Comparative Transcriptome Analysis Reveals the Molecular Basis of Brassica napus in Response to Aphid Stress
Abstract
1. Introduction
2. Results
2.1. Differences in Response to Aphid Stress between APL01 and Holly Varieties
2.2. Transcriptomic Analysis of APL01 and Holly in Response to Aphid Stress
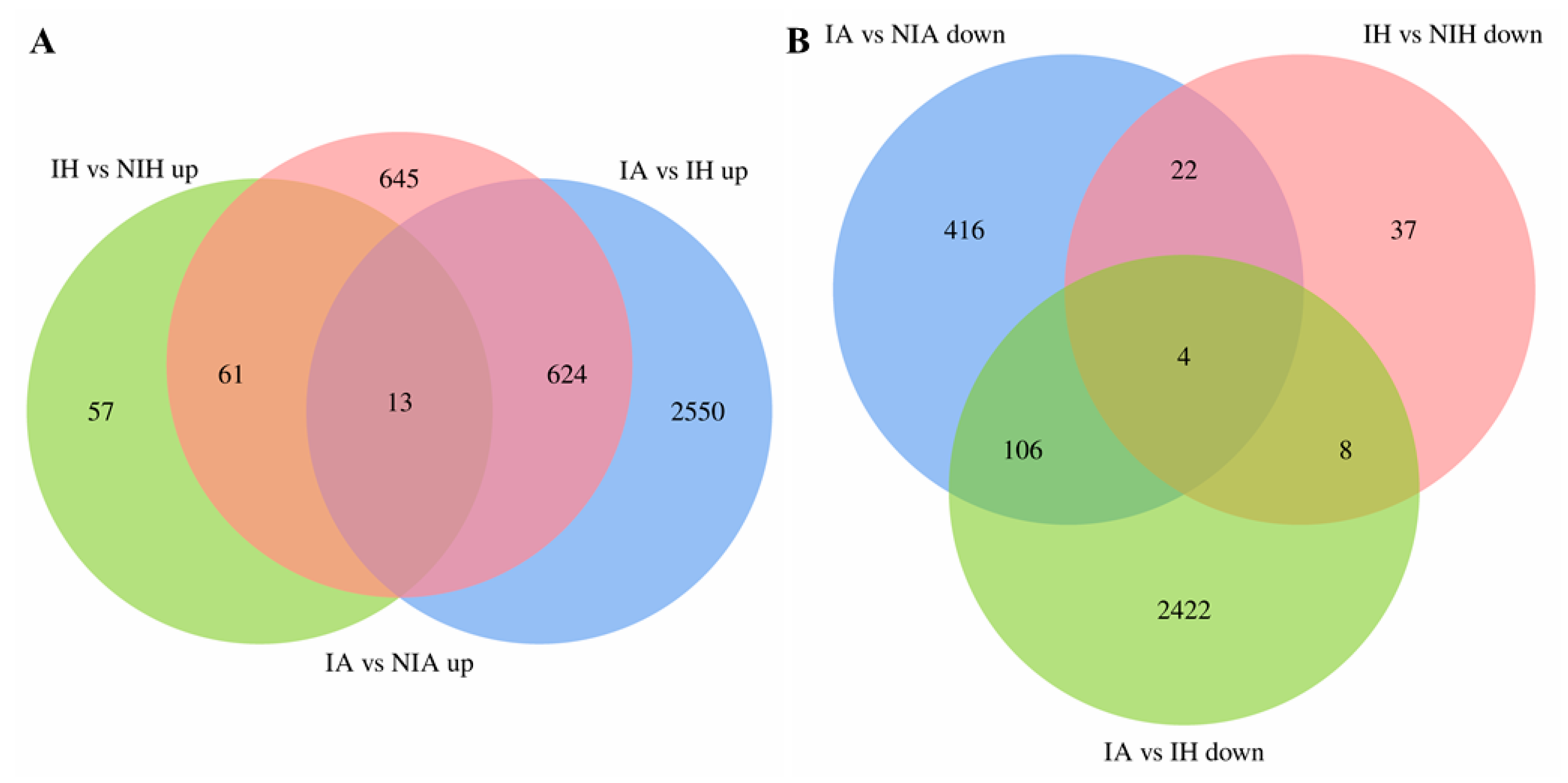
2.3. Common Transcriptomic Characteristics of APL01 and Holly Involved in Aphid Stress Defense
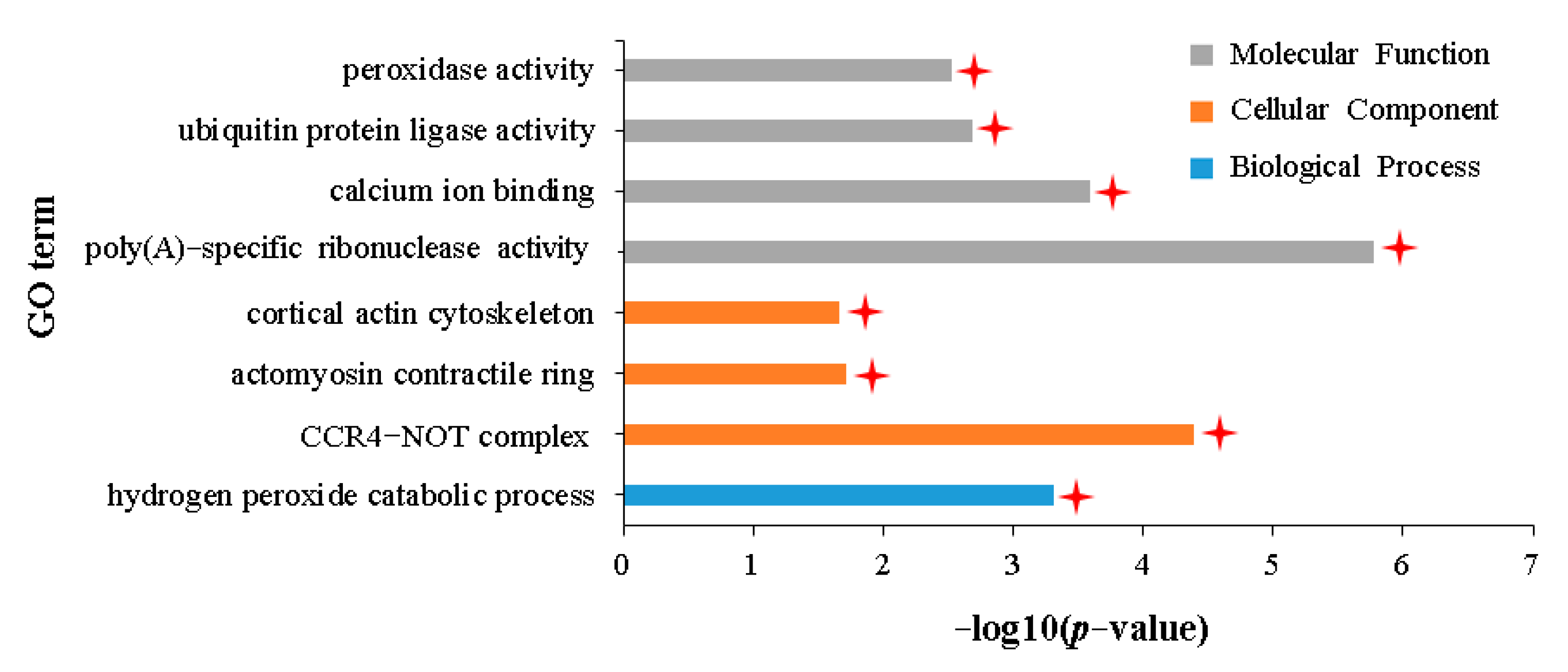
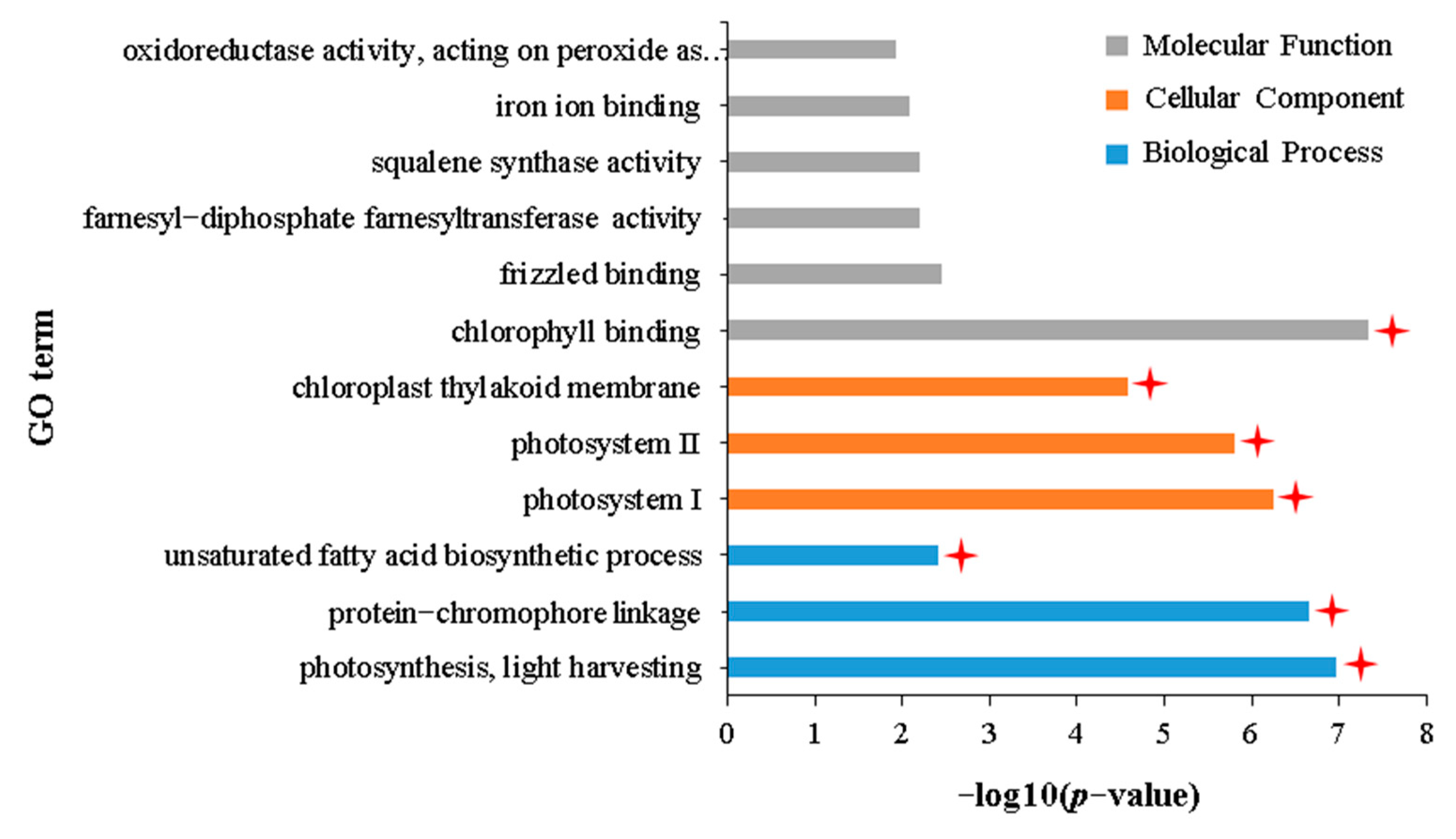
2.4. Transcriptomic Characteristics of APL01 with Stronger Inhibitory Effects on Aphid Proliferation Compared with Holly
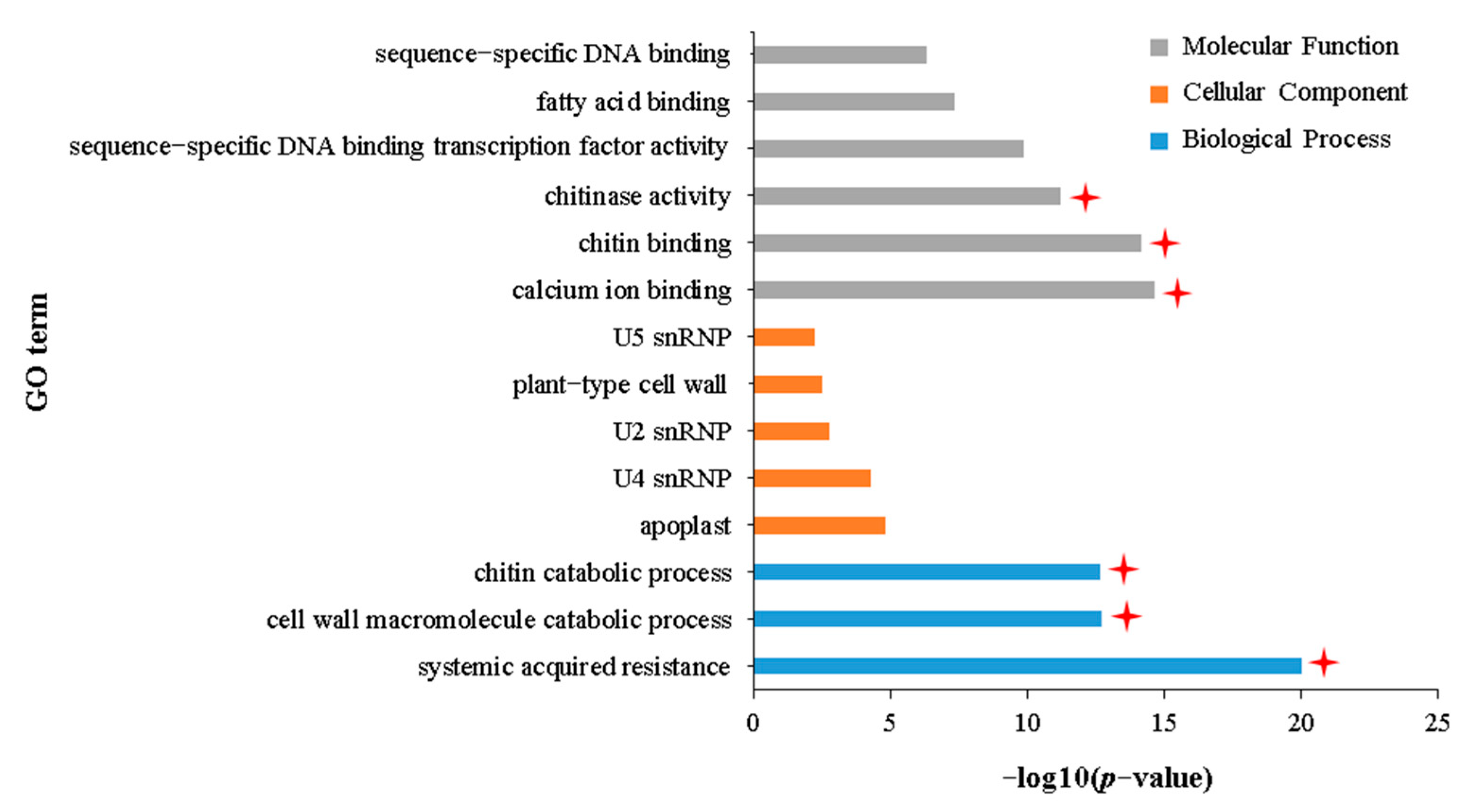
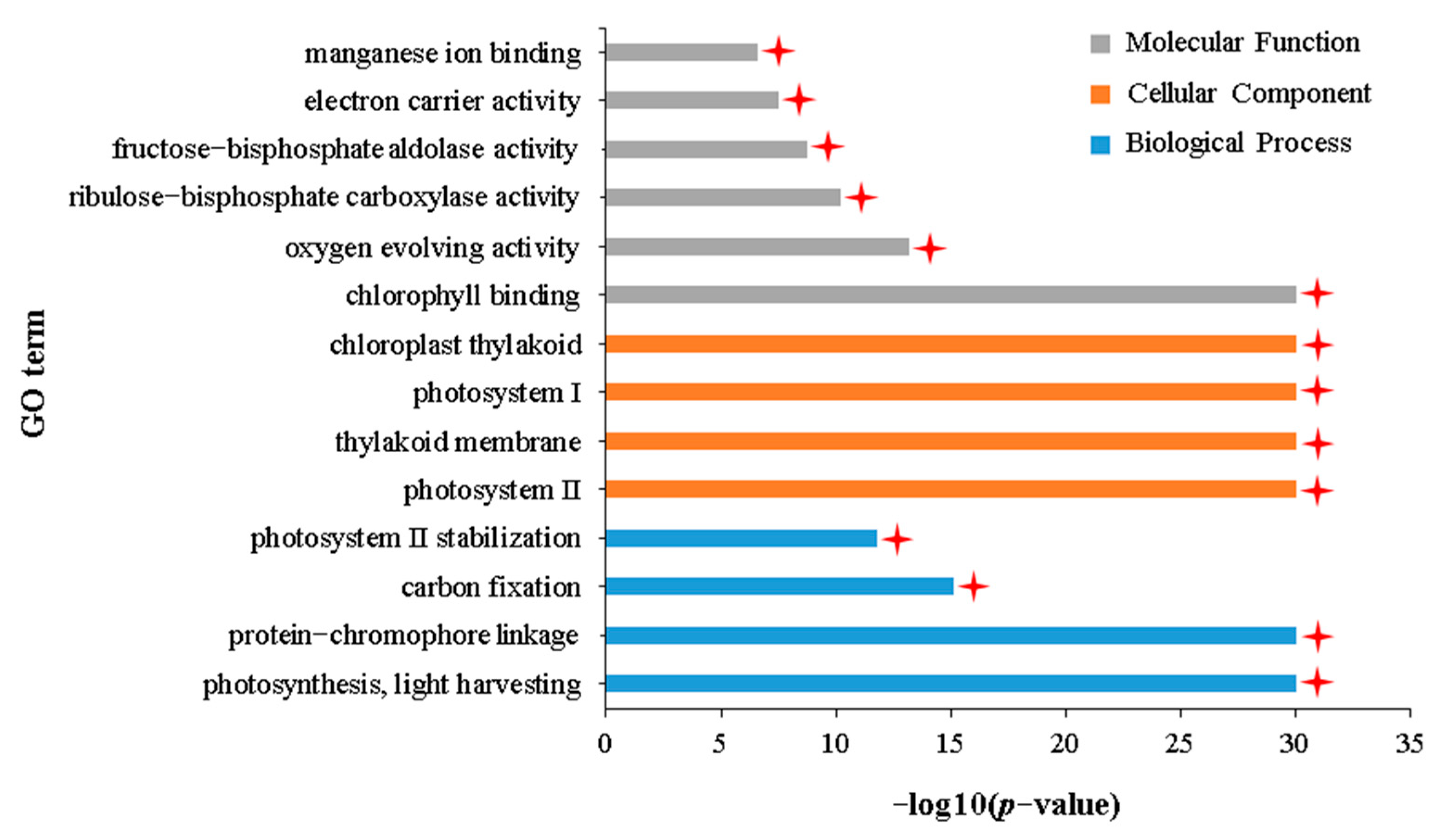
2.5. Common Transcriptomic Characteristics of Growth Inhibition in APL01 and Holly under Aphid Stress
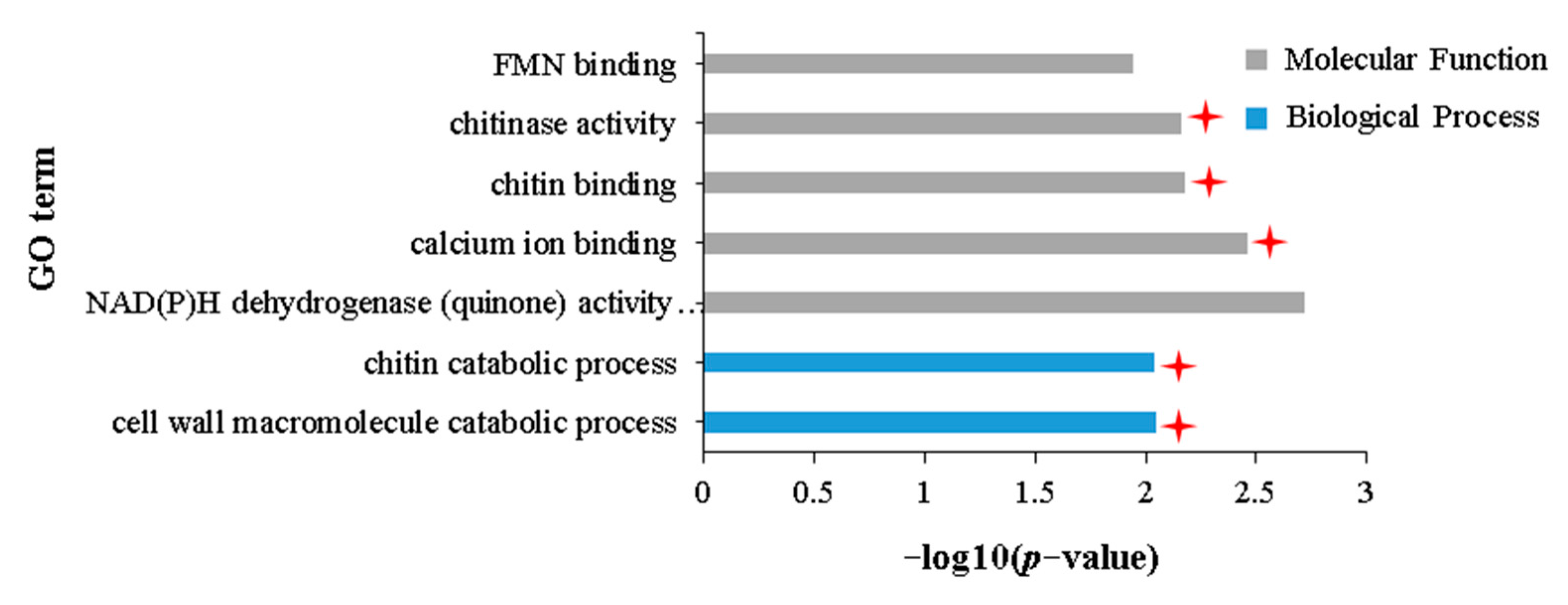
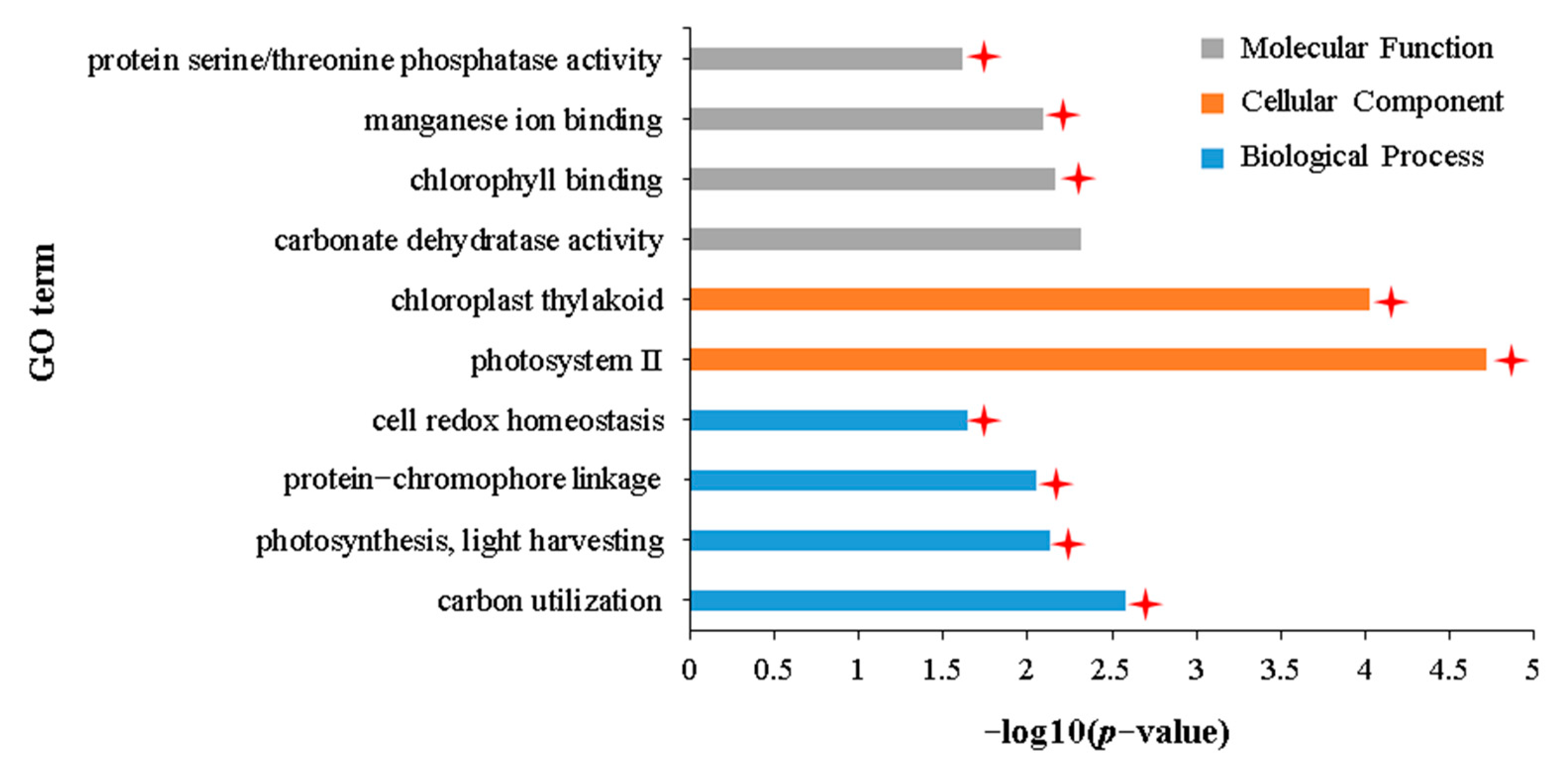
2.6. Transcriptomic Characteristics of the Stronger Tolerance of Holly to Aphid Stress Compared with APL01
2.7. Physiological Characteristics of APL01 and Holly in Response to Aphid Stress
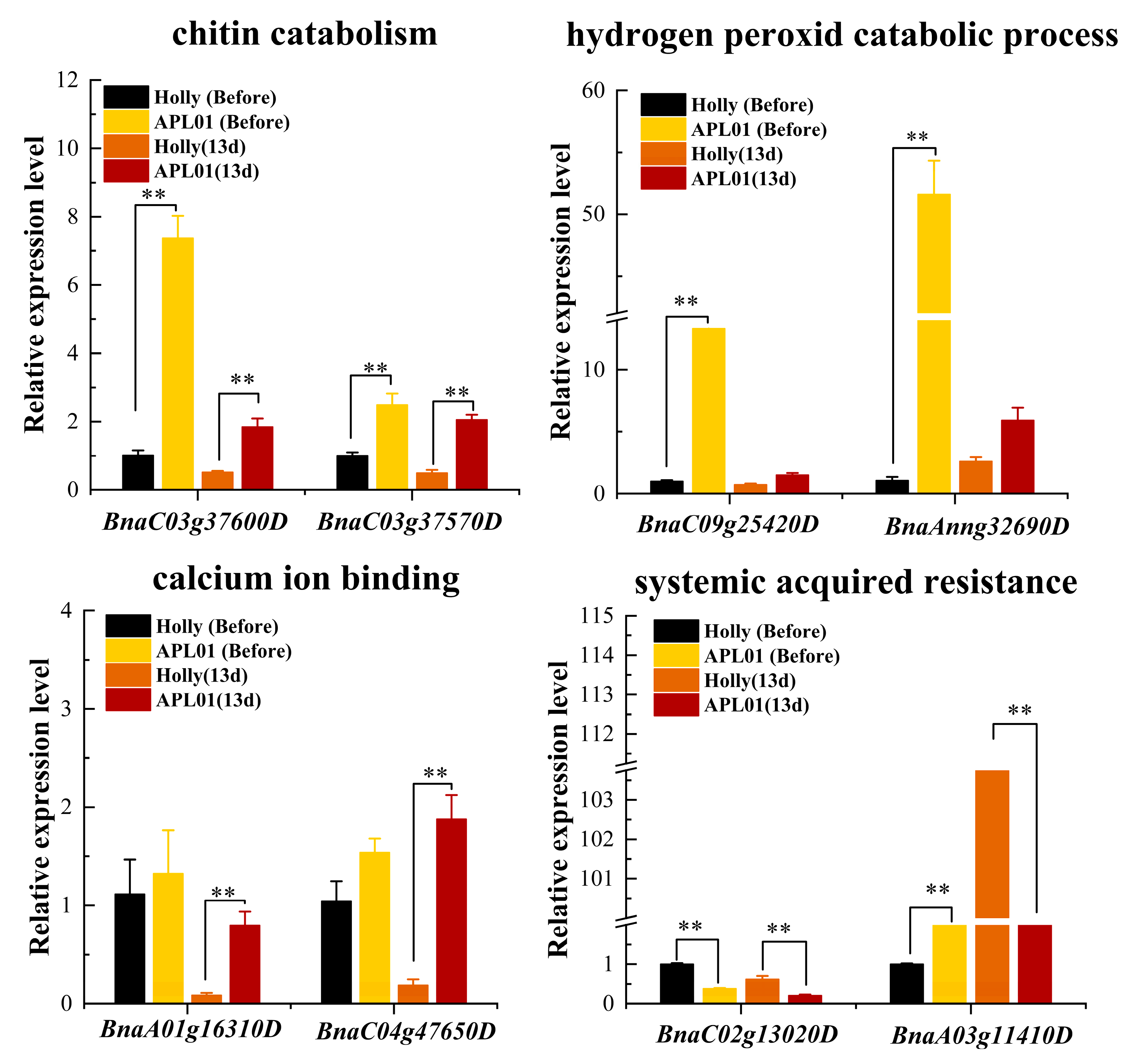
2.8. Screening of Candidate Genes Related to the Rapeseed Defense to Aphid Stress
3. Discussion
3.1. Antibiosis, Antixenosis, and Tolerance all Have a Role in the Response of Brassica napus to Aphid Stress
3.2. Potential Correlation between the Enzyme Activity of Scavenging ROS and the Defense of Brassica napus to Aphid Stress
3.3. Role of Chitinase Activity in the Antibiosis of Brassica napus to Aphid
3.4. The Degree of Inhibition of Photosynthesis under Aphid Stress Directly Determines the Tolerance of Brassica napus to Aphid Stress
3.5. Other Factors Potentially Related to the Defense of Brassica napus to Aphid Stress
3.6. Potential Correlation between Host Plant Defense against Chewing Insects and Piercing–Sucking Insects
4. Materials and Methods
4.1. Plant Material and Aphid Inoculation Experiment
4.2. Toxicity Test
4.3. RNA Sequencing and Bioinformatics Analysis
4.4. Determination of Photosynthetic Characteristics
4.5. Determination of Peroxidase, Catalase, and Chitinase Activities
4.6. Determination of Ribulose-Bisphosphate Carboxylase and Fructose-Bisphosphate Aldolase Activities
4.7. Quantitative Real-Time Reverse Transcription PCR Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Banga, S. Breeding for aphid resistance in rapeseed mustard. In Breeding Insect Resistant Crops for Sustainable Agriculture; Arora, R., Sandhu, S., Eds.; Springer: Berlin, Germany, 2017; pp. 171–199. [Google Scholar]
- Blackman, R.L.; Eastop, V.F. Aphids on the World’s Crops: An Identification and Information Guide, 2nd ed.; John Wiley and Sons: Chichester, UK, 2000; pp. 6–9. [Google Scholar]
- Singh, R.; Singh, G. Aphids and their biocontrol. In Ecofriendly Pest Management for Food Security; Omkar, Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 63–108. [Google Scholar]
- Yates, A.D.; Michel, A. Mechanisms of aphid adaptation to host plant resistance. Curr. Opin. Insect Sci. 2018, 26, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Elzinga, D.A.; Jander, G. The role of protein effectors in plant-aphid interactions. Curr. Opin. Plant Biol. 2013, 16, 451–456. [Google Scholar] [CrossRef]
- Berlandier, F.A. Alkaloid level in narrow-leafed lupin, Lupinus angustifolius, influences green peach aphid reproductive performance. Entomol. Exp. Appl. 1996, 79, 19–24. [Google Scholar] [CrossRef]
- Havlickova, H.; Cvikrova, M.; Eder, J. Changes in the pattem of phenolic adds induced by aphid infestation in two winter wheat cultivars. Bull. OILB SROP 1996, 19, 106–110. [Google Scholar]
- Shepherd, T.; Robertson, G.W.; Griffiths, D.W.; Birch, A.N.E. Epicuticular wax composition in relation to aphid infestation and resistance in red raspberry (Rubus idaeus L.). Phytochemistry 1999, 17, 1866–1875. [Google Scholar]
- Lattanzio, V.; Arpaia, S.; Cardinali, A.; Di Venere, D.; Linsalata, V. Role of endogenous flavonoids in resistance mechanism of Vigna to aphids. J. Agric. Food Chem. 2000, 48, 5316–5320. [Google Scholar] [CrossRef]
- Moran, P.J.; Thompson, G.A. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001, 125, 1074–1085. [Google Scholar] [CrossRef]
- Mohase, L.; van der Westhuizen, A.J. Salicylic acid is involved in resistance responses in the Russian wheat aphid-wheat interaction. J. Plant Physiol. 2002, 159, 585–590. [Google Scholar] [CrossRef]
- Chapman, K.M.; Marchi-Werle, L.; Hunt, T.E.; Heng-Moss, T.M.; Louis, J. Abscisic and jasmonic acids contribute to soybean tolerance to the soybean aphid (Aphis glycines matsumura). Sci. Rep. 2018, 8, 15112–15148. [Google Scholar] [CrossRef]
- Jaouannet, M.; Rodriguez, P.A.; Thorpe, P.; Lenoir, C.J.; MacLeod, R.; Escudero-Martinez, C.; Bos, J.I. Plant immunity in plant-aphid interactions. Front. Plant Sci. 2014, 5, 663. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Boyko, E.V. The molecular bases of plant resistance and defense responses to aphid feeding: Current status. Entomol. Exp. Appl. 2007, 122, 1–16. [Google Scholar] [CrossRef]
- Prado, E.; Tjallingii, W.F. Behavioral evidence for local reduction of aphid-induced resistance. J. Insect Sci. 2007, 7, 48. [Google Scholar] [CrossRef]
- Babikova, Z.; Gilbert, L.; Bruce, T.J.; Birkett, M.; Caulfield, J.C.; Woodcock, C.; Pickett, J.A.; Johnson, D. Underground signals carried through common mycelial networks warn neighbouring plants of aphid attack. Ecol. Lett. 2013, 16, 835–843. [Google Scholar] [CrossRef]
- Turcotte, M.M.; Lochab, A.K.; Turley, N.E.; Johnson, M.T. Plant domestication slows pest evolution. Ecol. Lett. 2015, 18, 907–915. [Google Scholar] [CrossRef]
- Agrawal, A.A. Induced responses to herbivory and increased plant performance. Science 1998, 279, 1201–1202. [Google Scholar] [CrossRef]
- Zust, T.; Agrawal, A.A. Mechanisms and evolution of plant resistance to aphids. Nat. Plants 2016, 2, 15206. [Google Scholar] [CrossRef]
- Prince, D.C.; Drurey, C.; Zipfel, C.; Hogenhout, S.A. The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiol. 2014, 164, 2207–2219. [Google Scholar] [CrossRef]
- Sauge, M.; Lacroze, J.; Poëssel, J.; Pascal, T.; Kervella, J. Induced resistance by Myzus persicae in the peach cultivar ‘Rubira’. Entomol. Exp. Appl. 2002, 102, 29–37. [Google Scholar] [CrossRef]
- Lee, J.S.; Yoo, M.H.; Jung, J.K.; Bilyeu, K.D.; Lee, J.D.; Kang, S. Detection of novel QTLs for foxglove aphid resistance in soybean. Theor. Appl. Genet. 2015, 128, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Huynh, B.L.; Ehlers, J.D.; Ndeve, A.; Wanamaker, S.; Lucas, M.R.; Close, T.J.; Roberts, P.A. Genetic mapping and legume synteny of aphid resistance in African cowpea (Vigna unguiculata L. Walp.) grown in California. Mol. Breed. 2015, 35, 36. [Google Scholar] [CrossRef] [PubMed]
- Jegadeeswaran, M.; Kadirvel, P.; Srinivas, P.S.; Senthilvel, S.; Selvaraj, V.M.; Mobeen, S.; Reddy, Y.R.; Kiran, B.U.; Mukta, N. Genetic mapping reveals a major QTL associated with tolerance to the aphid, Uroleucon compositae (Theobald) in safflower (Carthamus tinctorius). Plant Breed. 2021, 140, 320–330. [Google Scholar] [CrossRef]
- Zhang, J.; Li, W.; Lv, P.; Yang, X.; Xu, W.; Ni, X.; Feng, H.; Zhao, G.; Pu, M.; Zhou, S.; et al. Whole-genome resequencing and transcriptome analysis provide insights on aphid-resistant quantitative trait loci/genes in Sorghum bicolor. Plant Breed. 2021, 140, 618–629. [Google Scholar] [CrossRef]
- Du, W. Map and Fine Map Aphid Resistance Genes in Soybean Plant Introduction (PI) 567597c, 567585a and 567537. Ph.D. Thesis, Michigan State University, East Lansing, MI, USA, 2016. [Google Scholar]
- Betsiashvili, M.; Ahern, K.R.; Jander, G. Additive effects of two quantitative trait loci that confer Rhopalosiphum maidis (corn leaf aphid) resistance in maize inbred line Mo17. J. Exp. Bot. 2015, 66, 571–578. [Google Scholar] [CrossRef]
- Zhang, G.; Gu, C.; Wang, D. Molecular mapping of soybean aphid resistance genes in PI 567541B. Theor. Appl. Genet. 2009, 118, 473–482. [Google Scholar] [CrossRef]
- Kim, K.S.; Hill, C.B.; Hartman, G.L.; Hyten, D.L.; Hudson, M.E.; Diers, B.W. Fine mapping of the soybean aphid-resistance gene Rag2 in soybean PI 200538. Theor. Appl. Genet. 2010, 121, 599–610. [Google Scholar] [CrossRef]
- Kim, K.S.; Chirumamilla, A.; Hill, C.B.; Hartman, G.L.; Diers, B.W. Identification and molecular mapping of two soybean aphid resistance genes in soybean PI 587732. Theor. Appl. Genet. 2014, 127, 1251–1259. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Z.; Wen, Z.; Gu, C.; An, Y.C.; Bales, C.; DiFonzo, C.; Song, Q.; Wang, D. Fine mapping of the soybean aphid-resistance genes Rag6 and Rag3c from Glycine soja 85-32. Theor. Appl. Genet. 2017, 130, 2601–2615. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Z.; Bales, C.; Gu, C.; DiFonzo, C.; Li, M.; Song, Q.; Cregan, P.; Yang, Z.; Wang, D. Mapping novel aphid resistance QTL from wild soybean, Glycine soja 85-32. Theor. Appl. Genet. 2017, 130, 1941–1952. [Google Scholar] [CrossRef]
- Bhusal, S.J.; Jiang, G.; Song, Q.; Cregan, P.B.; Wright, D.; Gonzalez-Hernandez, J.L. Genome-wide detection of genetic loci associated with soybean aphid resistance in soybean germplasm PI 603712. Euphytica 2017, 213, 144. [Google Scholar] [CrossRef]
- LaMantia, J.M.; Mian, A.R.; Redinbaugh, M.G. Genetic mapping of soybean aphid biotype 3 and 4 resistance in PI 606390A. Mol. Breed. 2019, 39, 53. [Google Scholar] [CrossRef]
- Kim, K.S.; Kim, J.M.; Jung, J.; Shin, I.; Park, S.; Lee, J.S.; Jeong, S.C.; Lee, J.D.; Jung, J.K.; Ha, B.K.; et al. Fine-mapping and candidate gene analysis for the foxglove aphid resistance gene Raso2 from wild soybean PI 366121. Theor. Appl. Genet. 2021, 134, 2687–2698. [Google Scholar] [CrossRef] [PubMed]
- Smigocki, A.; Neal, J.W.; McCanna, I.; Douglass, L. Cytokinin-mediated insect resistance in Nicotiana plants transformed with the ipt gene. Plant Mol. Biol. 1993, 23, 325–335. [Google Scholar] [CrossRef]
- Broekgaarden, C.; Snoeren, T.A.; Dicke, M.; Vosman, B. Exploiting natural variation to identify insect-resistance genes. Plant Biotechnol. J. 2011, 9, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Nakasu, E.Y.; Edwards, M.G.; Fitches, E.; Gatehouse, J.A.; Gatehouse, A.M. Transgenic plants expressing ω-ACTX-Hv1a and snowdrop lectin (GNA) fusion protein show enhanced resistance to aphids. Front. Plant Sci. 2014, 5, 673. [Google Scholar] [CrossRef]
- Losvik, A.; Beste, L.; Mehrabi, S.; Jonsson, L. The protease inhibitor CI2c gene induced by bird cherry-oat aphid in barley inhibits green peach aphid fecundity in transgenic Arabidopsis. Int. J. Mol. Sci. 2017, 18, 1317. [Google Scholar] [CrossRef]
- Stoger, E.; Williams, S.; Christou, P.; Down, R.E.; Gatehouse, J.A. Expression of the insecticidal lectin from snowdrop (Galanthus nivalis agglutinin; GNA) in transgenic wheat plants: Effects on predation by the grain aphid Sitobion avenae. Mol. Breed. 1999, 5, 65–73. [Google Scholar] [CrossRef]
- Wang, W. Regulatory Roles of Jasmonate Receptor Protein COl1 in Nicotiana tabacum and Brassica napus. Ph.D. Thesis, Southwest University, Chongqing, China, 2015. [Google Scholar]
- Yan, J.; Zhang, C.; Gu, M.; Bai, Z.; Zhang, W.; Qi, T.; Cheng, Z.; Peng, W.; Luo, H.; Nan, F.; et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 2009, 21, 2220–2236. [Google Scholar] [CrossRef]
- Jung, H.W.; Panigrahi, G.K.; Jung, G.Y.; Lee, Y.J.; Shin, K.H.; Sahoo, A.; Choi, E.S.; Lee, E.; Man, K.K.; Yang, S.H.; et al. Pathogen-associated molecular pattern-triggered immunity involves proteolytic degradation of core nonsense-mediated mRNA decay factors during the early defense response. Plant Cell 2020, 32, 1081–1101. [Google Scholar] [CrossRef]
- Li, W.; Xiong, Y.; Lai, L.B.; Zhang, K.; Li, Z.; Kang, H.; Dai, L.; Gopalan, V.; Wang, G.L.; Liu, W. The rice RNase P protein subunit Rpp30 confers broad-spectrum resistance to fungal and bacterial pathogens. Plant Biotechnol. J. 2021, 19, 1988–1999. [Google Scholar] [CrossRef]
- Zhang, Z.J.; Gao, Q.; Fang, X.D.; Ding, Z.H.; Gao, D.M.; Xu, W.Y.; Cao, Q.; Qiao, J.H.; Yang, Y.Z.; Han, C.; et al. CCR4, a RNA decay factor, is hijacked by a plant cytorhabdovirus phosphoprotein to facilitate virus replication. eLife 2020, 9, e53753. [Google Scholar] [CrossRef]
- Roman, P. Efficacy of naphthoquinones as insecticides against the house fly. Ind. Crop. Prod. 2013, 43, 745–750. [Google Scholar]
- Pathipati, U.R.; Sambangi, P. Defensive role of Gossypium hirsutum L. anti-oxidative enzymes and phenolic acids in response to Spodoptera litura F. feeding. J. Asia-Pac. Entomol. 2013, 16, 131–136. [Google Scholar]
- Han, S.; Schroeder, E.A.; Silva-Garcia, C.G.; Hebestreit, K.; Mair, W.B.; Brunet, A. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 2017, 544, 185–190. [Google Scholar] [CrossRef]
- Yang, Y.; Luo, K.; Jiang, C.; Wu, J.W.; Zhu, G.M. Accumulation and metabolism of polyunsaturated fatty acids from food in Musca domestica larvae and the effects on their growth. Acta Entomol. Sin. 2019, 62, 578–585. [Google Scholar]
- Jones, D.P.; Sies, H. The redox code. Antioxid. Redox Sign. 2015, 23, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Painter, R.H. Insect Resistance in Crop Plants; The University Press of Kansas: Lawrence, KS, USA, 1951; pp. 154–156. [Google Scholar]
- Ray, P.D.; Huang, B.W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012, 24, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Apel, K. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Koramutla, M.K.; Kaur, A.; Negi, M.; Venkatachalam, P.; Bhattacharya, R. Elicitation of jasmonate-mediated host defense in Brassica juncea (L.) attenuates population growth of mustard aphid Lipaphis erysimi (Kalt.). Planta 2014, 240, 177–194. [Google Scholar] [CrossRef]
- Nagpure, A.; Choudhary, B.; Gupta, R.K. Chitinases: In agriculture and human healthcare. Crit. Rev. Biotechnol. 2014, 34, 215–232. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, C.; Yokoyama, A.; Itoh, Y.; Hashimoto, M.; Watanabe, T.; Fukamizo, T. Comparative study of the reaction mechanism of family 18 chitinases from plants and microbes. J. Biochem. 2002, 131, 557–564. [Google Scholar] [CrossRef]
- Chandra, S.; Dutta, A.K.; Chandrashekara, K.N.; Acharya, K. In silico characterization, homology modeling of Camellia sinensis chitinase and its evolutionary analyses with other plant chitinases. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2017, 87, 685–695. [Google Scholar] [CrossRef]
- Jopcik, M.; Moravcikova, J.; Matusikova, I.; Bauer, M.; Rajninec, M.; Libantova, J. Structural and functional characterisation of a class I endochitinase of the carnivorous sundew (Drosera rotundifolia L.). Planta 2017, 245, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Hurej, M.; Van Der Werf, W. The influence of black bean aphid, Aphis fabae Scop., and its honeydew on the photosynthesis of sugar beet. Ann. Appl. Biol. 1993, 122, 189–200. [Google Scholar] [CrossRef]
- Shannag, H.K. Effect of black bean aphid, Aphis fabae, on transpiration, stomatal conductance and crude protein content of faba bean. Ann. Appl. Biol. 2007, 151, 183–188. [Google Scholar] [CrossRef]
- Koch, K.G.; Chapman, K.; Louis, J.; Heng-Moss, T.; Sarath, G. Plant tolerance: A unique approach to control hemipteran pests. Front. Plant Sci. 2016, 7, 1363. [Google Scholar] [CrossRef]
- Macedo, T.B. Physiological Responses of Plants to Piercing-Sucking Arthropods. Ph.D. Thesis, The University of Nebraska-Lincoln, Lincoln, NE, USA, 2003. [Google Scholar]
- Luo, J.; Wei, K.; Wang, S.; Zhao, W.; Ma, C.; Hettenhausen, C.; Wu, J.; Cao, G.; Sun, G.; Baldwin, I.T.; et al. COI1-regulated hydroxylation of jasmonoyl-L-isoleucine impairs Nicotiana attenuate’s resistance to the generalist herbivore Spodoptera litura. J. Agric. Food Chem. 2016, 64, 2822–2831. [Google Scholar] [CrossRef]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, X.; Du, T.; Shu, Y.; Tan, F.; Wang, J. Effects of exogenous salicylic acid application to aboveground part on the defense responses in Bt (Bacillus thuringiensis) and non-Bt corn (Zea mays L.) seedlings. Plants 2022, 11, 2162. [Google Scholar] [CrossRef]
- Wu, G.; Shortt, B.J.; Lawrence, E.B.; Leon, J.; Fitzsimmons, K.C.; Levine, E.B.; Raskin, I.; Shah, D.M. Activation of host defense mechanisms by elevated production of H2O2 in transgenic plants. Plant Physiol. 1997, 115, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Malamy, J.; Henning, J.; Conrath, U.; Sanchez-Casas, P.; Silva, H.; Ricigliano, J.; Klessig, D.K. Induction, modification, and transduction of the salicylic acid signal in plant defense responses. Proc. Natl. Acad. Sci. USA 1995, 92, 4134–4137. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Moeder, W.; Yoshioka, K.; Shan, L. A tale of many families: Calcium channels in plant immunity. Plant Cell 2022, 34, 1551–1567. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J.; et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Kachroo, A.; Kachroo, P. Mobile signals in systemic acquired resistance. Curr. Opin. Plant Biol. 2020, 58, 41–47. [Google Scholar] [CrossRef]
- Vlot, A.C.; Sales, J.H.; Lenk, M.; Bauer, K.; Brambilla, A.; Sommer, A.; Chen, Y.; Wenig, M.; Nayem, S. Systemic propagation of immunity in plants. New Phytol. 2021, 229, 1234–1250. [Google Scholar] [CrossRef]
- Wan, J.; He, M.; Hou, Q.; Zou, L.; Yang, Y.; Wei, Y.; Chen, X. Cell wall associated immunity in plants. Stress Biol. 2021, 1, 3. [Google Scholar] [CrossRef]
- Kessler, A.; Baldwin, I.T. Plant responses to insect herbivory: The emerging molecular analysis. Annu. Rev. Plant Biol. 2002, 53, 299–328. [Google Scholar] [CrossRef]
- Smith, C.M.; Clement, S.L. Molecular bases of plant resistance to arthropods. Annu. Rev. Entomol. 2012, 57, 309–328. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, S.; Lu, A.; Chen, F.; Tang, F.; Guan, Z.; Teng, N. Production and characterisation of the intergeneric hybrids between Dendranthema morifolium and Artemisia vulgaris exhibiting enhanced resistance to chrysanthemum aphid (Macrosiphoniella sanbourni). Planta 2010, 231, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Peng, F.; Yin, Z.; Wang, J.; Zhang, Y.; Zhang, H.; Fan, Y.; Xu, N.; Huang, H.; Ni, K.; et al. Secondary metabolite pathway of SDG (secoisolariciresinol) was observed to trigger ROS scavenging system in response to Ca2+ stress in cotton. Genomics 2022, 114, 110398. [Google Scholar] [CrossRef]
- Young, M.; Wakefield, M.; Smyth, G.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cao, C.; Zheng, S.; Zhang, H.; Liu, P.; Ge, Q.; Li, J.; Ren, Z. Transcriptomic analysis of short-fruit 1 (sf1) reveals new insights into the variation of fruit-related traits in Cucumis sativus. Sci. Rep. 2017, 7, 2950. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenfuhrer, J. TopGO: Enrichment analysis for Gene Ontology; R Package Version 2.36.0; R Foundation: Vienna, Austria, 2010. [Google Scholar]
- Wang, Y.; Liang, C.; Meng, Z.; Li, Y.; Abid, M.A.; Askari, M.; Wang, P.; Wang, Y.; Sun, G.; Cai, Y.; et al. Leveraging Atriplex hortensis choline monooxygenase to improve chilling tolerance in cotton. Environ. Exp. Bot. 2019, 162, 364–373. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, H.; Sun, C.; Ma, Q.; Bu, H.; Chong, K.; Xu, Y. A C2H2 zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice. J. Plant Physiol. 2018, 229, 100–110. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Variety | 7 d | 10 d | 13 d | 16 d | Wilting Rate 2 |
|---|---|---|---|---|---|
| APL01 | 11 ± 4.0 a | 22 ± 8.96 a | 59 ± 9.61 a | 93 ± 13.23 a | 66.7% a |
| Holly | 8 ± 3.51 b | 30 ± 11.15 b | 66 ± 23.07 a | 165 ± 73.50 b | 28.6% b |
| Control 1 | 51 ± 39.88 c | 109 ± 73.17 c | 236 ± 167.94 b | 398 ± 338.06 c | 93.5% c |
| Variety | No. Aphids Inoculated | No. Dead Aphids | Mortality of Aphids |
|---|---|---|---|
| APL01 | 20 | 13.67 ± 2.08 a | 68.3% a |
| Holly | 20 | 12.67 ± 2.51 a | 63.3% a |
| Control 1 | 20 | 1.33 ± 1.21 b | 6.7% b |
| Comparison Group | Up 1 | Down 2 |
|---|---|---|
| IA vs. NIA | 1314 | 548 |
| IH vs. NIH | 131 | 71 |
| IA vs. IH | 3187 | 2540 |
| NIA vs. NIH | 2255 | 2217 |
| GO ID | Term | Annotated a | Significant_A b | Significant_H c |
|---|---|---|---|---|
| GO:0042744 | hydrogen peroxide catabolic process | 258 | 11 (4.3%) | 4 (1.6%) |
| GO:0005509 | calcium ion binding | 819 | 53 (6.5%) | 5 (0.6%) |
| GO:0061630 | ubiquitin protein ligase activity | 360 | 8 (2.2%) | 3 (0.8%) |
| GO:0004601 | peroxidase activity | 409 | 13 (3.2%) | 4 (0.9%) |
| GO:0009627 | systemic acquired resistance | 97 | 21 (21.6%) | 1 (1.0%) |
| GO:0016998 | cell wall macromolecule catabolic process | 59 | 14 (23.7%) | 1 (1.7%) |
| GO:0006032 | chitin catabolic process | 60 | 14 (23.3%) | 1 (1.7%) |
| GO:0008061 | chitin binding | 58 | 16 (27.6%) | 1 (1.7%) |
| GO:0004568 | chitinase activity | 61 | 14 (22.9%) | 1 (1.6%) |
| GO:0080032 | methyl jasmonate esterase activity | 11 | 0 | 2 (18.2%) |
| GO:0010333 | terpene synthase activity | 97 | 2 (2.1%) | 2 (2.1%) |
| GO:0080031 | methyl salicylate esterase activity | 7 | 0 | 1 (14.3%) |
| GO ID | Term | Annotated 1 | Significant_A 2 | Significant_H 3 |
|---|---|---|---|---|
| GO:0009765 | photosynthesis, light harvesting | 82 | 44 (53.7%) | 9 (10.9%) |
| GO:0009523 | photosystem II | 177 | 81 (45.8%) | 10 (5.6%) |
| GO:0009522 | photosystem I | 137 | 73 (53.3%) | 9 (6.6%) |
| GO:0009535 | chloroplast thylakoid membrane | 362 | 72 (19.9%) | 9 (2.5%) |
| GO:0016168 | chlorophyll binding | 90 | 44 (48.9%) | 9 (10%) |
| GO:0030145 | manganese ion binding | 107 | 8 (7.5%) | 1 (0.9%) |
| GO:0010242 | oxygen evolving activity | 11 | 7 (63.6%) | 0 |
| GO:0016984 | ribulose-bisphosphate carboxylase activity | 24 | 7 (29.2%) | 0 |
| GO:0004332 | fructose-bisphosphate aldolase activity | 21 | 6 (28.6%) | 0 |
| GO:0009055 | electron carrier activity | 549 | 17 (3.1%) | 0 |
| GO:0015977 | carbon fixation | 57 | 13 (22.8%) | 0 |
| GO:0042549 | photosystem II stabilization | 12 | 7 (58.3%) | 0 |
| Enzyme | Variety | Before | 7 d | 13 d |
|---|---|---|---|---|
| Peroxidase | APL01 | 81.62 ± 5.47 a | 106.45 ± 3.90 a | 214.05 ± 7.00 a |
| Holly | 60.46 ± 6.59 b | 151.12 ± 19.85 b | 128.57 ± 10.03 b | |
| Catalase | APL01 | 99.76 ± 5.99 a | 367.42 ± 5.35 a | 330.61 ± 10.35 a |
| Holly | 174.34 ± 5.11 b | 368.70 ± 4.10 a | 249.89 ± 6.46 b | |
| Chitinase | APL01 | 0.01505 ± 0.00001 a | 0.01497 ± 0.00001 a | 0.01493 ± 0.00001a |
| Holly | 0.01491 ± 0.00001 b | 0.01490 ± 0.00001 a | 0.01490 ± 0.00001a |
| Index | Variety | Before | 7d | 13d |
|---|---|---|---|---|
| Net photosynthetic rate (μmol(CO2)m−2 s−1) | APL01 | 2.24 ± 1.08 a | 7.75 ± 1.12 a | 5.00 ± 1.03 a |
| Holly | 3.72 ± 1.34 a | 13.43 ± 1.56 b | 37.9 ± 1.87 b | |
| Chlorophyll relative content (SPAD) | APL01 | 39.97 ± 2.69 a | 40.77 ± 2.81 a | 29.37 ± 2.54 a |
| Holly | 38.37 ± 2.10 a | 36.03 ± 2.17 b | 31.43 ± 2.79 a | |
| Ribulose-bisphosphate carboxylase | APL01 | 85.51 ± 6.44 a | 58.78 ± 4.43 a | 51.45 ± 2.41 a |
| Holly | 47.16 ± 4.29 b | 134.61 ± 6.52 b | 330.79 ± 7.66 b | |
| Fructose-bisphosphate aldolase | APL01 | 501.37 ± 82.45 a | 292.42 ± 39.80 a | 236.48 ± 28.39 a |
| Holly | 125.20 ± 22.41 b | 267.68 ± 22.25 a | 835.51 ± 21.01 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Cai, L.; Ding, T.; Tian, E.; Yan, X.; Wang, X.; Zhang, J.; Yu, K.; Chen, Z. Comparative Transcriptome Analysis Reveals the Molecular Basis of Brassica napus in Response to Aphid Stress. Plants 2023, 12, 2855. https://doi.org/10.3390/plants12152855
Li Y, Cai L, Ding T, Tian E, Yan X, Wang X, Zhang J, Yu K, Chen Z. Comparative Transcriptome Analysis Reveals the Molecular Basis of Brassica napus in Response to Aphid Stress. Plants. 2023; 12(15):2855. https://doi.org/10.3390/plants12152855
Chicago/Turabian StyleLi, Yuanhong, Lei Cai, Ting Ding, Entang Tian, Xiaohong Yan, Xiaodong Wang, Jiefu Zhang, Kunjiang Yu, and Zhuo Chen. 2023. "Comparative Transcriptome Analysis Reveals the Molecular Basis of Brassica napus in Response to Aphid Stress" Plants 12, no. 15: 2855. https://doi.org/10.3390/plants12152855
APA StyleLi, Y., Cai, L., Ding, T., Tian, E., Yan, X., Wang, X., Zhang, J., Yu, K., & Chen, Z. (2023). Comparative Transcriptome Analysis Reveals the Molecular Basis of Brassica napus in Response to Aphid Stress. Plants, 12(15), 2855. https://doi.org/10.3390/plants12152855








