by Influencing Protein Expression in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 8354. [Google Scholar] [CrossRef]
- Chen, Y.; Shahid, M.Q.; Wu, J.; Deng, R.; Chen, Z.; Wang, L.; Liu, G.; Zhou, H.; Liu, X. Thermo-Sensitive Genic Male Sterile Lines of Neo-Tetraploid Rice Developed through Gene Editing Technology Revealed High Levels of Hybrid Vigor. Plants 2022, 11, 1390. [Google Scholar] [CrossRef]
- Pak, H.; Wang, H.; Kim, Y.; Song, U.; Tu, M.; Wu, D.; Jiang, L. Creation of male-sterile lines that can be restored to fertility by exogenous methyl jasmonate for the establishment of a two-line system for the hybrid production of rice (Oryza sativa L.). Plant Biotechnol. J. 2021, 19, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wang, T.; Li, Y.; Hu, J.; Kan, R.; Qiu, M.; Deng, Y.; Liu, P.; Zhang, L.; Dong, H.; et al. A novel strategy for creating a new system of third-generation hybrid rice technology using a cytoplasmic sterility gene and a genic male-sterile gene. Plant Biotechnol. J. 2021, 19, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Shen, L.; Hu, P.; Liu, Q.; Zhu, X.; Qian, Q.; Wang, K.; Wang, Y. Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing. J. Integr. Plant Biol. 2019, 61, 1201–1205. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, D.; Chen, M.; Liang, W.; Wei, J.; Qi, Y.; Yuan, Z. Development of japonica Photo-Sensitive Genic Male Sterile Rice Lines by Editing Carbon Starved Anther Using CRISPR/Cas9. J. Genet. Genom. 2016, 43, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Z.; He, G.; Ma, L.; Deng, X.W. CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterility in bread wheat. J. Genet. Genom. 2020, 47, 263–272. [Google Scholar] [CrossRef]
- Okada, A.; Arndell, T.; Borisjuk, N.; Sharma, N.; Watson-Haigh, N.S.; Tucker, E.J.; Baumann, U.; Langridge, P.; Whitford, R. CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 2019, 17, 1905–1913. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, M.; Albertsen, M.C.; Young, J.K.; Cigan, A.M. Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.). Plant Mol. Biol. 2018, 97, 371–383. [Google Scholar] [CrossRef]
- Zhang, W.; Qi, X.; Zhi, H.; Ren, Y.; Zhang, L.; Gao, Y.; Sui, Y.; Zhang, H.; Tang, S.; Jia, G.; et al. A straight-forward seed production technology system for foxtail millet (Setaria italica). J. Integr. Plant Biol. 2023. early view. [Google Scholar] [CrossRef]
- Nadeem, M.; Chen, A.; Hong, H.; Li, D.; Li, J.; Zhao, D.; Wang, W.; Wang, X.; Qiu, L. GmMs1 encodes a kinesin-like protein essential for male fertility in soybean (Glycine max L.). J. Integr. Plant Biol. 2021, 63, 1054–1064. [Google Scholar] [CrossRef]
- Li, H.; You, C.; Yoshikawa, M.; Yang, X.; Gu, H.; Li, C.; Cui, J.; Chen, X.; Ye, N.; Zhang, J.; et al. A spontaneous thermo-sensitive female sterility mutation in rice enables fully mechanized hybrid breeding. Cell Res. 2022, 32, 931–945. [Google Scholar] [CrossRef]
- Mahlandt, A.; Singh, D.K.; Mercier, R. Engineering apomixis in crops. Theor. Appl. Genet. 2023, 136, 131. [Google Scholar] [CrossRef] [PubMed]
- Fei, X.; Shi, J.; Liu, Y.; Niu, J.; Wei, A. The steps from sexual reproduction to apomixis. Planta 2019, 249, 1715–1730. [Google Scholar] [CrossRef] [PubMed]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Q.; Shen, Y.; Hua, Y.; Wang, J.; Lin, J.; Wu, M.; Sun, T.; Cheng, Z.; Mercier, R.; et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 2019, 37, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; He, Z.; Zhang, Y.; Hu, F.; Li, M.; Liu, Q.; Huang, Y.; Wang, J.; Zhang, W.; Wang, C.; et al. Synthetic apomixis enables stable transgenerational transmission of heterotic phenotypes in hybrid rice. Plant Commun. 2023, 4, 100470. [Google Scholar] [CrossRef] [PubMed]
- Vernet, A.; Meynard, D.; Lian, Q.; Mieulet, D.; Gibert, O.; Bissah, M.; Rivallan, R.; Autran, D.; Leblanc, O.; Meunier, A.C.; et al. High-frequency synthetic apomixis in hybrid rice. Nat. Commun. 2022, 13, 7963. [Google Scholar] [CrossRef] [PubMed]
- Eliby, S.; Bekkuzhina, S.; Kishchenko, O.; Iskakova, G.; Kylyshbayeva, G.; Jatayev, S.; Soole, K.; Langridge, P.; Borisjuk, N.; Shavrukov, Y. Developments and prospects for doubled haploid wheat. Biotechnol. Adv. 2022, 60, 108007. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Britt, A.B.; Tripathi, L.; Sharma, S.; Upadhyaya, H.D.; Ortiz, R. Haploids: Constraints and opportunities in plant breeding. Biotechnol. Adv. 2015, 33, 812–829. [Google Scholar] [CrossRef]
- Shen, K.; Qu, M.; Zhao, P. The Roads to Haploid Embryogenesis. Plants 2023, 12, 243. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, D.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Li, Y.; Lin, Z.; Yue, Y.; Zhao, H.; Fei, X.; Lizhu, E.; Liu, C.; Chen, S.; Lai, J.; Song, W. Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nat. Plants 2021, 7, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Sun, J.; Li, R.; Yan, S.; Chen, W.; Guo, L.; Qin, G.; Wang, P.; Luo, C.; Huang, W.; et al. A reactive oxygen species burst causes haploid induction in maize. Mol. Plant 2022, 15, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liang, D.; Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Zhou, H.; Kelliher, T.; Zhang, X.; et al. Rice Haploid Inducer Development by Genome Editing. Methods Mol. Biol. 2021, 2238, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shi, C.; Li, S.; Zhang, B.; Luo, P.; Peng, X.; Zhao, P.; Dresselhaus, T.; Sun, M.X. A female in vivo haploid-induction system via mutagenesis of egg cell-specific peptidases. Mol. Plant 2023, 16, 471–480. [Google Scholar] [CrossRef]
- Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Liang, D.; Liu, J.; Sahoo, G.; Kelliher, T. OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 2018, 4, 530–533. [Google Scholar] [CrossRef]
- Xie, E.; Li, Y.; Tang, D.; Lv, Y.; Shen, Y.; Cheng, Z. A strategy for generating rice apomixis by gene editing. J. Integr. Plant Biol. 2019, 61, 911–916. [Google Scholar] [CrossRef]
- Zhao, X.; Yuan, K.; Liu, Y.; Zhang, N.; Yang, L.; Zhang, Y.; Wang, Y.; Ji, J.; Fang, Z.; Han, F.; et al. In vivo maternal haploid induction based on genome editing of DMP in Brassica oleracea. Plant Biotechnol. J. 2022, 20, 2242–2244. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Y.; Chen, B.; Liu, J.; Wang, D.; Li, M.; Qi, X.; Liu, C.; Boutilier, K.; Chen, S. Establishment of a dmp based maternal haploid induction system for polyploid Brassica napus and Nicotiana tabacum. J. Integr. Plant Biol. 2022, 64, 1281–1294. [Google Scholar] [CrossRef]
- Wang, N.; Xia, X.; Jiang, T.; Li, L.; Zhang, P.; Niu, L.; Cheng, H.; Wang, K.; Lin, H. In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol. J. 2022, 20, 22–24. [Google Scholar] [CrossRef]
- Liu, C.; Zhong, Y.; Qi, X.; Chen, M.; Liu, Z.; Chen, C.; Tian, X.; Li, J.; Jiao, Y.; Wang, D.; et al. Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat. Plant Biotechnol. J. 2020, 18, 316–318. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Yu, K.; Wei, J.; Gui, H.; Liu, C.; Liang, D.; Wang, Y.; Zhou, H.; Carlin, R.; Rich, R.; et al. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat. Biotechnol. 2020, 38, 1397–1401. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Geng, S.; Zhang, H.; Jia, M.; Wang, Z.; Deng, Z.; Tao, S.; Liao, R.; Wang, F.; Kong, X.; et al. Matrilineal empowers wheat pollen with haploid induction potency by triggering postmitosis reactive oxygen species activity. New Phytol. 2022, 233, 2405–2414. [Google Scholar] [CrossRef] [PubMed]
- Kelliher, T.; Starr, D.; Su, X.J.; Tang, G.Z.; Chen, Z.Y.; Carter, J.; Wittich, P.E.; Dong, S.J.; Green, J.; Burch, E.; et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef]
- Dong, L.; Li, L.; Liu, C.; Liu, C.; Geng, S.; Li, X.; Huang, C.; Mao, L.; Chen, S.; Xie, C. Genome Editing and Double-Fluorescence Proteins Enable Robust Maternal Haploid Induction and Identification in Maize. Mol. Plant 2018, 11, 1214–1217. [Google Scholar] [CrossRef]
- Wang, B.B.; Zhu, L.; Zhao, B.B.; Zhao, Y.P.; Xie, Y.R.; Zheng, Z.G.; Li, Y.Y.; Sun, J.; Wang, H.Y. Development of a Haploid-Inducer Mediated Genome Editing System for Accelerating Maize Breeding. Mol. Plant 2019, 12, 597–602. [Google Scholar] [CrossRef]
- Qi, X.; Gao, H.; Lv, R.; Mao, W.; Zhu, J.; Liu, C.; Mao, L.; Li, X.; Xie, C. CRISPR/dCas-mediated gene activation toolkit development and its application for parthenogenesis induction in maize. Plant Commun. 2023, 4, 100449. [Google Scholar] [CrossRef]
- Li, Y.; Li, D.; Xiao, Q.; Wang, H.; Wen, J.; Tu, J.; Shen, J.; Fu, T.; Yi, B. An in planta haploid induction system in Brassica napus. J. Integr. Plant Biol. 2022, 64, 1140–1144. [Google Scholar] [CrossRef]
- Cheng, Z.; Sun, Y.; Yang, S.; Zhi, H.; Yin, T.; Ma, X.; Zhang, H.; Diao, X.; Guo, Y.; Li, X.; et al. Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica). Plant Biotechnol. J. 2021, 19, 1089–1091. [Google Scholar] [CrossRef]
- Tang, H.; Wang, K.; Zhang, S.; Han, Z.; Chang, Y.; Qiu, Y.; Yu, M.; Du, L.; Ye, X. A fast technique for visual screening of wheat haploids generated from TaMTL-edited mutants carrying anthocyanin markers. Plant Commun. 2023, 4, 100569. [Google Scholar] [CrossRef]
- Wang, D.; Zhong, Y.; Feng, B.; Qi, X.; Yan, T.; Liu, J.; Guo, S.; Wang, Y.; Liu, Z.; Cheng, D.; et al. The RUBY reporter enables efficient haploid identification in maize and tomato. Plant Biotechnol. J. 2023, 21, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. GABA-enriched tomato is first CRISPR-edited food to enter market. Nat. Biotechnol. 2022, 40, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing Crop Transformation in the Era of Genome Editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef]
- Wang, N.; Ryan, L.; Sardesai, N.; Wu, E.; Lenderts, B.; Lowe, K.; Che, P.; Anand, A.; Worden, A.; van Dyk, D.; et al. Leaf transformation for efficient random integration and targeted genome modification in maize and sorghum. Nat. Plants 2023, 9, 255–270. [Google Scholar] [CrossRef]
- Gaillochet, C.; Pena Fernandez, A.; Goossens, V.; D’Halluin, K.; Drozdzecki, A.; Shafie, M.; Van Duyse, J.; Van Isterdael, G.; Gonzalez, C.; Vermeersch, M.; et al. Systematic optimization of Cas12a base editors in wheat and maize using the ITER platform. Genome Biol. 2023, 24, 6. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Q.; Wang, X.; Jin, X.; Peng, J.; Zhang, H.; Wang, Y. CRISPR/Cas Technology Revolutionizes Crop Breeding. Plants 2023, 12, 3119. https://doi.org/10.3390/plants12173119
Tang Q, Wang X, Jin X, Peng J, Zhang H, Wang Y. CRISPR/Cas Technology Revolutionizes Crop Breeding. Plants. 2023; 12(17):3119. https://doi.org/10.3390/plants12173119
Chicago/Turabian StyleTang, Qiaoling, Xujing Wang, Xi Jin, Jun Peng, Haiwen Zhang, and Youhua Wang. 2023. "CRISPR/Cas Technology Revolutionizes Crop Breeding" Plants 12, no. 17: 3119. https://doi.org/10.3390/plants12173119
APA StyleTang, Q., Wang, X., Jin, X., Peng, J., Zhang, H., & Wang, Y. (2023). CRISPR/Cas Technology Revolutionizes Crop Breeding. Plants, 12(17), 3119. https://doi.org/10.3390/plants12173119





