Improvements in Protoplast Isolation Protocol and Regeneration of Different Cabbage (Brassica oleracea var. capitata L.) Cultivars
Abstract
1. Introduction
2. Results
2.1. Optimization of Protoplast Isolation Protocol
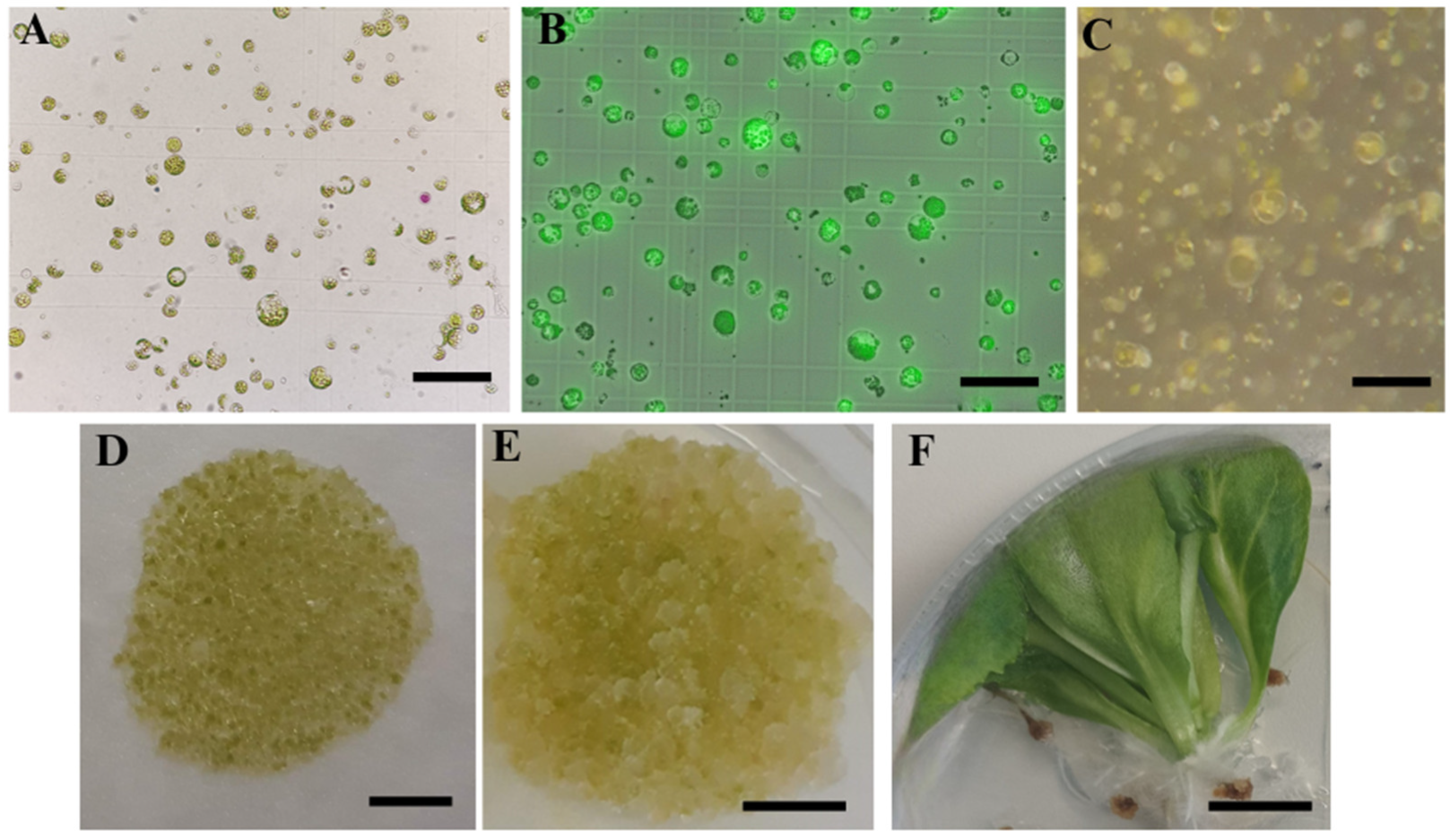
2.2. Protoplast Cultivation and Regeneration
3. Discussion
4. Materials and Methods
4.1. Donor Plant Material
4.2. Protoplast Isolation
4.3. Protoplast Regeneration
| Medium Name | Medium Composition | Reference | Protocol |
|---|---|---|---|
| CPP | Kao and Michayluk [28] macro- and microelements and organic acids, B5 (Gamborg et al. [53]) vitamins, 74 g/L glucose, 250 mg/L casein enzymatic hydrolysate, 0.1 mg/L 2,4-D, 0.2 mg/L zeatin; pH 5.6 | [23] | 1, 2 |
| CPPD | Kao and Michayluk [28] macro- and microelements and 0.4X organic acids, B5 (Gamborg et al. [53]) vitamins, 20 g/L mannitol, 30 g/L sucrose, 250 mg/L casein enzymatic hydrolysate, 0.1 mg/L NAA, 0.2 mg/L zeatin; pH 5.6 | [49] | 2 |
| J1 | MS medium NH4NO3 free (#M0238, Duchefa), 30 g/L sucrose, 60 g/L myo-inositol, 0.4 mg/L thiamine-HCl, 2 mg/L 2,4-D, 0.5 mg/L BAP; pH 5.8 | [33] | 3 |
| J2 | MS basal macro- and micronutrients (#M0221, Duchefa), 30 g/L sucrose, 60 g/L myo-inositol, 0.4 mg/L thiamine-HCl, 2 mg/L 2,4-D, 0.5 mg/L BAP; pH 5.8 | [33] | 3 |
5. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Šamec, D.; Pavlović, I.; Salopek-Sondi, B. White Cabbage (Brassica oleracea Var. Capitata f. Alba): Botanical, Phytochemical and Pharmacological Overview. Phytochem. Rev. 2017, 16, 117–135. [Google Scholar] [CrossRef]
- Le, T.N.; Luong, H.Q.; Li, H.-P.; Chiu, C.-H.; Hsieh, P.-C. Broccoli (Brassica oleracea L. Var. Italica) Sprouts as the Potential Food Source for Bioactive Properties: A Comprehensive Study on In Vitro Disease Models. Foods 2019, 8, 532. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.R.; Anthony, P.; Power, J.B.; Lowe, K.C. Plant Protoplasts: Status and Biotechnological Perspectives. Biotechnol. Adv. 2005, 23, 131–171. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.-J.; Yuan, J.-L.; Wu, F.-H.; Yuan, Y.-H.; Cheng, Q.-W.; Hsu, C.-T.; Lin, C.-S. Protoplasts: From Isolation to CRISPR/Cas Genome Editing Application. Front. Genome Ed. 2021, 3, 717017. [Google Scholar] [CrossRef] [PubMed]
- Earle, E.D.; Cardi, T.; Dickson, M.H.; Hansen, L.N.; Heath, D.W.; Ren, J.-P.; Sigareva, M. Contributions of Protoplast Fusion to Improvement of Brassica Crops. In Plant Biotechnology and In Vitro Biology in the 21st Century: Proceedings of the IXth International Congress of the International Association of Plant Tissue Culture and Biotechnology, Jerusalem, Israel, 14–19 June 1998; Altman, A., Ziv, M., Izhar, S., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 131–134. ISBN 978-9-40114-661-6. [Google Scholar]
- Christey, M.C.; Makaroff, C.A.; Earle, E.D. Atrazine-Resistant Cytoplasmic Male-Sterile-Nigra Broccoli Obtained by Protoplast Fusion between Cytoplasmic Male-Sterile Brassica oleracea and Atrazine-Resistant Brassica Campestris. Theor. Appl. Genet. 1991, 83, 201–208. [Google Scholar] [CrossRef]
- Ren, J.P.; Dickson, M.H.; Earle, E.D. Improved Resistance to Bacterial Soft Rot by Protoplast Fusion between Brassica Rapa and B. Oleracea. Theor. Appl. Genet. 2000, 100, 810–819. [Google Scholar] [CrossRef]
- Sahab, S.; Hayden, M.J.; Mason, J.; Spangenberg, G. Mesophyll Protoplasts and PEG-Mediated Transfections: Transient Assays and Generation of Stable Transgenic Canola Plants. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; Volume 1864, pp. 131–152. ISBN 978-1-49398-778-8. [Google Scholar]
- Ran, Y.; Liang, Z.; Gao, C. Current and Future Editing Reagent Delivery Systems for Plant Genome Editing. Sci. China Life Sci. 2017, 60, 490–505. [Google Scholar] [CrossRef]
- Li, W.; Teng, F.; Li, T.; Zhou, Q. Simultaneous Generation and Germline Transmission of Multiple Gene Mutations in Rat Using CRISPR-Cas Systems. Nat. Biotechnol. 2013, 31, 684–686. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome Editing in Rice and Wheat Using the CRISPR/Cas System. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef]
- Subburaj, S.; Chung, S.J.; Lee, C.; Ryu, S.M.; Kim, D.H.; Kim, J.S.; Bae, S.; Lee, G.J. Site-Directed Mutagenesis in Petunia × Hybrida Protoplast System Using Direct Delivery of Purified Recombinant Cas9 Ribonucleoproteins. Plant Cell Rep. 2016, 35, 1535–1544. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-Free Genome Editing in Plants with Preassembled CRISPR-Cas9 Ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Viola, R.; Jung, M.; Koo, O.; Kim, S.; Kim, J.; Velasco, R.; Kanchiswamy, C.N. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.-S.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P. Genome Editing in Potato via CRISPR-Cas9 Ribonucleoprotein Delivery. Physiol. Plant. 2018, 164, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Cocking, E.C. A Method for the Isolation of Plant Protoplasts and Vacuoles. Nature 1960, 187, 962–963. [Google Scholar] [CrossRef]
- Duarte, P.; Ribeiro, D.; Carqueijeiro, I.; Bettencourt, S.; Sottomayor, M. Chapter 13 Protoplast Transformation as a Plant-Transferable Transient Expression System. In Biotechnology of Plant Secondary Metabolism: Methods and Protocols; Humana Press: New York, NY, USA, 2016; Volume 1405, pp. 137–148. [Google Scholar] [CrossRef]
- Navrátilová, B. Protoplast Cultures and Protoplast Fusion Focused on Brassicaceae: A Review. Hortic. Sci. 2004, 31, 140–157. [Google Scholar] [CrossRef]
- Davey, M.R.; Anthony, P.; Power, J.B.; Lowe, K.C. Plant Protoplast Technology: Current Status. Acta Physiol. Plant. 2005, 27, 117–130. [Google Scholar] [CrossRef]
- Chen, L.P.; Zhang, M.F.; Xiao, Q.B.; Wu, J.G.; Hirata, Y. Plant Regeneration from Hypocotyl Protoplasts of Red Cabbage (Brassica oleracea) by Using Nurse Cultures. Plant Cell Tissue Organ Cult. 2004, 77, 133–138. [Google Scholar] [CrossRef]
- Kaur, N.; Vyvadilová, M.; Klíma, M.; Bechyně, M. A Simple Procedure for Mesophyll Protoplast Culture and Plant Regeneration in Brassica oleracea L. and Brassica napus L. Czech J. Genet. Plant Breed. 2018, 42, 103–110. [Google Scholar] [CrossRef]
- Chikkala, V.R.N.; Nugent, G.D.; Dix, P.J.; Stevenson, T.W. Regeneration from Leaf Explants and Protoplasts of Brassica oleracea Var. Botrytis (Cauliflower). Sci. Hortic. 2009, 119, 330–334. [Google Scholar] [CrossRef]
- Kiełkowska, A.; Adamus, A. An Alginate-Layer Technique for Culture of Brassica oleracea L. Protoplasts. Vitr. Cell. Dev. Biol.–Plant 2012, 48, 265–273. [Google Scholar] [CrossRef]
- Lin, C.S.; Hsu, C.T.; Yang, L.H.; Lee, L.Y.; Fu, J.Y.; Cheng, Q.W.; Wu, F.H.; Hsiao, H.C.W.; Zhang, Y.; Zhang, R.; et al. Application of Protoplast Technology to CRISPR/Cas9 Mutagenesis: From Single-Cell Mutation Detection to Mutant Plant Regeneration. Plant Biotechnol. J. 2018, 16, 1295–1310. [Google Scholar] [CrossRef]
- Reed, K.M.; Bargmann, B.O.R. Protoplast Regeneration and Its Use in New Plant Breeding Technologies. Front. Genome Ed. 2021, 3, 734951. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sandgrind, S.; Moss, O.; Guan, R.; Ivarson, E.; Wang, E.S.; Kanagarajan, S.; Zhu, L.-H. Efficient Protoplast Regeneration Protocol and CRISPR/Cas9-Mediated Editing of Glucosinolate Transporter (GTR) Genes in Rapeseed (Brassica napus L.). Front. Plant Sci. 2021, 12, 680859. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Kao, K.N.; Michayluk, M.R. Nutritional Requirements for Growth of Vicia Hajastana Cells and Protoplasts at a Very Low Population Density in Liquid Media. Planta 1975, 126, 105–110. [Google Scholar] [CrossRef]
- Kiełkowska, A.; Adamus, A. Embedding in Filter-Sterilized Alginate Enhances Brassica oleracea L. Protoplast Culture. Acta Biol. Cracoviensia Ser. Bot. 2014, 56, 20–26. [Google Scholar] [CrossRef]
- Kiełkowska, A.; Adamus, A. Early Studies on the Effect of Peptide Growth Factor Phytosulfokine-α on Brassica oleracea var. Capitata L. Protoplasts. Acta Soc. Bot. Pol. 2017, 86, 1–11. [Google Scholar] [CrossRef]
- Kiełkowska, A.; Adamus, A. Peptide Growth Factor Phytosulfokine-α Stimulates Cell Divisions and Enhances Regeneration from B. oleracea Var. Capitata L. Protoplast Culture. J. Plant Growth Regul. 2019, 38, 931–944. [Google Scholar] [CrossRef]
- Kiełkowska, A.; Adamus, A. Exogenously Applied Polyamines Reduce Reactive Oxygen Species, Enhancing Cell Division and the Shoot Regeneration from Brassica oleracea L. Var. Capitata Protoplasts. Agronomy 2021, 11, 735. [Google Scholar] [CrossRef]
- Jie, E.-Y.; Kim, S.-W.; Jang, H.-R.; In, D.-S.; Liu, J.-R. Myo-Inositol Increases the Plating Efficiency of Protoplast Derived from Cotyledon of Cabbage (Brassica oleracea Var. Capitata). J. Plant Biotechnol. 2011, 38, 69–76. [Google Scholar] [CrossRef][Green Version]
- Yu, J.; Tu, L.; Subburaj, S.; Bae, S.; Lee, G.-J. Simultaneous Targeting of Duplicated Genes in Petunia Protoplasts for Flower Color Modification via CRISPR-Cas9 Ribonucleoproteins. Plant Cell Rep. 2021, 40, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.-S.; Samuelsson, M.; Hofvander, P. Efficient Targeted Multiallelic Mutagenesis in Tetraploid Potato (Solanum Tuberosum) by Transient CRISPR-Cas9 Expression in Protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Dovzhenko, A.; Bergen, U.; Koop, H.-U. Thin-Alginate-Layer Technique for Protoplast Culture of Tobacco Leaf Protoplasts: Shoot Formation in Less than Two Weeks. Protoplasma 1998, 204, 114–118. [Google Scholar] [CrossRef]
- Hu, Q.; Andersen, S.B.; Hansen, L.N. Plant Regeneration Capacity of Mesophyll Protoplasts from Brassica napus and Related Species. Plant Cell Tissue Organ Cult. 1999, 59, 189–196. [Google Scholar] [CrossRef]
- Simmonds, D.H.; Long, N.E.; Keller, W.A. High Plating Efficiency and Plant Regeneration Frequency in Low Density Protoplast Cultures Derived from an Embryogenic Brassica napus Cell Suspension. Plant Cell Tissue Organ Cult. 1991, 27, 231–241. [Google Scholar] [CrossRef]
- Hansen, L.N.; Earle, E.D. Novel Flowering and Fatty Acid Characters in Rapid Cycling Brassica napus L. Resynthesized by Protoplast Fusion. Plant Cell Rep. 1994, 14, 151–156. [Google Scholar] [CrossRef]
- Chakrabarty, R.; Viswakarma, N.; Bhat, S.R.; Kirti, P.B.; Singh, B.D.; Chopra, V.L. Agrobacterium-Mediated Transformation of Cauliflower: Optimization of Protocol and Development of Bt-Transgenic Cauliflower. J. Biosci. 2002, 27, 495–502. [Google Scholar] [CrossRef]
- Fidler, J.; Żyła, N.; Babula-Skowrońska, D. Impact of the Brassica oleracea Genome on Breeding and Crop Improvement. In The Brassica oleracea Genome; Liu, S., Snowdon, R., Kole, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 107–133. ISBN 978-3-03031-005-9. [Google Scholar]
- Beard, K.M.; Boling, A.W.H.; Bargmann, B.O.R. Protoplast Isolation, Transient Transformation, and Flow-Cytometric Analysis of Reporter-Gene Activation in Cannabis sativa L. Ind. Crops Prod. 2021, 164, 113360. [Google Scholar] [CrossRef]
- Moon, K.-B.; Park, J.-S.; Park, S.-J.; Lee, H.-J.; Cho, H.-S.; Min, S.-R.; Park, Y.-I.; Jeon, J.-H.; Kim, H.-S. A More Accessible, Time-Saving, and Efficient Method for In Vitro Plant Regeneration from Potato Protoplasts. Plants 2021, 10, 781. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, G.; Chen, Z.; Han, J.; Hu, Y.; Wang, K. Optimization of Protoplast Isolation, Transformation and Its Application in Sugarcane (Saccharum spontaneum L). Crop J. 2021, 9, 133–142. [Google Scholar] [CrossRef]
- Ren, R.; Gao, J.; Yin, D.; Li, K.; Lu, C.; Ahmad, S.; Wei, Y.; Jin, J.; Zhu, G.; Yang, F. Highly Efficient Leaf Base Protoplast Isolation and Transient Expression Systems for Orchids and Other Important Monocot Crops. Front. Plant Sci. 2021, 12, 626015. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.-H.; Shen, S.-C.; Lee, L.-Y.; Lee, S.-H.; Chan, M.-T.; Lin, C.-S. Tape-Arabidopsis Sandwich—A Simpler Arabidopsis Protoplast Isolation Method. Plant Methods 2009, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A Highly Efficient Rice Green Tissue Protoplast System for Transient Gene Expression and Studying Light/Chloroplast-Related Processes. Plant Methods 2011, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Maćkowska, K.; Jarosz, A.; Grzebelus, E. Plant Regeneration from Leaf-Derived Protoplasts within the Daucus Genus: Effect of Different Conditions in Alginate Embedding and Phytosulfokine Application. Plant Cell Tissue Organ Cult. 2014, 117, 241–252. [Google Scholar] [CrossRef]
- Grzebelus, E.; Szklarczyk, M.; Baranski, R. An Improved Protocol for Plant Regeneration from Leaf- and Hypocotyl-Derived Protoplasts of Carrot. Plant Cell Tissue Organ Cult. 2012, 109, 101–109. [Google Scholar] [CrossRef]
- Glimelius, K. High Growth Rate and Regeneration Capacity of Hypocotyl Protoplasts in Some Brassicaceae. Physiol. Plant. 1984, 61, 38–44. [Google Scholar] [CrossRef]
- Nugent, G.D.; Coyne, S.; Nguyen, T.T.; Kavanagh, T.A.; Dix, P.J. Nuclear and Plastid Transformation of Brassica oleracea Var. Botrytis (Cauliflower) Using PEG-Mediated Uptake of DNA into Protoplasts. Plant Sci. 2006, 170, 135–142. [Google Scholar] [CrossRef]
- Widholm, J.M. The Use of Fluorescein Diacetate and Phenosafranine for Determining Viability of Cultured Plant Cells. Stain Technol. 1972, 47, 189–194. [Google Scholar] [CrossRef]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient Requirements of Suspension Cultures of Soybean Root Cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
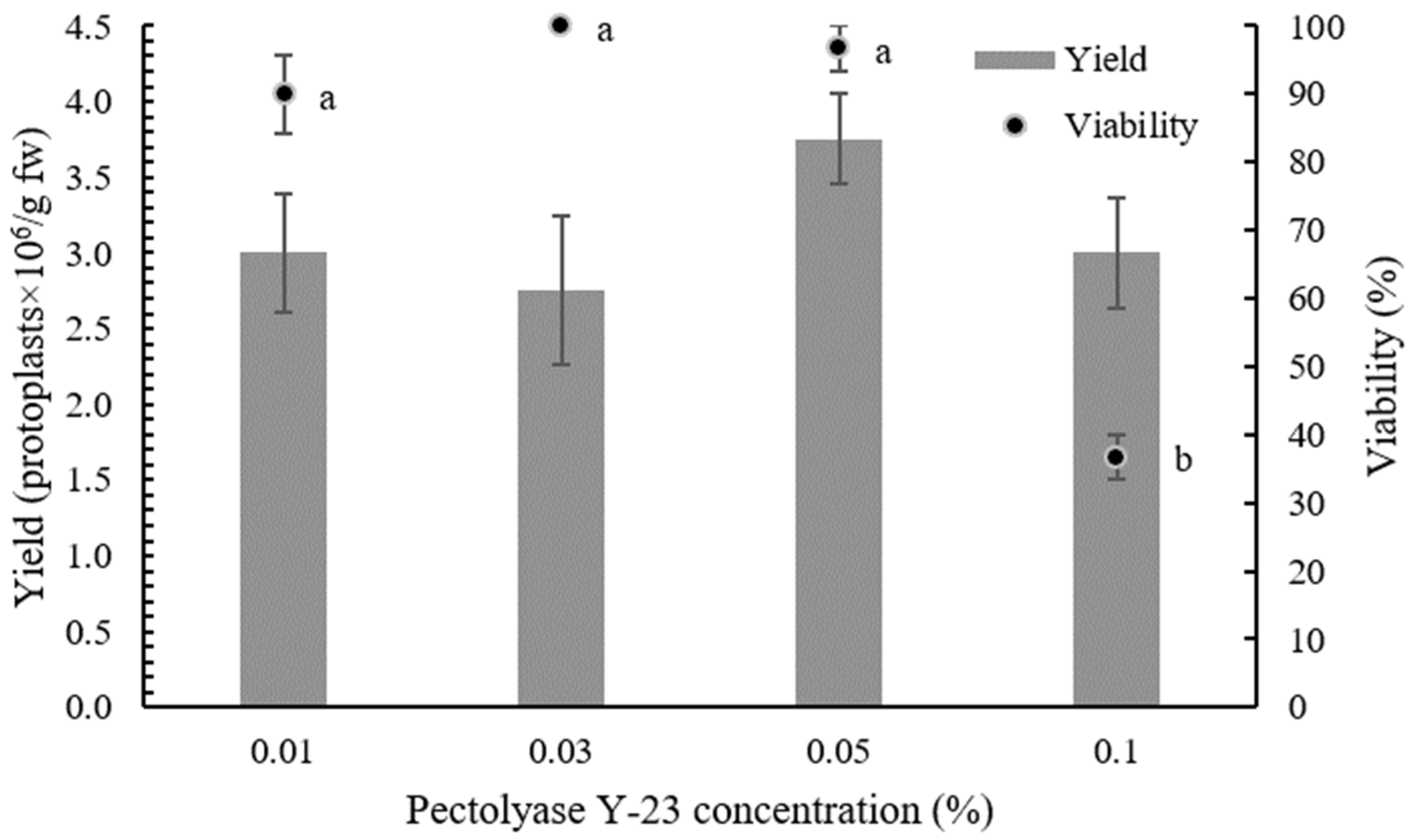
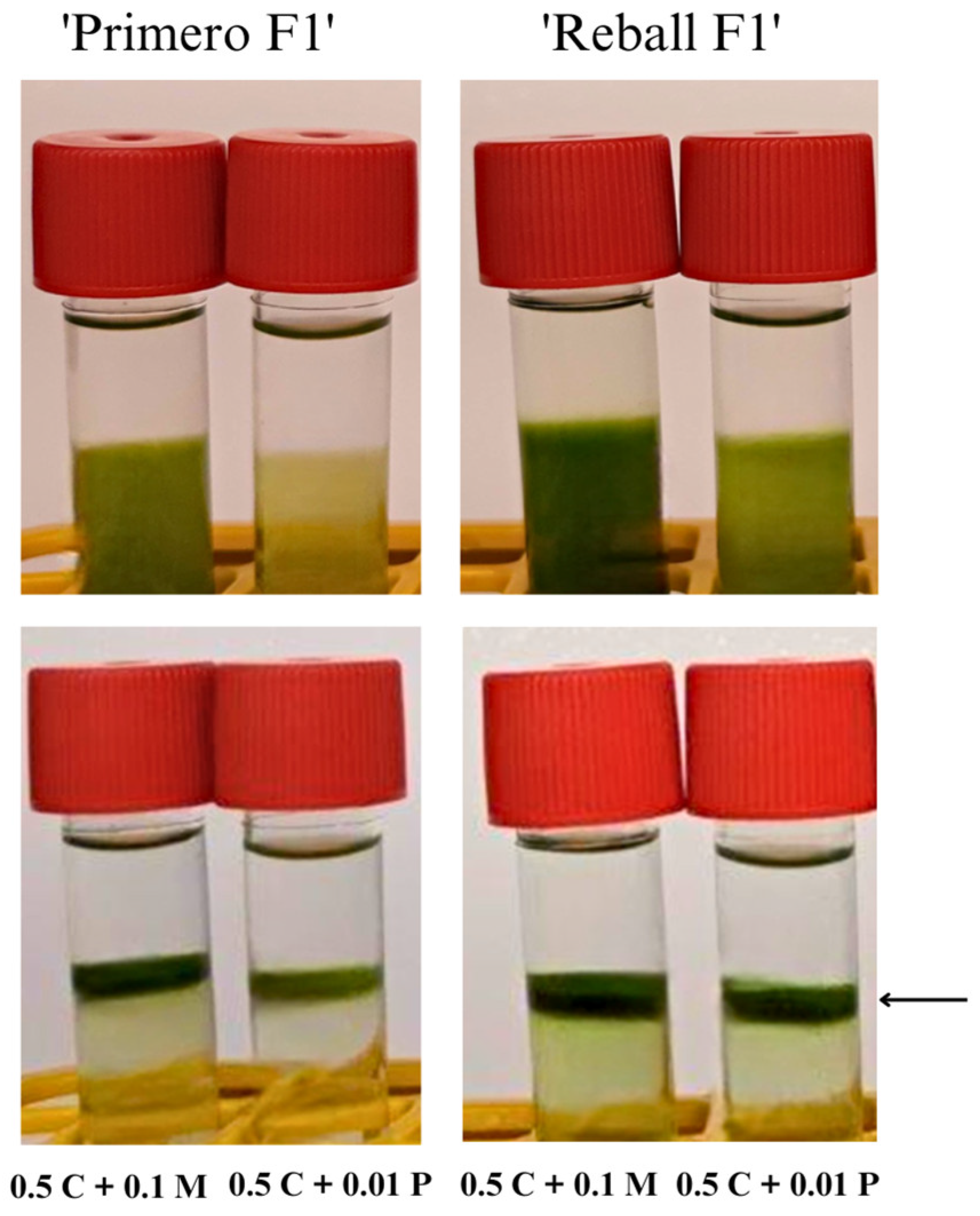
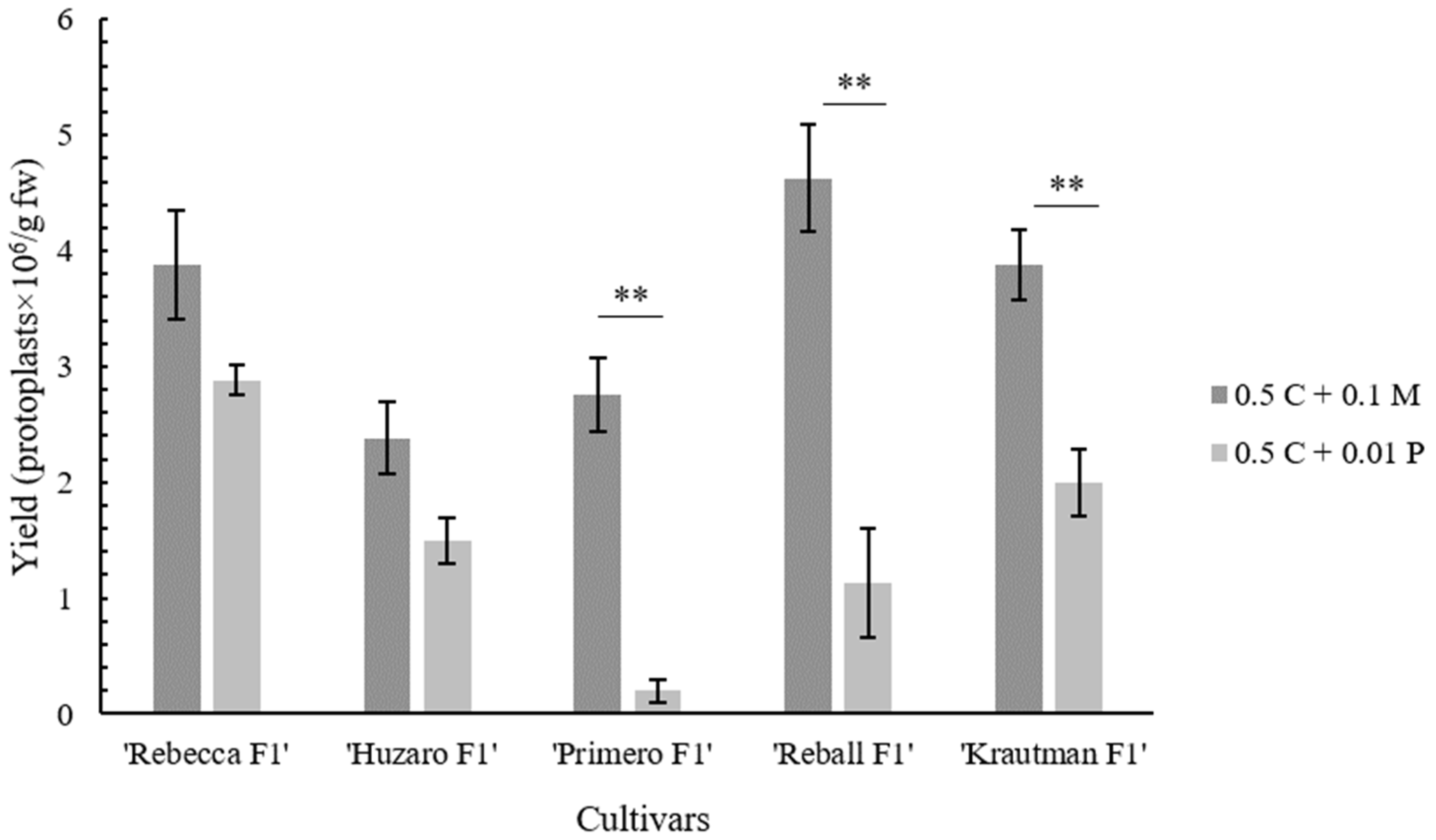
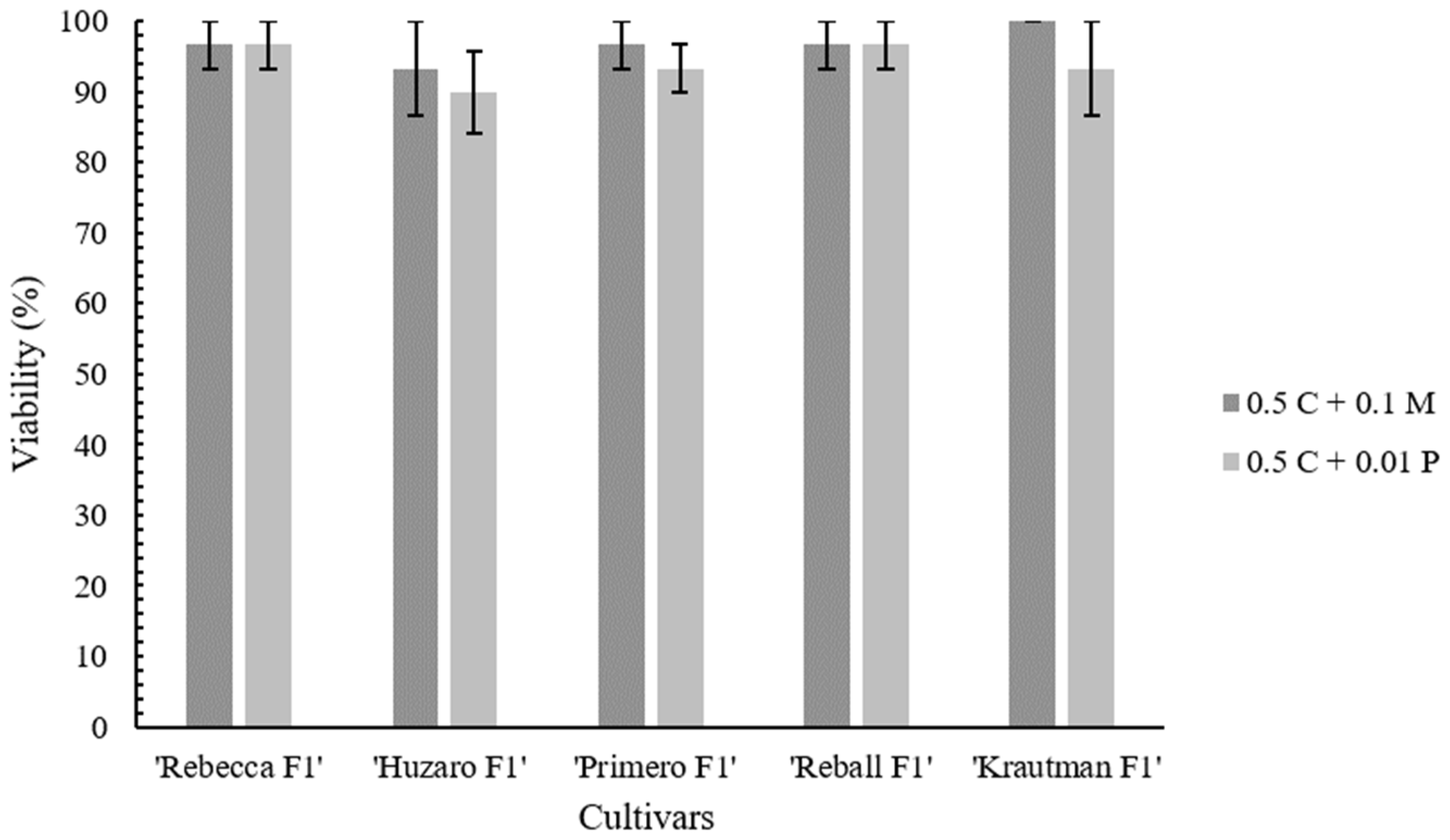
| Cellulase Onozuka RS Concentration (%) | Pectolyase Y-23 Concentration (%) | Yield ± SE (Protoplasts × 106/g fw) a |
|---|---|---|
| 0.5 | 0.01 | 3.00 ± 0.39 a |
| 0.5 | 0.03 | 2.75 ± 0.49 a |
| 0.5 | 0.05 | 3.75 ± 0.30 a |
| 0.5 | 0.1 | 3.00 ± 0.37 a |
| Cultivar | Enzyme Solution Composition | |
|---|---|---|
| 0.5 C + 0.1 M | 0.5 C + 0.01 P | |
| ‘Rebecca F1’ | 3.88 ± 0.47 | 2.88 ± 0.13 |
| ‘Primero F1’ | 2.75 ± 0.32 | 0.20 ± 0.10 |
| ‘Reball F1’ | 4.63 ± 0.47 | 1.13 ± 0.47 |
| ‘Krautman F1’ | 3.88 ± 0.31 | 2.00 ± 0.29 |
| ‘Huzaro F1’ | 2.38 ± 0.31 | 1.50 ± 0.20 |
| Cultivar | Protocol 1 | Protocol 2 | Protocol 3 |
|---|---|---|---|
| ‘Rebecca F1’ | + | + | − |
| ‘Reball F1’ | ++ | ++ | + |
| ‘Krautman F1’ | + | + | − |
| ‘Primero F1’ | − | − | + |
| ‘Huzaro F1’ | ++ | ++ | + |
| Protocol | Medium | PGR Concentration (mg/L) | Regeneration (%) | |
|---|---|---|---|---|
| 1 | RM11 | BAP 1 | 5.9 | |
| 1 | RM2 | BAP 1 | NAA 0.2 | 23.5 |
| 2 | RM1 | BAP 1 | 0 | |
| 2 | RM2 | BAP 1 | NAA 0.2 | 0 |
| 3 | RM3 | BAP 0.5 | 2,4-D 2 | 0 |
| Medium | PGR Concentration (mg/L) | Protocol | |
|---|---|---|---|
| RM1 | BAP 1 | 1, 2 | |
| RM2 | BAP 1 | NAA 0.2 | 1, 2 |
| RM3 | BAP 0.5 | 2,4-D 2 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stajič, E. Improvements in Protoplast Isolation Protocol and Regeneration of Different Cabbage (Brassica oleracea var. capitata L.) Cultivars. Plants 2023, 12, 3074. https://doi.org/10.3390/plants12173074
Stajič E. Improvements in Protoplast Isolation Protocol and Regeneration of Different Cabbage (Brassica oleracea var. capitata L.) Cultivars. Plants. 2023; 12(17):3074. https://doi.org/10.3390/plants12173074
Chicago/Turabian StyleStajič, Ester. 2023. "Improvements in Protoplast Isolation Protocol and Regeneration of Different Cabbage (Brassica oleracea var. capitata L.) Cultivars" Plants 12, no. 17: 3074. https://doi.org/10.3390/plants12173074
APA StyleStajič, E. (2023). Improvements in Protoplast Isolation Protocol and Regeneration of Different Cabbage (Brassica oleracea var. capitata L.) Cultivars. Plants, 12(17), 3074. https://doi.org/10.3390/plants12173074






