The Role of Anthocyanins in Plant Tolerance to Drought and Salt Stresses
Abstract
1. Introduction
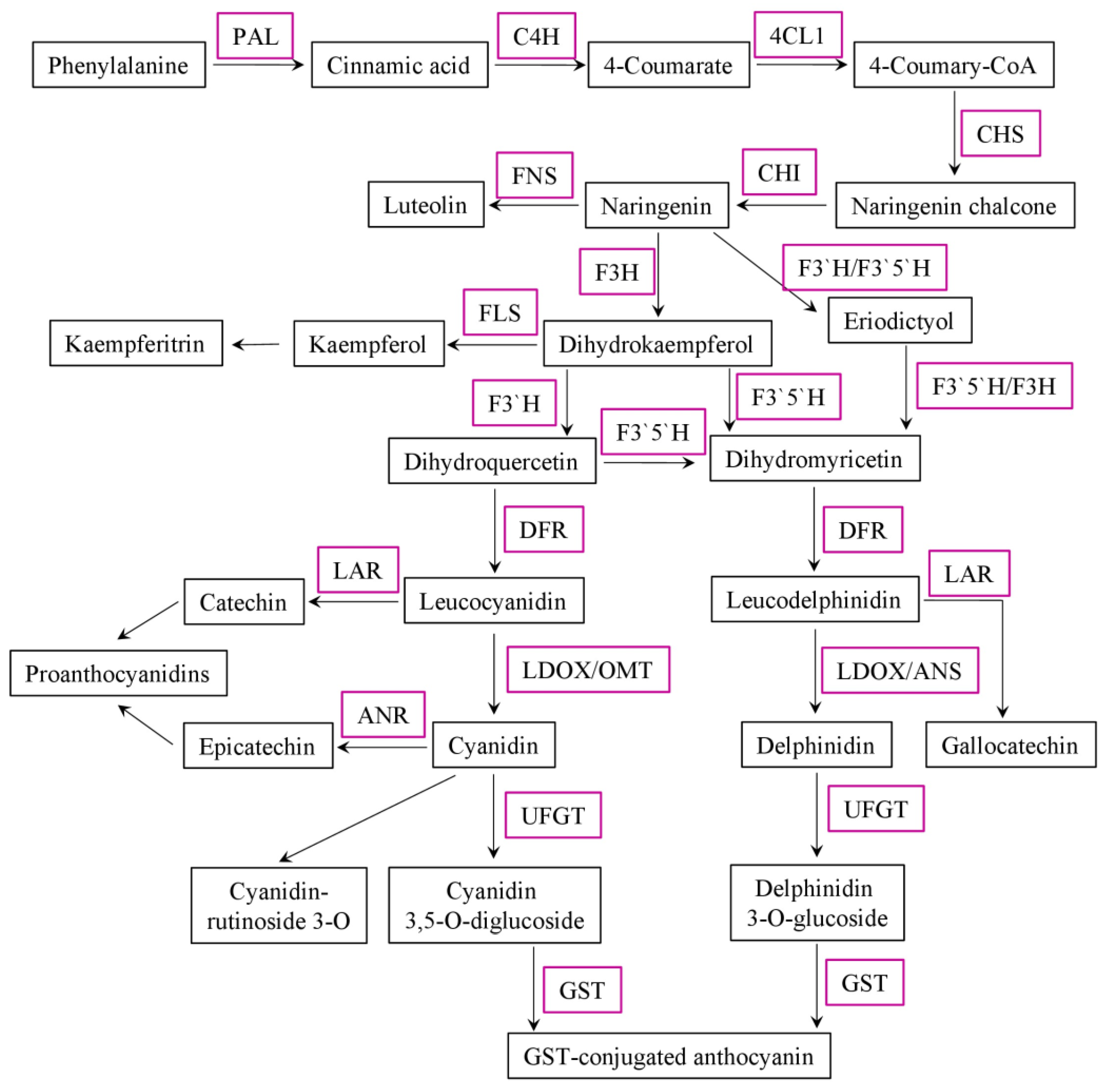
2. Anthocyanins and Drought Tolerance
2.1. Anthocyanins Induction by Drought
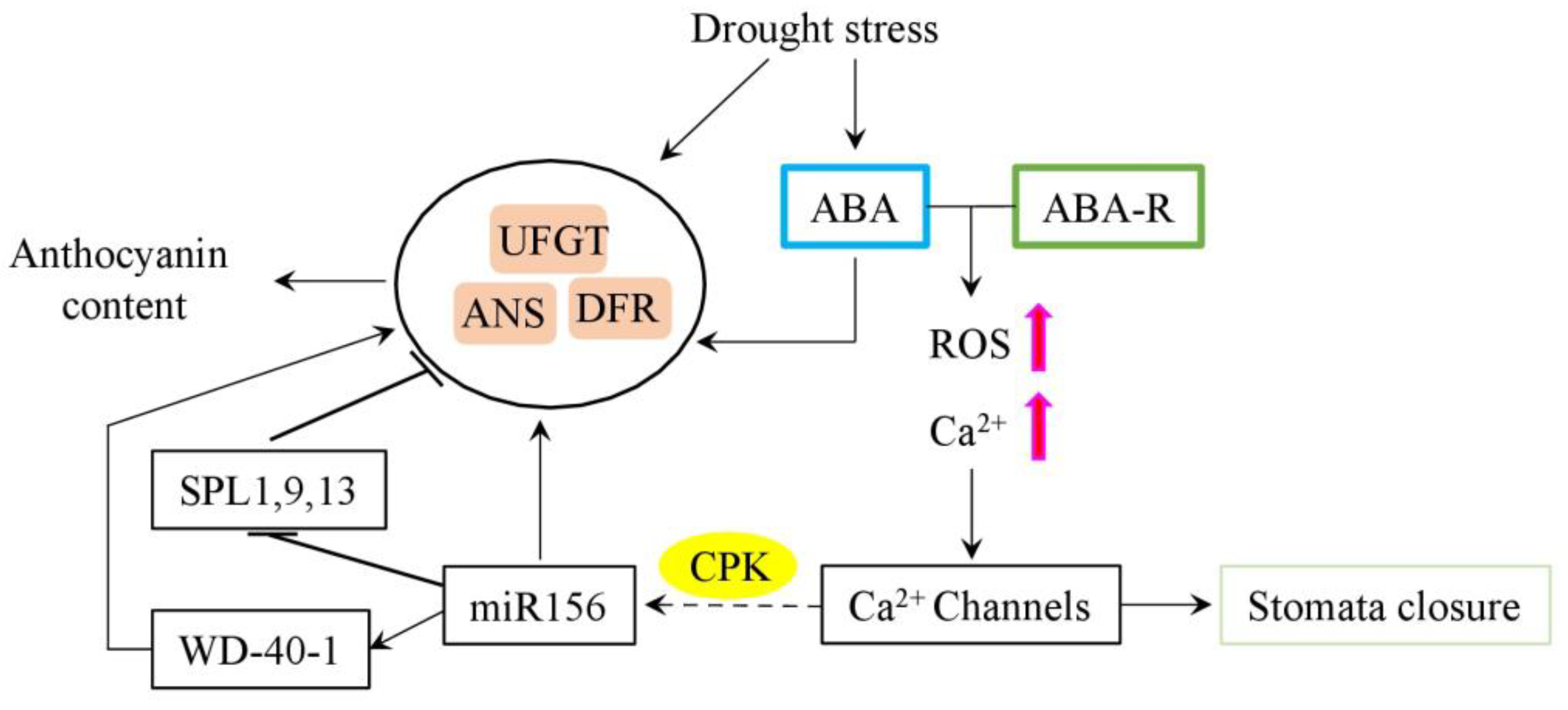
2.2. The Role of Abscisic Acid in the Regulation of Anthocyanin Biosynthesis
Abscisic Acid-microRNA Regulatory Module
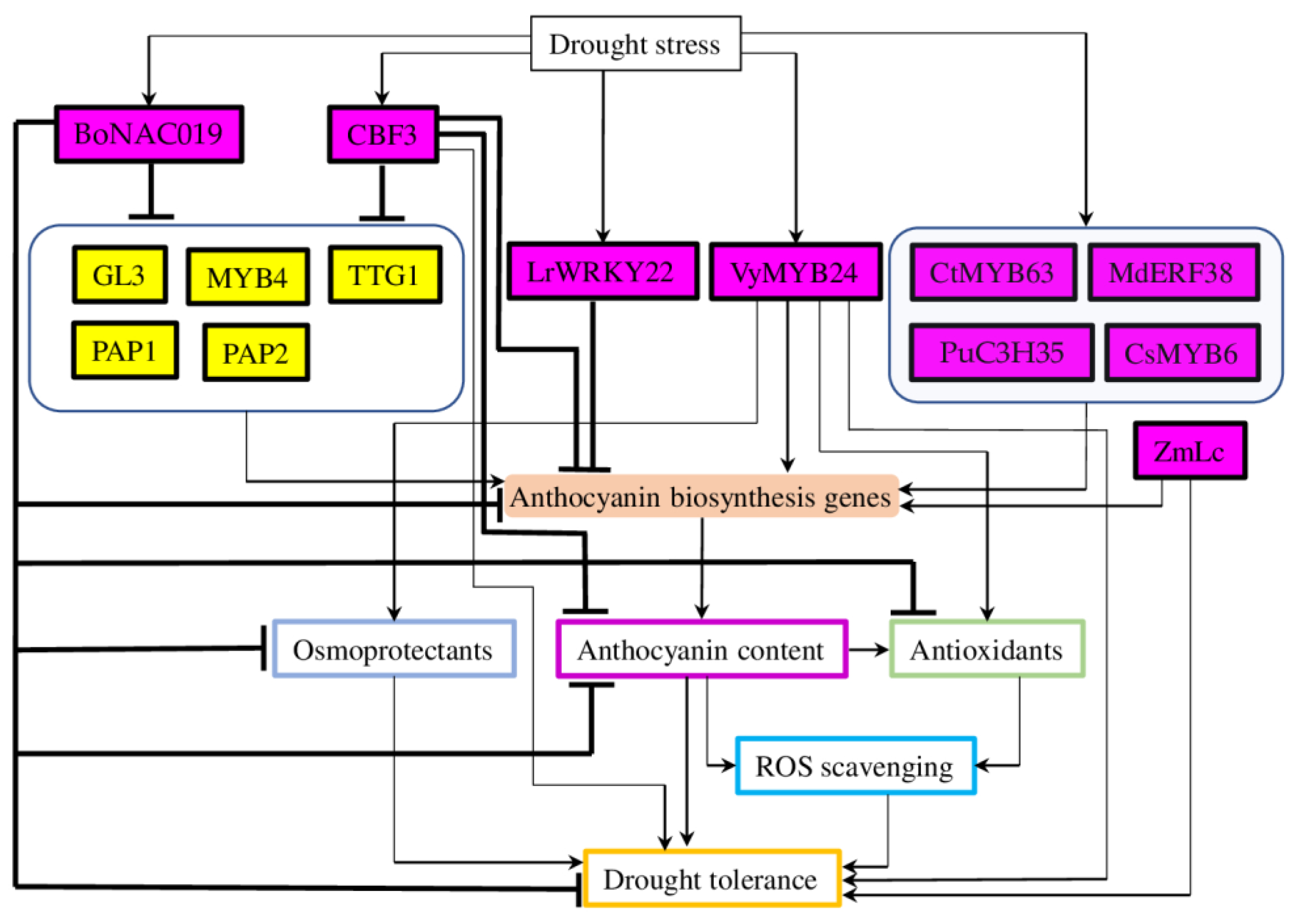
2.3. Transcription Factors Regulating Drought-Mediated Anthocyanin Biosynthesis
2.3.1. Myeloblastosis Transcription Factors
2.3.2. Basic Helix-Loop-Helix Transcription Factors
2.3.3. Apetala2/Ethylene Responsive Factor
2.3.4. Other Transcription Factor Families
2.4. Other Types of Regulators
3. Anthocyanins and Salinity Stress
3.1. Anthocyanin Induction by Salinity Stress
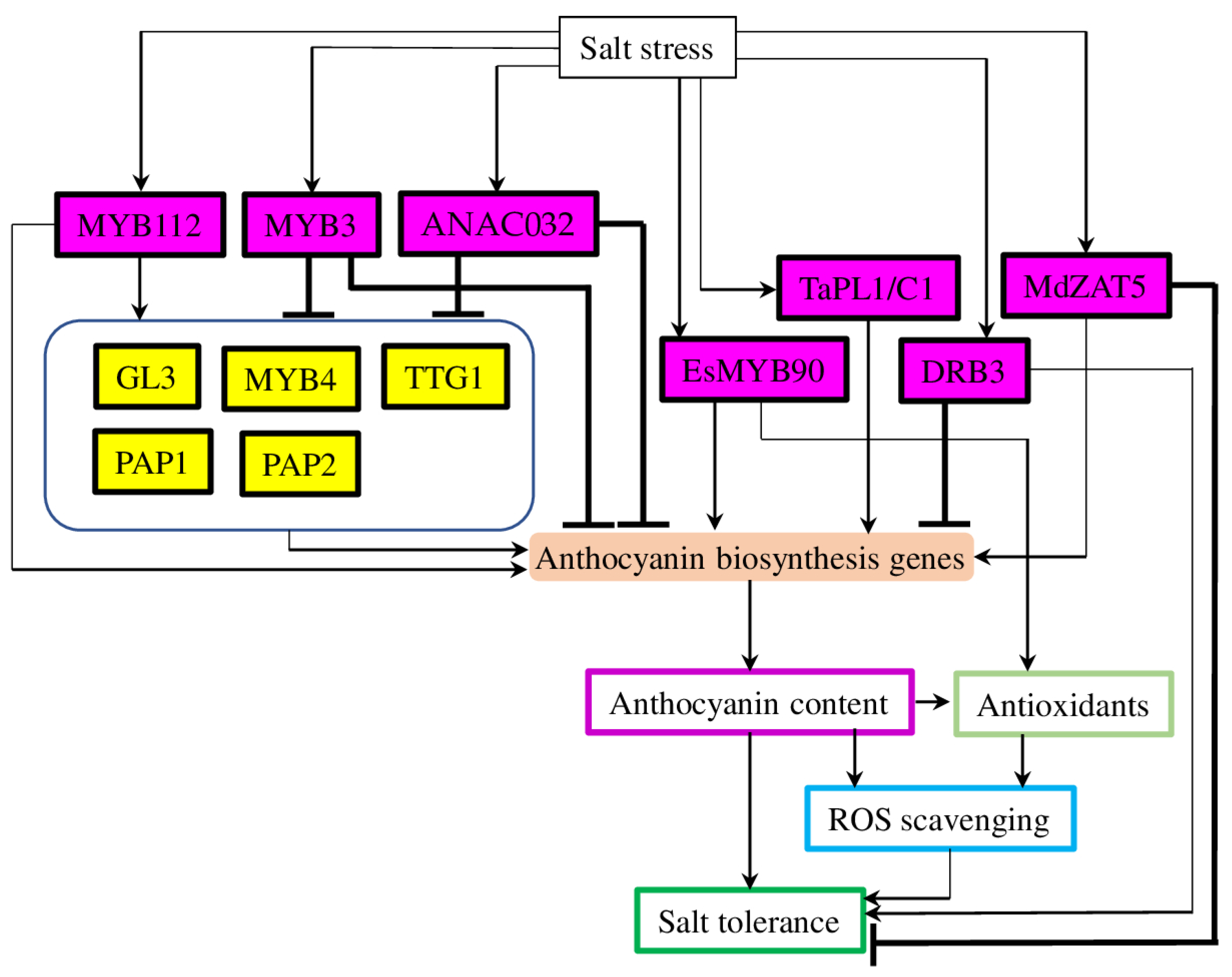
3.2. Transcription Factors Involved in Salt-Stress-Induced Anthocyanin Biosynthesis
3.2.1. Myeloblastosis Transcription Factors
3.2.2. Other Types of Transcription Factors
3.3. Manipulation with the Anthocyanin Biosynthesis Genes to Improve Salt Stress Tolerance
3.4. Other Factors, Regulating Anthocyanin Biosynthesis in Salt Tolerance
4. Anthocyanin Biosynthesis and Tolerance to Several Stressors
4.1. Manipulation of Anthocyanin Biosynthesis Genes to Improve Both Drought and Salt Stress
4.2. Transcription Factors Regulating Anthocyanin Biosynthesis and Resistance to Several Stressors
5. Conclusions and Future Prospective Studies
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and Anthocyanins: Colored Pigments as Food, Pharmaceutical Ingredients, and the Potential Health Benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed]
- Alappat, B.; Alappat, J. Anthocyanin Pigments: Beyond Aesthetics. Molecules 2020, 25, 5500. [Google Scholar] [CrossRef] [PubMed]
- Saigo, T.; Wang, T.; Watanabe, M.; Tohge, T. Diversity of Anthocyanin and Proanthocyanin Biosynthesis in Land Plants. Curr. Opin. Plant Biol. 2020, 55, 93–99. [Google Scholar] [CrossRef]
- Zhao, X.; Dai, X.; Gao, L.; Guo, L.; Zhuang, J.; Liu, Y.; Ma, X.; Wang, R.; Xia, T.; Wang, Y. Functional Analysis of an Uridine Diphosphate Glycosyltransferase Involved in the Biosynthesis of Polyphenolic Glucoside in Tea Plants (Camellia sinensis). J. Agric. Food Chem. 2017, 65, 10993–11001. [Google Scholar] [CrossRef]
- Ku, Y.-S.; Cheng, S.-S.; Cheung, M.-Y.; Niu, Y.; Liu, A.; Chung, G.; Lam, H.-M. The Poly-Glutamate Motif of GmMATE4 Regulates Its Isoflavone Transport Activity. Membranes 2022, 12, 206. [Google Scholar] [CrossRef]
- Wang, R.; Lu, N.; Liu, C.; Dixon, R.A.; Wu, Q.; Mao, Y.; Yang, Y.; Zheng, X.; He, L.; Zhao, B.; et al. MtGSTF7, a TT19-like GST Gene, Is Essential for Accumulation of Anthocyanins, but Not Proanthocyanins in Medicago truncatula. J. Exp. Bot. 2022, 73, 4129–4146. [Google Scholar] [CrossRef] [PubMed]
- Panara, F.; Passeri, V.; Lopez, L.; Porceddu, A.; Calderini, O.; Paolocci, F. Functional Characterization of MtrGSTF7, a Glutathione S-Transferase Essential for Anthocyanin Accumulation in Medicago truncatula. Plants 2022, 11, 1318. [Google Scholar] [CrossRef]
- Zhao, Y.; Jiang, C.; Lu, J.; Sun, Y.; Cui, Y. Research Progress of Proanthocyanidins and Anthocyanidins. Phytother. Res. 2023, 37, 2552–2577. [Google Scholar] [CrossRef]
- Cui, D.; Zhao, S.; Xu, H.; Allan, A.C.; Zhang, X.; Fan, L.; Chen, L.; Su, J.; Shu, Q.; Li, K. The Interaction of MYB, BHLH and WD40 Transcription Factors in Red Pear (Pyrus pyrifolia) Peel. Plant Mol. Biol 2021, 106, 407–417. [Google Scholar] [CrossRef]
- Khusnutdinov, E.; Sukhareva, A.; Panfilova, M.; Mikhaylova, E. Anthocyanin Biosynthesis Genes as Model Genes for Genome Editing in Plants. Int. J. Mol. Sci. 2021, 22, 8752. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Bashir, S.S.; Hussain, A.; Hussain, S.J.; Wani, O.A.; Zahid Nabi, S.; Dar, N.A.; Baloch, F.S.; Mansoor, S. Plant Drought Stress Tolerance: Understanding Its Physiological, Biochemical and Molecular Mechanisms. Biotechnol. Biotechnol. Equip. 2021, 35, 1912–1925. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Do Anthocyanins Function as Osmoregulators in Leaf Tissues? In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2002; Volume 37, pp. 103–127. ISBN 978-0-12-005937-9. [Google Scholar]
- Kaku, S.; Iwaya-Inoue, M.; Toki, K. Anthocyanin Influence on Water Proton NMR Relaxation Times and Water Contents in Leaves of Evergreen Woody Plants During the Winter. Plant Cell Physiol. 1992, 33, 131–137. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef]
- Eryılmaz, F. The Relationships between Salt Stress and Anthocyanin Content in Higher Plants. Biotechnol. Biotechnol. Equip. 2006, 20, 47–52. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative Stress, Antioxidants and Stress Tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Kaur, S.; Tiwari, V.; Kumari, A.; Chaudhary, E.; Sharma, A.; Ali, U.; Garg, M. Protective and Defensive Role of Anthocyanins under Plant Abiotic and Biotic Stresses: An Emerging Application in Sustainable Agriculture. J. Biotechnol. 2023, 361, 12–29. [Google Scholar] [CrossRef]
- Hinojosa-Gómez, J.; San Martín-Hernández, C.; Heredia, J.B.; León-Félix, J.; Osuna-Enciso, T.; Muy-Rangel, M.D. Anthocyanin Induction by Drought Stress in the Calyx of Roselle Cultivars. Molecules 2020, 25, 1555. [Google Scholar] [CrossRef]
- Chen, W.; Miao, Y.; Ayyaz, A.; Hannan, F.; Huang, Q.; Ulhassan, Z.; Zhou, Y.; Islam, F.; Hong, Z.; Farooq, M.A.; et al. Purple Stem Brassica napus Exhibits Higher Photosynthetic Efficiency, Antioxidant Potential and Anthocyanin Biosynthesis Related Genes Expression against Drought Stress. Front. Plant Sci. 2022, 13, 936696. [Google Scholar] [CrossRef]
- Cao, X.; Hu, Y.; Song, J.; Feng, H.; Wang, J.; Chen, L.; Wang, L.; Diao, X.; Wan, Y.; Liu, S.; et al. Transcriptome Sequencing and Metabolome Analysis Reveals the Molecular Mechanism of Drought Stress in Millet. Int. J. Mol. Sci. 2022, 23, 10792. [Google Scholar] [CrossRef]
- Maritim, T.K.; Korir, R.K.; Nyabundi, K.W.; Wachira, F.N.; Kamunya, S.M.; Muoki, R.C. Molecular Regulation of Anthocyanin Discoloration under Water Stress and High Solar Irradiance in Pluckable Shoots of Purple Tea Cultivar. Planta 2021, 254, 85. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Y.; Zhen, X.; Zhang, J.; Zhang, Q.; Liu, Z.; Hou, S.; Han, Y.; Zhang, B. Uncovering the Mechanism of Anthocyanin Accumulation in a Purple-Leaved Variety of Foxtail Millet (Setaria italica) by Transcriptome Analysis. PeerJ 2022, 10, e14099. [Google Scholar] [CrossRef]
- Kapoor, P.; Sharma, S.; Tiwari, A.; Kaur, S.; Kumari, A.; Sonah, H.; Goyal, A.; Krishania, M.; Garg, M. Genome–Transcriptome Transition Approaches to Characterize Anthocyanin Biosynthesis Pathway Genes in Blue, Black and Purple Wheat. Genes 2023, 14, 809. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Parihar, P.; Singh, S.; Mishra, R.K.; Singh, V.P.; Prasad, S.M. Reactive Oxygen Species Signaling and Stomatal Movement: Current Updates and Future Perspectives. Redox Biol. 2017, 11, 213–218. [Google Scholar] [CrossRef]
- González-Villagra, J.; Rodrigues-Salvador, A.; Nunes-Nesi, A.; Cohen, J.D.; Reyes-Díaz, M.M. Age-Related Mechanism and Its Relationship with Secondary Metabolism and Abscisic Acid in Aristotelia chilensis Plants Subjected to Drought Stress. Plant Physiol. Biochem. 2018, 124, 136–145. [Google Scholar] [CrossRef]
- González-Villagra, J.; Cohen, J.D.; Reyes-Díaz, M.M. Abscisic Acid Is Involved in Phenolic Compounds Biosynthesis, Mainly Anthocyanins, in Leaves of Aristotelia chilensis Plants (Mol.) Subjected to Drought Stress. Physiol. Plant. 2019, 165, 855–866. [Google Scholar] [CrossRef]
- Gai, Z.; Wang, Y.; Ding, Y.; Qian, W.; Qiu, C.; Xie, H.; Sun, L.; Jiang, Z.; Ma, Q.; Wang, L.; et al. Exogenous Abscisic Acid Induces the Lipid and Flavonoid Metabolism of Tea Plants under Drought Stress. Sci. Rep. 2020, 10, 12275. [Google Scholar] [CrossRef]
- Begum, Y. Regulatory Role of MicroRNAs (MiRNAs) in the Recent Development of Abiotic Stress Tolerance of Plants. Gene 2022, 821, 146283. [Google Scholar] [CrossRef] [PubMed]
- Jerome Jeyakumar, J.M.; Ali, A.; Wang, W.-M.; Thiruvengadam, M. Characterizing the Role of the MiR156-SPL Network in Plant Development and Stress Response. Plants 2020, 9, 1206. [Google Scholar] [CrossRef] [PubMed]
- González-Villagra, J.; Kurepin, L.V.; Reyes-Díaz, M.M. Evaluating the Involvement and Interaction of Abscisic Acid and MiRNA156 in the Induction of Anthocyanin Biosynthesis in Drought-Stressed Plants. Planta 2017, 246, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Xia, X.; Wei, M.; Sun, J.; Meng, J.; Tao, J. Overexpression of Herbaceous Peony MiR156e-3p Improves Anthocyanin Accumulation in Transgenic Arabidopsis thaliana Lateral Branches. 3 Biotech 2017, 7, 379. [Google Scholar] [CrossRef] [PubMed]
- Hanly, A.; Karagiannis, J.; Lu, Q.S.M.; Tian, L.; Hannoufa, A. Characterization of the Role of SPL9 in Drought Stress Tolerance in Medicago sativa. Int. J. Mol. Sci. 2020, 21, 6003. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 Improves Drought Stress Tolerance in Alfalfa (Medicago sativa) by Silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Feyissa, B.A.; Arshad, M.; Gruber, M.Y.; Kohalmi, S.E.; Hannoufa, A. The Interplay between MiR156/SPL13 and DFR/WD40–1 Regulate Drought Tolerance in Alfalfa. BMC Plant Biol. 2019, 19, 434. [Google Scholar] [CrossRef]
- Li, S.; Cheng, Z.; Li, Z.; Dong, S.; Yu, X.; Zhao, P.; Liao, W.; Yu, X.; Peng, M. MeSPL9 Attenuates Drought Resistance by Regulating JA Signaling and Protectant Metabolite Contents in Cassava. Theor. Appl. Genet. 2022, 135, 817–832. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Wang, X.; Yang, R.; Wu, Z.; Wang, H.; Wang, L.; Hu, Z.; Guo, S.; Zhang, H.; et al. MiR156 Regulates Anthocyanin Biosynthesis through SPL Targets and Other MicroRNAs in Poplar. Hortic. Res. 2020, 7, 118. [Google Scholar] [CrossRef]
- Li, C.; Yu, W.; Xu, J.; Lu, X.; Liu, Y. Anthocyanin Biosynthesis Induced by MYB Transcription Factors in Plants. Int. J. Mol. Sci. 2022, 23, 11701. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zeng, Y.; Li, Y.; Liu, Z.; Lin-Wang, K.; Espley, R.V.; Allan, A.C.; Zhang, J. Genomic Survey and Gene Expression Analysis of the MYB-Related Transcription Factor Superfamily in Potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 164, 2450–2464. [Google Scholar] [CrossRef]
- Hong, Y.; Ahmad, N.; Zhang, J.; Lv, Y.; Zhang, X.; Ma, X.; Xiuming, L.; Na, Y. Genome-Wide Analysis and Transcriptional Reprogrammings of MYB Superfamily Revealed Positive Insights into Abiotic Stress Responses and Anthocyanin Accumulation in Carthamus tinctorius L. Mol. Genet. Genom. 2022, 297, 125–145. [Google Scholar] [CrossRef]
- Zhu, Z.; Quan, R.; Chen, G.; Yu, G.; Li, X.; Han, Z.; Xu, W.; Li, G.; Shi, J.; Li, B. An R2R3-MYB Transcription Factor VyMYB24, Isolated from Wild Grape Vitis yanshanesis J. X. Chen., Regulates the Plant Development and Confers the Tolerance to Drought. Front. Plant Sci. 2022, 13, 966641. [Google Scholar] [CrossRef]
- Zhang, S.; Ren, Y.; Zhao, Q.; Wu, Y.; Zhuo, Y.; Li, H. Drought-induced CsMYB6 Interacts with CsbHLH111 to Regulate Anthocyanin Biosynthesis in Chaenomeles speciosa. Physiol. Plant. 2023, 175, e13859. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, S.R.; Habera, L.F.; Dellaporta, S.L.; Wessler, S.R. Lc, a Member of the Maize R Gene Family Responsible for Tissue-Specific Anthocyanin Production, Encodes a Protein Similar to Transcriptional Activators and Contains the Myc-Homology Region. Proc. Natl. Acad. Sci. USA 1989, 86, 7092–7096. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.; Ding, W.; Wu, C.; Wang, W.; Ye, S.; Cai, C.; Hu, X.; Wang, N.; Bai, W.; Tang, X.; et al. Production of Purple Ma Bamboo (Dendrocalamus latiflorus Munro) with Enhanced Drought and Cold Stress Tolerance by Engineering Anthocyanin Biosynthesis. Planta 2021, 254, 50. [Google Scholar] [CrossRef]
- Wu, M.; Upreti, S.; Yan, A.; Wakeel, A.; Wu, J.; Ge, S.; Liu, Y.; Liu, B.; Gan, Y. SPATULA Regulates Floral Transition and Photomorphogenesis in a PHYTOCHROME B-Dependent Manner in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 503, 2380–2385. [Google Scholar] [CrossRef] [PubMed]
- Reymond, M.C.; Brunoud, G.; Chauvet, A.; Martínez-Garcia, J.F.; Martin-Magniette, M.-L.; Monéger, F.; Scutt, C.P. A Light-Regulated Genetic Module Was Recruited to Carpel Development in Arabidopsis following a Structural Change to SPATULA. Plant Cell 2012, 24, 2812–2825. [Google Scholar] [CrossRef]
- Bernal-Gallardo, J.J.; Zuñiga-Mayo, V.M.; Marsch-Martinez, N.; De Folter, S. Novel Roles of SPATULA in the Control of Stomata and Trichome Number, and Anthocyanin Biosynthesis. Plants 2023, 12, 596. [Google Scholar] [CrossRef]
- Li, D.; He, Y.; Li, S.; Shi, S.; Li, L.; Liu, Y.; Chen, H. Genome-Wide Characterization and Expression Analysis of AP2/ERF Genes in Eggplant (Solanum melongena L.). Plant Physiol. Biochem. 2021, 167, 492–503. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of Oxidative and Drought Tolerance in Arabidopsis by Overaccumulation of Antioxidant Flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- An, J.; Zhang, X.; Bi, S.; You, C.; Wang, X.; Hao, Y. The ERF Transcription Factor MdERF38 Promotes Drought Stress-induced Anthocyanin Biosynthesis in Apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef]
- Wang, N.; Song, G.; Zhang, F.; Shu, X.; Cheng, G.; Zhuang, W.; Wang, T.; Li, Y.; Wang, Z. Characterization of the WRKY Gene Family Related to Anthocyanin Biosynthesis and the Regulation Mechanism under Drought Stress and Methyl Jasmonate Treatment in Lycoris radiata. Int. J. Mol. Sci. 2023, 24, 2423. [Google Scholar] [CrossRef]
- An, D.; Ma, Q.; Yan, W.; Zhou, W.; Liu, G.; Zhang, P. Divergent Regulation of CBF Regulon on Cold Tolerance and Plant Phenotype in Cassava Overexpressing Arabidopsis CBF3 Gene. Front. Plant Sci. 2016, 7, 1866. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lian, W.; Cao, Y.; Wang, X.; Wang, G.; Qi, C.; Liu, L.; Qin, S.; Yuan, X.; Li, X.; et al. Overexpression of BoNAC019, a NAC Transcription Factor from Brassica oleracea, Negatively Regulates the Dehydration Response and Anthocyanin Biosynthesis in Arabidopsis. Sci. Rep. 2018, 8, 13349. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yang, J.; Pak, S.; Zeng, M.; Sun, J.; Yu, S.; He, Y.; Li, C. PuC3H35 Confers Drought Tolerance by Enhancing Lignin and Proanthocyanidin Biosynthesis in the Roots of Populus ussuriensis. New Phytol. 2022, 233, 390–408. [Google Scholar] [CrossRef]
- Wan, D.; Li, R.; Zou, B.; Zhang, X.; Cong, J.; Wang, R.; Xia, Y.; Li, G. Calmodulin-Binding Protein CBP60g Is a Positive Regulator of Both Disease Resistance and Drought Tolerance in Arabidopsis. Plant Cell Rep. 2012, 31, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Wan, D.; Li, R.; Han, X.; Li, G.; Wang, R. Calmodulin-Binding Protein CBP60g Functions as a Negative Regulator in Arabidopsis Anthocyanin Accumulation. PLoS ONE 2017, 12, e0173129. [Google Scholar] [CrossRef]
- Yu, Z.; Dong, W.; Teixeira Da Silva, J.A.; He, C.; Si, C.; Duan, J. Ectopic Expression of DoFLS1 from Dendrobium Officinale Enhances Flavonol Accumulation and Abiotic Stress Tolerance in Arabidopsis thaliana. Protoplasma 2021, 258, 803–815. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, M.; Li, X.; Zhou, R.; Xue, X.; Zhang, J.; Liu, N.; Xue, R.; Qi, X. Overexpression of the Wheat TaPsb28 Gene Enhances Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2023, 24, 5226. [Google Scholar] [CrossRef]
- Gutha, L.R.; Casassa, L.F.; Harbertson, J.F.; Naidu, R.A. Modulation of Flavonoid Biosynthetic Pathway Genes and Anthocyanins Due to Virus Infection in Grapevine (Vitis vinifera L.) Leaves. BMC Plant Biol. 2010, 10, 187. [Google Scholar] [CrossRef]
- Cui, Z.-H.; Bi, W.-L.; Hao, X.-Y.; Li, P.-M.; Duan, Y.; Walker, M.A.; Xu, Y.; Wang, Q.-C. Drought Stress Enhances Up-Regulation of Anthocyanin Biosynthesis in Grapevine leafroll-associated virus 3-Infected in Vitro Grapevine (Vitis vinifera) Leaves. Plant Dis. 2017, 101, 1606–1615. [Google Scholar] [CrossRef]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant Salt Stress: Adaptive Responses, Tolerance Mechanism and Bioengineering for Salt Tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Jahantigh, O.; Najafi, F.; Badi, H.N.; Khavari-Nejad, R.A.; Sanjarian, F. Changes in Antioxidant Enzymes Activities and Proline, Total Phenol and Anthocyanine Contents in Hyssopus officinalis L. Plants under Salt Stress. Acta Biol. Hung. 2016, 67, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Loconsole, D.; Cristiano, G.; De Lucia, B. Glassworts: From Wild Salt Marsh Species to Sustainable Edible Crops. Agriculture 2019, 9, 14. [Google Scholar] [CrossRef]
- Patel, S. Salicornia: Evaluating the Halophytic Extremophile as a Food and a Pharmaceutical Candidate. 3 Biotech 2016, 6, 104. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.-S.; Seo, Y. Antiadipogenic Activity of Isohamnetin 3-O-β-D-Glucopyranoside from Salicornia herbacea. Immunopharmacol. Immunotoxicol. 2012, 34, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, J.; Guo, H.; Zong, J.; Chen, J.; Wang, Y.; Li, D.; Li, L.; Wang, J.; Liu, J. Growth and Physiological Responses of Two Phenotypically Distinct Accessions of Centipedegrass (Eremochloa ophiuroides (Munro) Hack.) to Salt Stress. Plant Physiol. Biochem. 2018, 126, 1–10. [Google Scholar] [CrossRef]
- Mbarki, S.; Sytar, O.; Zivcak, M.; Abdelly, C.; Cerda, A.; Brestic, M. Anthocyanins of Coloured Wheat Genotypes in Specific Response to SalStress. Molecules 2018, 23, 1518. [Google Scholar] [CrossRef]
- Lee, W.J.; Jeong, C.Y.; Kwon, J.; Van Kien, V.; Lee, D.; Hong, S.-W.; Lee, H. Drastic Anthocyanin Increase in Response to PAP1 Overexpression in Fls1 Knockout Mutant Confers Enhanced Osmotic Stress Tolerance in Arabidopsis thaliana. Plant Cell Rep. 2016, 35, 2369–2379. [Google Scholar] [CrossRef]
- Truong, H.A.; Lee, W.J.; Jeong, C.Y.; Trịnh, C.S.; Lee, S.; Kang, C.-S.; Cheong, Y.-K.; Hong, S.-W.; Lee, H. Enhanced Anthocyanin Accumulation Confers Increased Growth Performance in Plants under Low Nitrate and High Salt Stress Conditions Owing to Active Modulation of Nitrate Metabolism. J. Plant Physiol. 2018, 231, 41–48. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, W.J.; Le, Q.T.; Hong, S.-W.; Lee, H. Growth Performance Can Be Increased under High Nitrate and High Salt Stress through Enhanced Nitrate Reductase Activity in Arabidopsis Anthocyanin Over-Producing Mutant Plants. Front. Plant Sci. 2021, 12, 644455. [Google Scholar] [CrossRef]
- Saad, K.R.; Kumar, G.; Mudliar, S.N.; Giridhar, P.; Shetty, N.P. Salt Stress-Induced Anthocyanin Biosynthesis Genes and MATE Transporter Involved in Anthocyanin Accumulation in Daucus carota Cell Culture. ACS Omega 2021, 6, 24502–24514. [Google Scholar] [CrossRef]
- Nounjan, N.; Theerakulpisut, P. Physiological Evaluation for Salt Tolerance in Green and Purple Leaf Color Rice Cultivars at Seedling Stage. Physiol. Mol. Biol. Plants 2021, 27, 2819–2832. [Google Scholar] [CrossRef]
- Qi, Y.; Li, C.; Duan, C.; Gu, C.; Zhang, Q. Integrated Metabolomic and Transcriptomic Analysis Reveals the Flavonoid Regulatory Network by Eutrema EsMYB90. Int. J. Mol. Sci. 2021, 22, 8751. [Google Scholar] [CrossRef]
- Qi, Y.; Gu, C.; Wang, X.; Gao, S.; Li, C.; Zhao, C.; Li, C.; Ma, C.; Zhang, Q. Identification of the Eutrema salsugineum EsMYB90 Gene Important for Anthocyanin Biosynthesis. BMC Plant Biol. 2020, 20, 186. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, Y.; Qi, Y.; Duan, C.; Zhang, H.; Zhang, Q. Eutrema EsMYB90 Gene Improves Growth and Antioxidant Capacity of Transgenic Wheat under Salinity Stress. Front. Plant Sci. 2022, 13, 856163. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.H.; Choi, M.-G.; Kang, C.-S.; Park, C.-S.; Choi, S.-B.; Park, Y.-I. A Wheat R2R3-MYB Protein PURPLE PLANT1 (TaPL1) Functions as a Positive Regulator of Anthocyanin Biosynthesis. Biochem. Biophys. Res. Commun. 2016, 469, 686–691. [Google Scholar] [CrossRef]
- Lotkowska, M.E.; Tohge, T.; Fernie, A.R.; Xue, G.-P.; Balazadeh, S.; Mueller-Roeber, B. The Arabidopsis Transcription Factor MYB112 Promotes Anthocyanin Formation during Salinity and under High Light Stress. Plant Physiol. 2015, 169, 1862–1880. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Jeon, S.J.; Yanders, S.; Park, S.; Kim, H.S.; Kim, S. MYB3 Plays an Important Role in Lignin and Anthocyanin Biosynthesis under Salt Stress Condition in Arabidopsis. Plant Cell Rep. 2022, 41, 1549–1560. [Google Scholar] [CrossRef]
- Li, C.; Shi, L.; Wang, Y.; Li, W.; Chen, B.; Zhu, L.; Fu, Y. Arabidopsis ECAP Is a New Adaptor Protein That Connects JAZ Repressors with the TPR2 Co-Repressor to Suppress Jasmonate-Responsive Anthocyanin Accumulation. Mol. Plant 2020, 13, 246–265. [Google Scholar] [CrossRef]
- Li, C.; Shi, L.; Li, X.; Wang, Y.; Bi, Y.; Li, W.; Ma, H.; Chen, B.; Zhu, L.; Fu, Y. ECAP Is a Key Negative Regulator Mediating Different Pathways to Modulate Salt Stress-induced Anthocyanin Biosynthesis in Arabidopsis. New Phytol. 2022, 233, 2216–2231. [Google Scholar] [CrossRef]
- Masliah, G.; Barraud, P.; Allain, F.H.-T. RNA Recognition by Double-Stranded RNA Binding Domains: A Matter of Shape and Sequence. Cell. Mol. Life Sci. 2012, 70, 1875–1895. [Google Scholar] [CrossRef] [PubMed]
- Sawano, H.; Matsuzaki, T.; Usui, T.; Tabara, M.; Fukudome, A.; Kanaya, A.; Tanoue, D.; Hiraguri, A.; Horiguchi, G.; Ohtani, M.; et al. Double-Stranded RNA-Binding Protein DRB3 Negatively Regulates Anthocyanin Biosynthesis by Modulating PAP1 Expression in Arabidopsis thaliana. J. Plant Res. 2017, 130, 45–55. [Google Scholar] [CrossRef] [PubMed]
- An, J.-P.; Qu, F.-J.; Yao, J.-F.; Wang, X.-N.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. The BZIP Transcription Factor MdHY5 Regulates Anthocyanin Accumulation and Nitrate Assimilation in Apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Meng, X.; Cai, J.; Li, G.; Dong, T.; Li, Z. Basic Leucine Zipper Transcription Factor SlbZIP1 Mediates Salt and Drought Stress Tolerance in Tomato. BMC Plant Biol. 2018, 18, 83. [Google Scholar] [CrossRef]
- Fan, L.; Xu, L.; Wang, Y.; Tang, M.; Liu, L. Genome- and Transcriptome-Wide Characterization of BZIP Gene Family Identifies Potential Members Involved in Abiotic Stress Response and Anthocyanin Biosynthesis in Radish (Raphanus sativus L.). Int. J. Mol. Sci. 2019, 20, 6334. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Wu, M.; Zhao, Y.; Zhang, A.; Liu, B.; Schiefelbein, J.; Gan, Y. Involvement of C2H2 Zinc Finger Proteins in the Regulation of Epidermal Cell Fate Determination in Arabidopsis: C2H2 Proteins in Regulation of Epidermal Cell Fate. J. Integr. Plant Biol. 2014, 56, 1112–1117. [Google Scholar] [CrossRef]
- Wang, D.-R.; Yang, K.; Wang, X.; Lin, X.-L.; Rui, L.; Liu, H.-F.; Liu, D.-D.; You, C.-X. Overexpression of MdZAT5, an C2H2-Type Zinc Finger Protein, Regulates Anthocyanin Accumulation and Salt Stress Response in Apple Calli and Arabidopsis. Int. J. Mol. Sci. 2022, 23, 1897. [Google Scholar] [CrossRef]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.A.; Rothstein, S.J. The Arabidopsis Transcription Factor ANAC032 Represses Anthocyanin Biosynthesis in Response to High Sucrose and Oxidative and Abiotic Stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Dang, Z.; Zheng, L.; Wang, J.; Gao, Z.; Wu, S.; Qi, Z.; Wang, Y. Transcriptomic Profiling of the Salt-Stress Response in the Wild Recretohalophyte Reaumuria trigyna. BMC Genom. 2013, 14, 29. [Google Scholar] [CrossRef]
- Zhang, H.; Du, C.; Wang, Y.; Wang, J.; Zheng, L.; Wang, Y. The Reaumuria trigyna Leucoanthocyanidin Dioxygenase (RtLDOX) Gene Complements Anthocyanidin Synthesis and Increases the Salt Tolerance Potential of a Transgenic Arabidopsis LDOX Mutant. Plant Physiol. Biochem. 2016, 106, 278–287. [Google Scholar] [CrossRef]
- Kim, J.; Lee, W.J.; Vu, T.T.; Jeong, C.Y.; Hong, S.-W.; Lee, H. High Accumulation of Anthocyanins via the Ectopic Expression of AtDFR Confers Significant Salt Stress Tolerance in Brassica napus L. Plant Cell Rep. 2017, 36, 1215–1224. [Google Scholar] [CrossRef]
- Wang, J.; Li, D.; Peng, Y.; Cai, M.; Liang, Z.; Yuan, Z.; Du, X.; Wang, J.; Schnable, P.S.; Gu, R.; et al. The Anthocyanin Accumulation Related ZmBZ1, Facilitates Seedling Salinity Stress Tolerance via ROS Scavenging. Int. J. Mol. Sci. 2022, 23, 16123. [Google Scholar] [CrossRef]
- Wu, Q.; Su, N.; Zhang, X.; Liu, Y.; Cui, J.; Liang, Y. Hydrogen Peroxide, Nitric Oxide and UV RESISTANCE LOCUS8 Interact to Mediate UV-B-Induced Anthocyanin Biosynthesis in Radish Sprouts. Sci. Rep. 2016, 6, 29164. [Google Scholar] [CrossRef]
- Sadat-Hosseini, M.; Naeimi, A.; Boroomand, N.; Aalifar, M.; Farajpour, M. Alleviating the Adverse Effects of Salinity on Roselle Plants by Green Synthesized Nanoparticles. Sci. Rep. 2022, 12, 18165. [Google Scholar] [CrossRef] [PubMed]
- Tisarum, R.; Theerawitaya, C.; Samphumphuang, T.; Polispitak, K.; Thongpoem, P.; Singh, H.P.; Cha-Um, S. Alleviation of Salt Stress in Upland Rice (Oryza sativa L. ssp. indica Cv. Leum Pua) Using Arbuscular Mycorrhizal Fungi Inoculation. Front. Plant Sci. 2020, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic Stress Responses in Plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Faqir Napar, W.P.; Kaleri, A.R.; Ahmed, A.; Nabi, F.; Sajid, S.; Ćosić, T.; Yao, Y.; Liu, J.; Raspor, M.; Gao, Y. The Anthocyanin-Rich Tomato Genotype LA-1996 Displays Superior Efficiency of Mechanisms of Tolerance to Salinity and Drought. J. Plant Physiol. 2022, 271, 153662. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.-J.; Zhang, F.-J.; Zhang, G.-Z.; Jiang, X.-Y.; Yu, H.-M.; Hou, B.-K. The Arabidopsis UDP-Glycosyltransferases UGT79B2 and UGT79B3, Contribute to Cold, Salt and Drought Stress Tolerance via Modulating Anthocyanin Accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, X.; Ma, B.; Wu, Z.; Zheng, L.; Qi, Z.; Wang, Y. A Leucoanthocyanidin Dioxygenase Gene (RtLDOX2) from the Feral Forage Plant Reaumuria trigyna Promotes the Accumulation of Flavonoids and Improves Tolerance to Abiotic Stresses. J. Plant Res. 2021, 134, 1121–1138. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Fan, F.; Yu, Q.; Zhang, P. A Moss 2-Oxoglutarate/Fe(II)-Dependent Dioxygenases (2-ODD) Gene of Flavonoids Biosynthesis Positively Regulates Plants Abiotic Stress Tolerance. Front. Plant Sci. 2022, 13, 850062. [Google Scholar] [CrossRef]
- Outchkourov, N.S.; Karlova, R.; Hölscher, M.; Schrama, X.; Blilou, I.; Jongedijk, E.; Simon, C.D.; Van Dijk, A.D.J.; Bosch, D.; Hall, R.D.; et al. Transcription Factor-Mediated Control of Anthocyanin Biosynthesis in Vegetative Tissues. Plant Physiol. 2018, 176, 1862–1878. [Google Scholar] [CrossRef]
- Naing, A.H.; Park, K.I.; Ai, T.N.; Chung, M.Y.; Han, J.S.; Kang, Y.-W.; Lim, K.B.; Kim, C.K. Overexpression of Snapdragon Delila (Del) Gene in Tobacco Enhances Anthocyanin Accumulation and Abiotic Stress Tolerance. BMC Plant Biol. 2017, 17, 65. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Ai, T.N.; Lim, K.B.; Lee, I.J.; Kim, C.K. Overexpression of Rosea1 from Snapdragon Enhances Anthocyanin Accumulation and Abiotic Stress Tolerance in Transgenic Tobacco. Front. Plant Sci. 2018, 9, 1070. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.; Zhang, Y.; Lyu, Y. A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 3195. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Deng, R.; Bai, Y.; Wu, H.; Li, C.; Wu, Q.; Zhao, H. Tartary Buckwheat R2R3-MYB Gene FtMYB3 Negatively Regulates Anthocyanin and Proanthocyanin Biosynthesis. Int. J. Mol. Sci. 2022, 23, 2775. [Google Scholar] [CrossRef]
- Ren, M.; Wang, Z.; Xue, M.; Wang, X.; Zhang, F.; Zhang, Y.; Zhang, W.; Wang, M. Constitutive Expression of an A-5 Subgroup Member in the DREB Transcription Factor Subfamily from Ammopiptanthus mongolicus Enhanced Abiotic Stress Tolerance and Anthocyanin Accumulation in Transgenic Arabidopsis. PLoS ONE 2019, 14, e0224296. [Google Scholar] [CrossRef]
- Fu, M.; Kang, H.K.; Son, S.-H.; Kim, S.-K.; Nam, K.H. A Subset of Arabidopsis RAV Transcription Factors Modulates Drought and Salt Stress Responses Independent of ABA. Plant Cell Physiol. 2014, 55, 1892–1904. [Google Scholar] [CrossRef]
- Liu, J.; Deng, Z.; Liang, C.; Sun, H.; Li, D.; Song, J.; Zhang, S.; Wang, R. Genome-Wide Analysis of RAV Transcription Factors and Functional Characterization of Anthocyanin-Biosynthesis-Related RAV Genes in Pear. Int. J. Mol. Sci. 2021, 22, 5567. [Google Scholar] [CrossRef]
- Strygina, K.V.; Khlestkina, E.K. Myc-like Transcriptional Factors in Wheat: Structural and Functional Organization of the Subfamily I Members. BMC Plant Biol. 2019, 19, 50. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dabravolski, S.A.; Isayenkov, S.V. The Role of Anthocyanins in Plant Tolerance to Drought and Salt Stresses. Plants 2023, 12, 2558. https://doi.org/10.3390/plants12132558
Dabravolski SA, Isayenkov SV. The Role of Anthocyanins in Plant Tolerance to Drought and Salt Stresses. Plants. 2023; 12(13):2558. https://doi.org/10.3390/plants12132558
Chicago/Turabian StyleDabravolski, Siarhei A., and Stanislav V. Isayenkov. 2023. "The Role of Anthocyanins in Plant Tolerance to Drought and Salt Stresses" Plants 12, no. 13: 2558. https://doi.org/10.3390/plants12132558
APA StyleDabravolski, S. A., & Isayenkov, S. V. (2023). The Role of Anthocyanins in Plant Tolerance to Drought and Salt Stresses. Plants, 12(13), 2558. https://doi.org/10.3390/plants12132558






