Expression of EPL1 from Trichoderma atroviride in Arabidopsis Confers Resistance to Bacterial and Fungal Pathogens
Abstract
1. Introduction
2. Results
2.1. Generation of Arabidopsis EPL1 Expressing Lines (35S::TaEPL1)
2.2. The Expression of the EPL1 Gene in Arabidopsis Results in Accelerated Growth
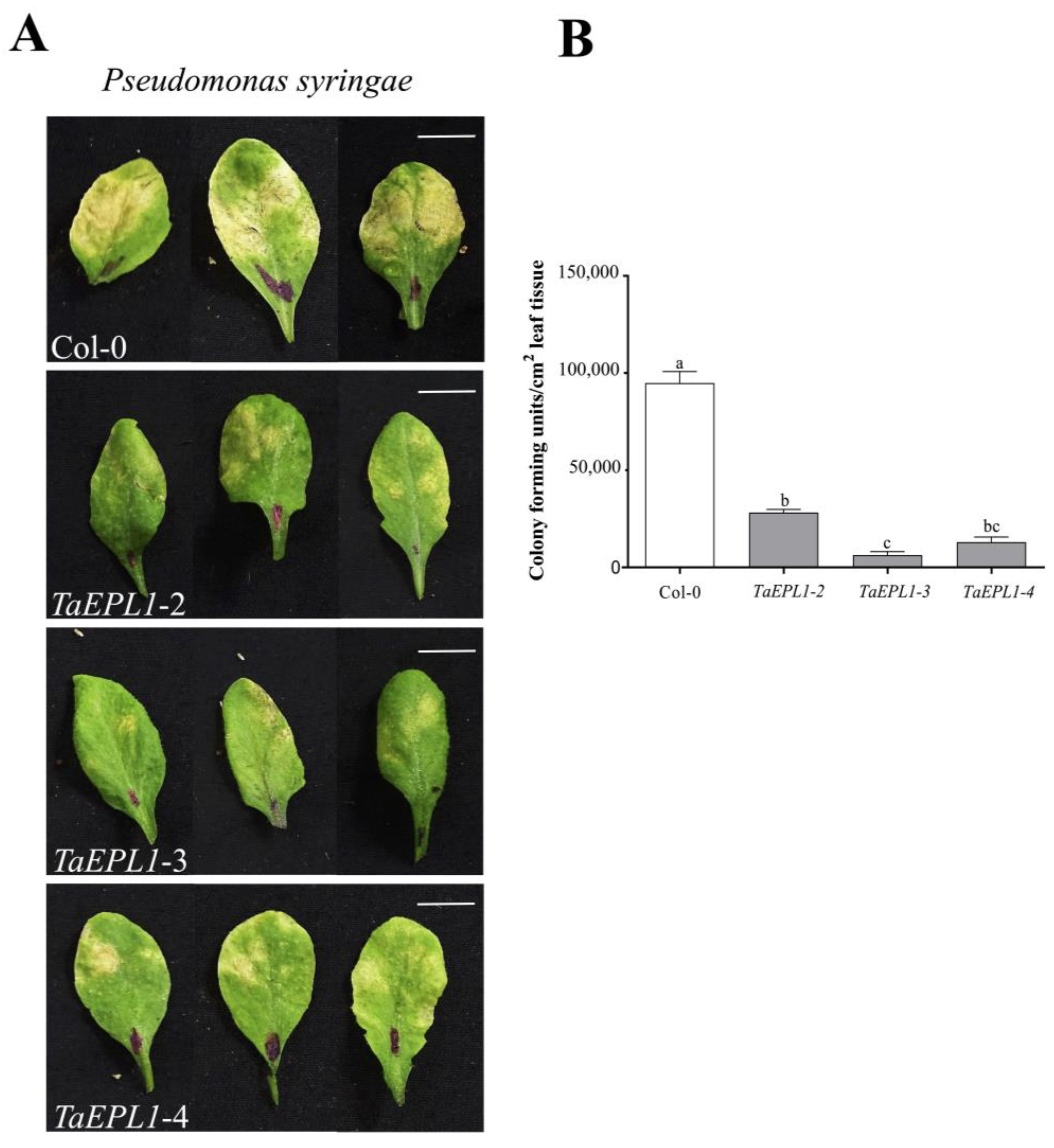
2.3. The Arabidopsis 35S::TaEPL1 Lines Are More Resistant to Infection by Pseudomonas Syringae
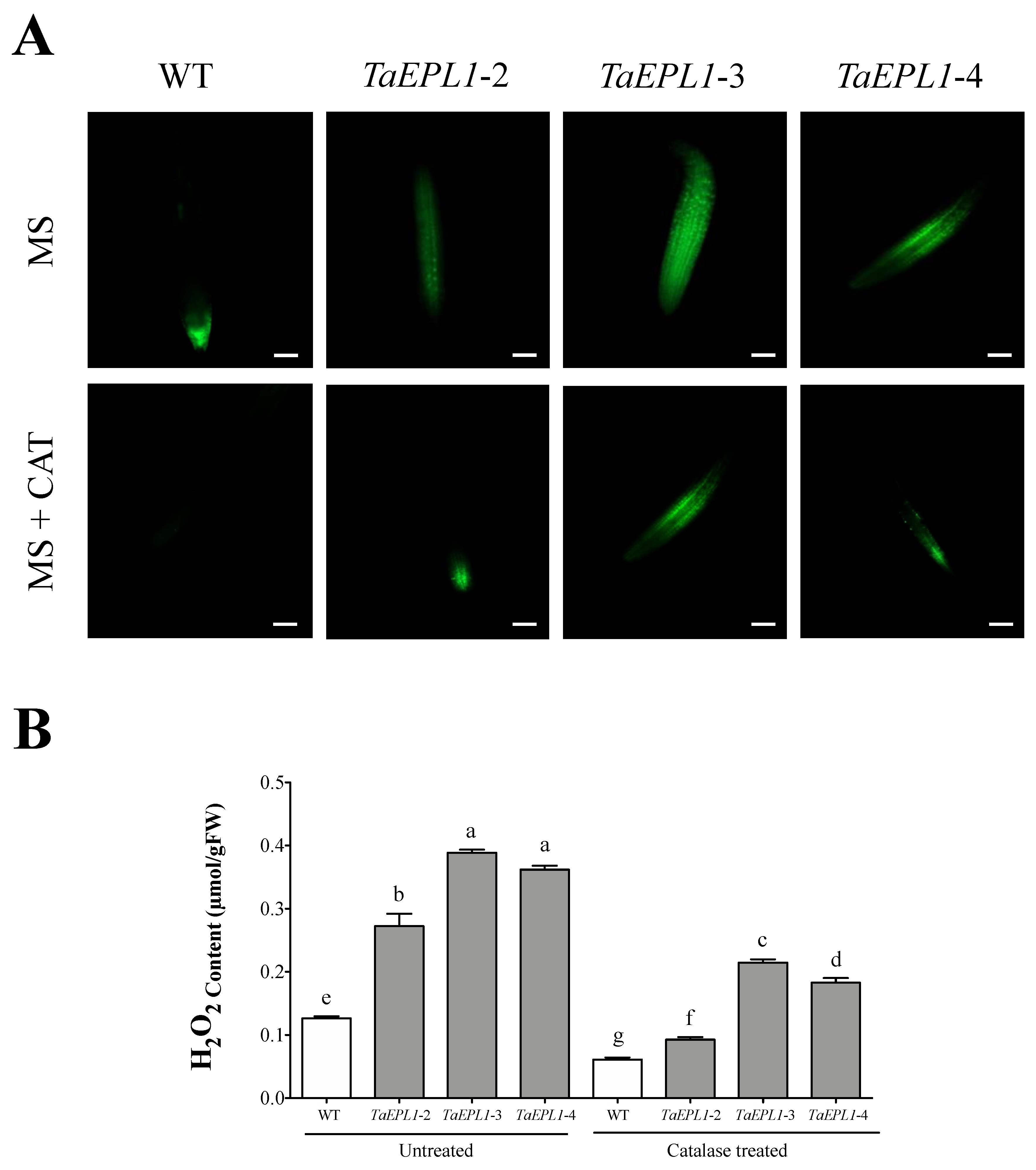
2.4. The Arabidopsis 35S::TaEPL1 Lines Display Increased Reactive Oxygen Species (ROS)
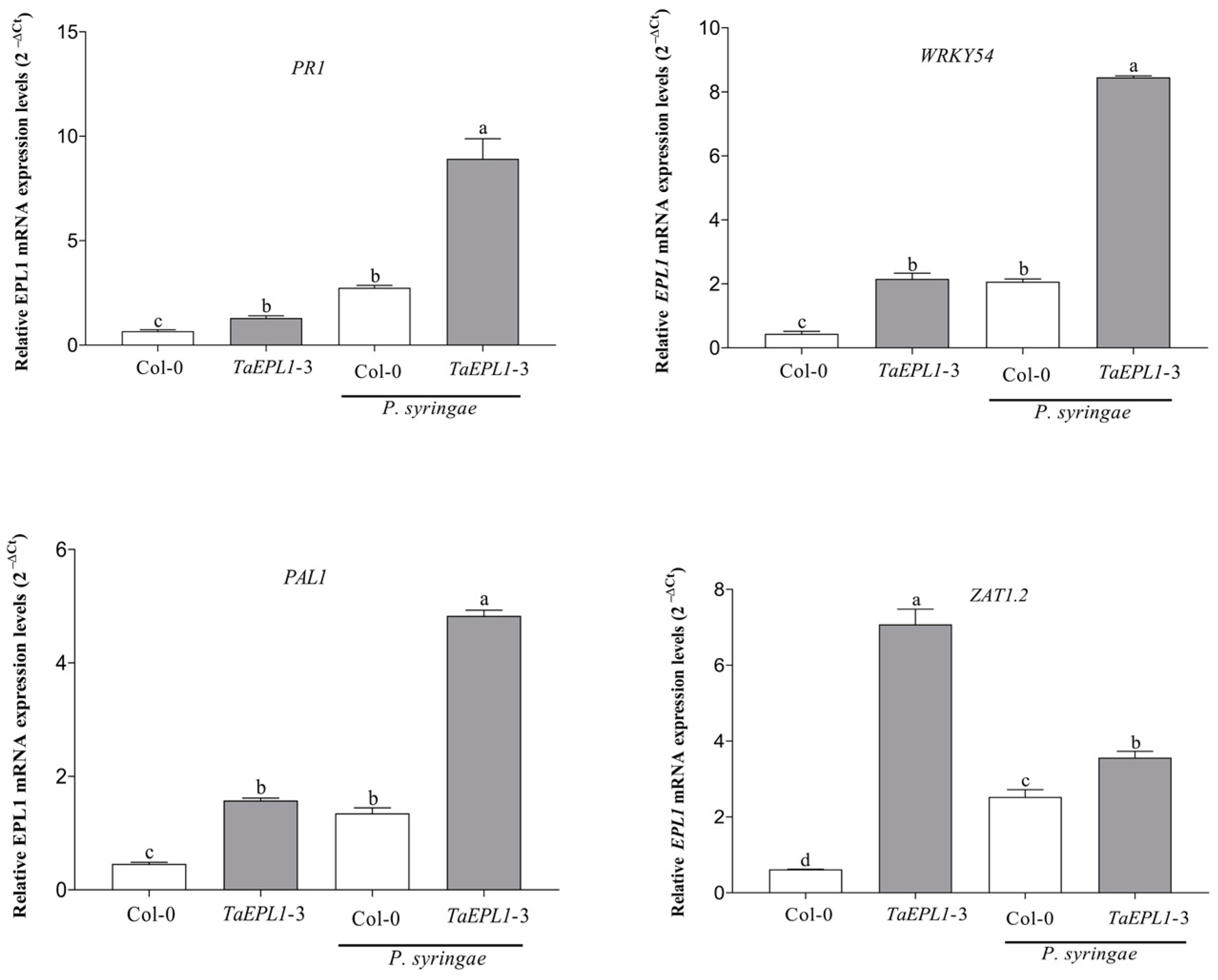
2.5. The 35S::TaEPL1-3 Line Shows a Higher Accumulation of Transcripts of SA-Related Genes under Pseudomonas Syringae Infection
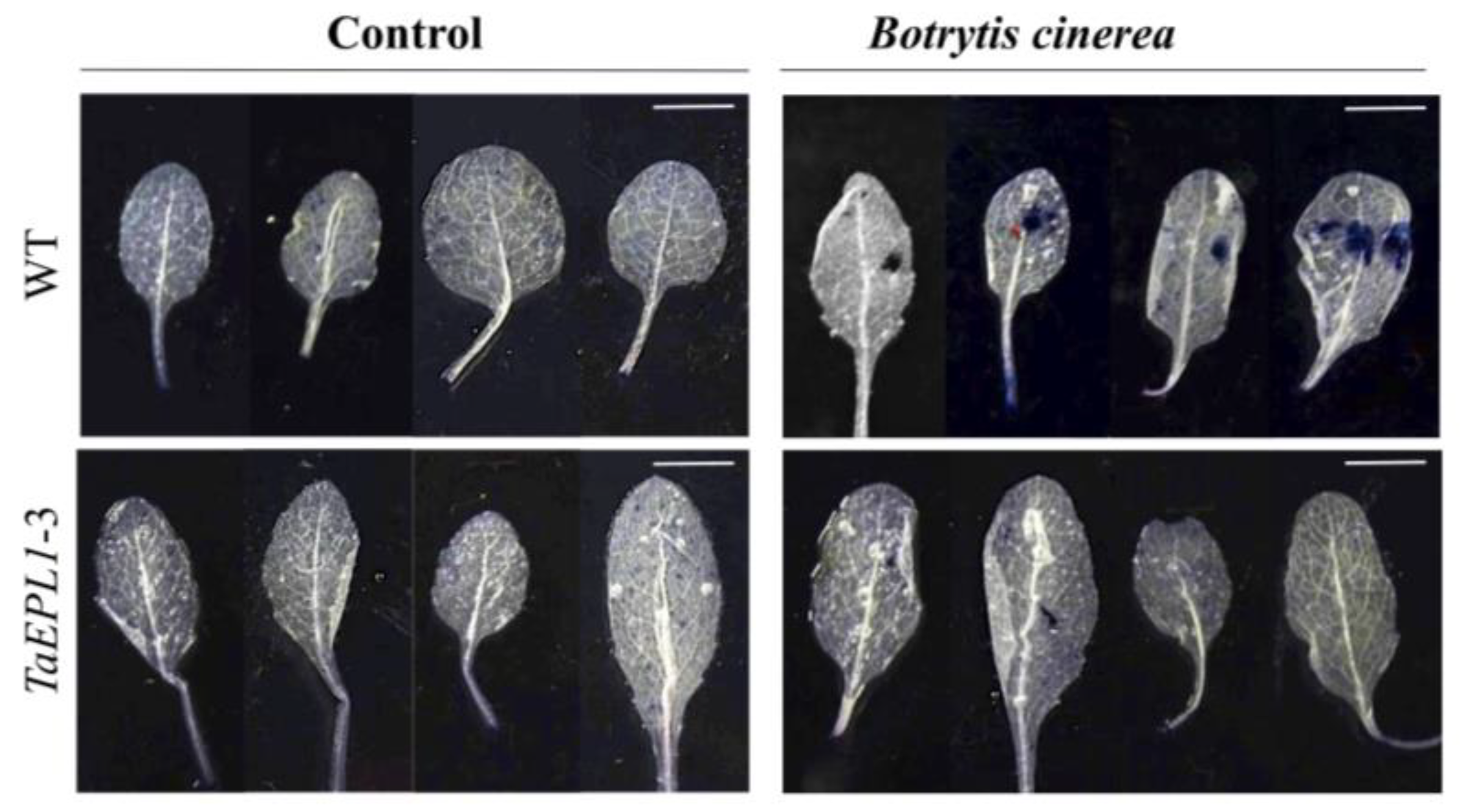
2.6. The Arabidopsis 35S::TaEPL1 Lines Were More Resistant to Botrytis cinerea Infection
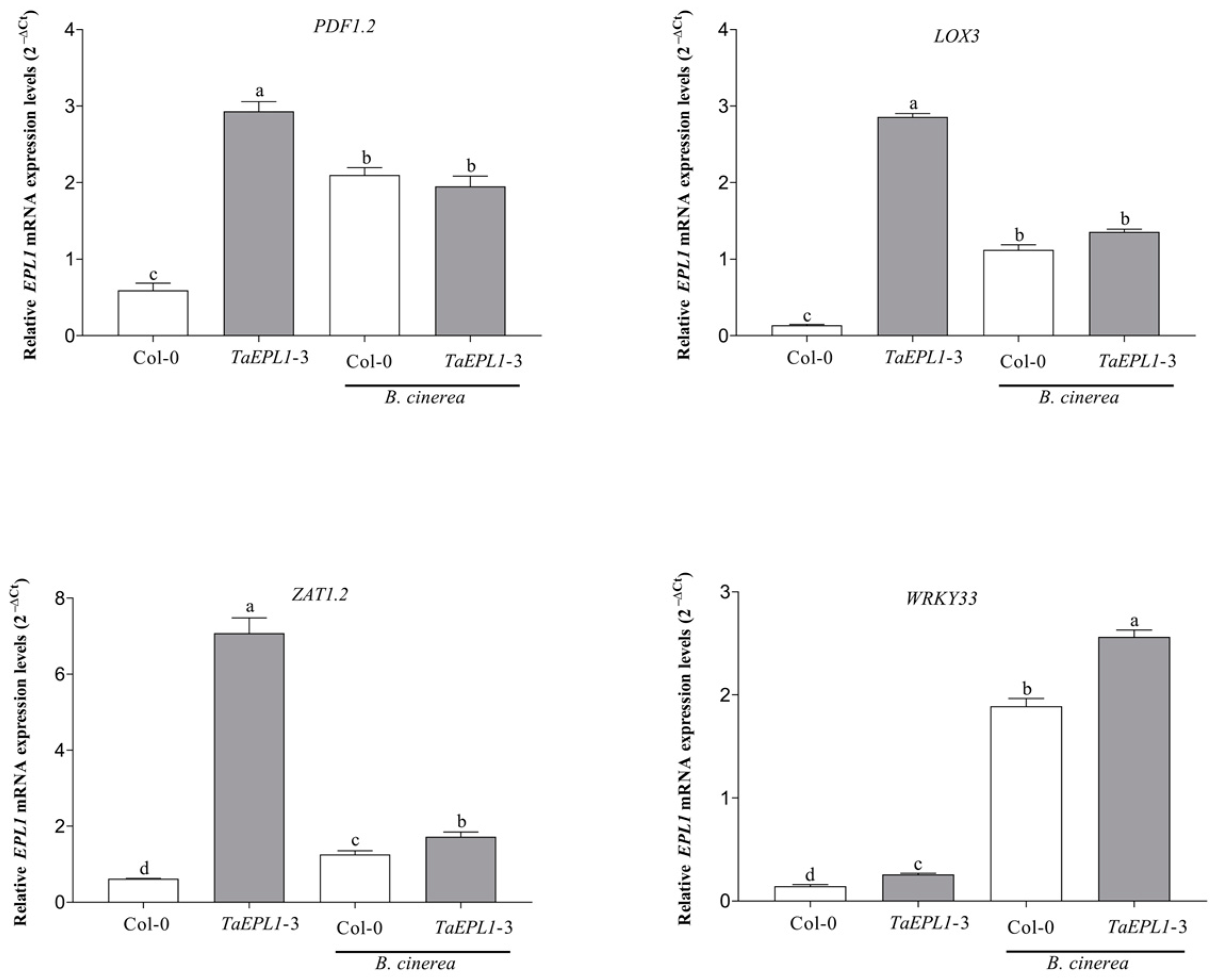
2.7. Expression Changes Plant Defense Genes in the 35S::TaEPL1-3 Line under Botrytis cinerea Infection
2.8. Determination of H2O2 Content in Arabidopsis WT and 35S::TaEPL1-3 Lines under B. cinerea and P. syringae Infection
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Generation of Arabidopsis thaliana 35S::TaEPL1 Lines
4.3. Measurement of Fresh Weight of 35S::TaEPL1 and WT Lines
4.4. Pathogen Inocula Preparation
4.5. Arabidopsis 35S::TaEPL1 Expression Lines—Pseudomonas syringae Inoculation Assays
4.6. Arabidopsis 35S::TaEPL1 Expression Lines—Botrytis cinerea Inoculation Assays
4.7. Plant Tissue Trypan Blue Staining after Botrytis cinerea Infection
4.8. Hydrogen Peroxide (H2O2) Detection by DCFH2-DA Labeling
4.9. Determination of Hydrogen Peroxide Content with Potassium Iodide (KI)
4.10. RNA Isolation and RT-qPCR Gene Expression Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species—Opportunistic, avirulent plant symbionts. Nat. Rev. Genet. 2004, 2, 43–56. [Google Scholar] [CrossRef]
- Khan, R.A.A.; Najeeb, S.; Hussain, S.; Xie, B.; Li, Y. Bioactive Secondary Metabolites from Trichoderma spp. against Phytopathogenic Fungi. Microorganisms 2020, 8, 817. [Google Scholar] [CrossRef]
- Guo, Y.; Fan, Z.; Yi, X.; Zhang, Y.; Khan, R.A.A.; Zhou, Z. Sustainable Management of Soil-Borne Bacterium Ralstonia solanacearum In Vitro and In Vivo through Fungal Metabolites of Different Trichoderma spp. Sustainability 2021, 13, 1491. [Google Scholar] [CrossRef]
- Ruan, J.; Yao, X.; Zhang, K.; Zhao, M. Trichoderma and Its Role in Biological Control of Plant Fungal and Nematode Disease. Front. Microbiol. 2023, 14, 1275. [Google Scholar] [CrossRef]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.; Marra, R.; Barbetti, M.; Li, H.; Woo, S.; Lorito, M. A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 2008, 72, 80–86. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Horwitz, B.A.; Singh, U.S.; Mukherjee, M.; Schmoll, M. Trichoderma in agriculture, industry and medicine: An overview. In Trichoderma Biology and Applications; CAB International: Boston, MA, USA, 2013; Volume 16, pp. 1–9. [Google Scholar] [CrossRef]
- Zin, N.A.; Badaluddin, N.A. Biological functions of Trichoderma spp. for agriculture applications. Ann. Agric. Sci. 2020, 65, 168–178. [Google Scholar] [CrossRef]
- Hermosa, R.; Viterbo, A.; Chet, I.; Monte, E. Plant-beneficial effects of Trichoderma and of its genes. Microbiology 2012, 158, 17–25. [Google Scholar] [CrossRef]
- Alfiky, A.; Weisskopf, L. Deciphering Trichoderma–Plant–Pathogen Interactions for Better Development of Biocontrol Applications. J. Fungi 2021, 7, 61. [Google Scholar] [CrossRef]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Ruocco, M.; Woo, S.; Lorito, M. Trichoderma Secondary Metabolites that Affect Plant Metabolism. Nat. Prod. Commun. 2012, 7, 1934578X1200701133. [Google Scholar] [CrossRef]
- Nieto-Jacobo, M.F.; Steyaert, J.M.; Salazar-Badillo, F.B.; Nguyen, D.V.; Rostás, M.; Braithwaite, M.; De Souza, J.T.; Jimenez-Bremont, J.F.; Ohkura, M.; Stewart, A.; et al. Environmental Growth Conditions of Trichoderma spp. Affects Indole Acetic Acid Derivatives, Volatile Organic Compounds, and Plant Growth Promotion. Front. Plant Sci. 2017, 8, 102. [Google Scholar] [CrossRef]
- González-Pérez, E.; Ortega-Amaro, M.A.; Salazar-Badillo, F.B.; Bautista, E.; Douterlungne, D.; Jiménez-Bremont, J.F. The Arabidopsis-Trichoderma interaction reveals that the fungal growth medium is an important factor in plant growth induction. Sci. Rep. 2018, 8, 16427. [Google Scholar] [CrossRef]
- Thakur, M.; Sohal, B.S. Role of Elicitors in Inducing Resistance in Plants against Pathogen Infection: A Review. Int. Sch. Res. Not. 2013, 2013, 762412. [Google Scholar] [CrossRef]
- Djonović, S.; Pozo, M.J.; Dangott, L.J.; Howell, C.R.; Kenerley, C.M. Sm1, a Proteinaceous Elicitor Secreted by the Biocontrol Fungus Trichoderma virens Induces Plant Defense Responses and Systemic Resistance. Mol. Plant-Microbe Interact. 2006, 19, 838–853. [Google Scholar] [CrossRef]
- Lombardi, L.; Faoro, F.; Luti, S.; Baccelli, I.; Martellini, F.; Bernardi, R.; Picciarelli, P.; Scala, A.; Pazzagli, L. Differential timing of defense-related responses induced by cerato-platanin and cerato-populin, two non-catalytic fungal elicitors. Physiol. Plant. 2013, 149, 408–421. [Google Scholar] [CrossRef]
- Luti, S.; Sella, L.; Quarantin, A.; Pazzagli, L.; Baccelli, I. Twenty years of research on cerato-platanin family proteins: Clues, conclusions, and unsolved issues. Fungal Biol. Rev. 2019, 34, 13–24. [Google Scholar] [CrossRef]
- Seidl, V.; Marchetti, M.; Schandl, R.; Allmaier, G.; Kubicek, C.P. Epl1, the major secreted protein of Hypocrea atroviridis on glucose, is a member of a strongly conserved protein family comprising plant defense response elicitors. FEBS J. 2006, 273, 4346–4359. [Google Scholar] [CrossRef]
- Buensanteai, N.; Mukherjee, P.K.; Horwitz, B.A.; Cheng, C.; Dangott, L.J.; Kenerley, C.M. Expression and purification of biologically active Trichoderma virens proteinaceous elicitor Sm1 in Pichia pastoris. Protein Expr. Purif. 2010, 72, 131–138. [Google Scholar] [CrossRef]
- Djonović, S.; Vargas, W.A.; Kolomiets, M.V.; Horndeski, M.; Wiest, A.; Kenerley, C.M. A Proteinaceous Elicitor Sm1 from the Beneficial Fungus Trichoderma virens Is Required for Induced Systemic Resistance in Maize. Plant Physiol. 2007, 145, 875–889. [Google Scholar] [CrossRef]
- Salas-Marina, M.A.; Isordia-Jasso, M.I.; Islas-Osuna, M.A.; Delgado-Sánchez, P.; Bremont, J.F.J.; Rodríguez-Kessler, M.; Rosales-Saavedra, M.T.; Herrera-Estrella, A.; Ecasas-Flores, S. The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum. Front. Plant Sci. 2015, 6, 77. [Google Scholar] [CrossRef]
- Vargas, W.A.; Djonović, S.; Sukno, S.A.; Kenerley, C.M. Dimerization Controls the Activity of Fungal Elicitors That Trigger Systemic Resistance in Plants. J. Biol. Chem. 2008, 283, 19804–19815. [Google Scholar] [CrossRef]
- Wang, Y.; Song, J.; Wu, Y.; Odeph, M.; Liu, Z.; Howlett, B.J.; Wang, S.; Yang, P.; Yao, L.; Zhao, L.; et al. Eplt4 Proteinaceous Elicitor Produced in Pichia pastoris Has a Protective Effect Against Cercosporidium sofinum Infections of Soybean Leaves. Appl. Biochem. Biotechnol. 2013, 169, 722–737. [Google Scholar] [CrossRef]
- Yu, W.; Mijiti, G.; Huang, Y.; Fan, H.; Wang, Y.; Liu, Z. Functional analysis of eliciting plant response protein Epl1-Tas from Trichoderma asperellum ACCC30536. Sci. Rep. 2018, 8, 7974. [Google Scholar] [CrossRef]
- Frías, M.; González, C.; Brito, N. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011, 192, 483–495. [Google Scholar] [CrossRef]
- Frías, M.; Brito, N.; González, C. The Botrytis cinerea cerato-platanin BcSpl1 is a potent inducer of systemic acquired resistance (SAR) in tobacco and generates a wave of salicylic acid expanding from the site of application. Mol. Plant Pathol. 2013, 14, 191–196. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, H.; Li, G.; Li, W.; Wang, X.; Song, F. Ectopic expression of MgSM1, a Cerato-platanin family protein from Magnaporthe grisea, confers broad-spectrum disease resistance in Arabidopsis. Plant Biotechnol. J. 2009, 7, 763–777. [Google Scholar] [CrossRef]
- Hong, Y.; Yang, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of MoSM1, encoding for an immunity-inducing protein from Magnaporthe oryzae, in rice confers broad-spectrum resistance against fungal and bacterial diseases. Sci. Rep. 2017, 7, srep41037. [Google Scholar] [CrossRef]
- Pazzagli, L.; Cappugi, G.; Manao, G.; Camici, G.; Santini, A.; Scala, A. Purification, Characterization, and Amino Acid Sequence of Cerato-platanin, a New Phytotoxic Protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 1999, 274, 24959–24964. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Kim, S.G.; Tsuda, K.; Gupta, R.; Park, S.-Y.; Kim, S.T.; Kang, K.Y. Magnaporthe oryzae-Secreted Protein MSP1 Induces Cell Death and Elicits Defense Responses in Rice. Mol. Plant-Microbe Interact. 2016, 29, 299–312. [Google Scholar] [CrossRef]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant. 2010, 138, 414–429. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Diezel, C.; Somssich, I.E. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 2012, 159, 266–285. [Google Scholar] [CrossRef]
- Salazar-Badillo, F.B.; Sánchez-Rangel, D.; Becerra-Flora, A.; López-Gómez, M.; Nieto-Jacobo, F.; Mendoza-Mendoza, A.; Jiménez-Bremont, J.F. Arabidopsis thaliana polyamine content is modified by the interaction with different Trichoderma species. Plant Physiol. Biochem. 2015, 95, 49–56. [Google Scholar] [CrossRef]
- Benítez, T.; Rincón, A.M.; Limón, M.C.; Codón, A.C. Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 2004, 7, 249–260. [Google Scholar]
- Sáenz-Mata, J.; Jiménez-Bremont, J.F. HR4 Gene is induced in the Arabidopsis-Trichoderma atroviride beneficial interaction. Int. J. Mol. Sci. 2012, 13, 9110–9128. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Horwitz, B.A.; Kenerley, C.M. Secondary metabolism in Trichoderma—A genomic perspective. Microbiology 2012, 158, 35–45. [Google Scholar] [CrossRef]
- Shoresh, M.; Harman, E.G. Differential expression of maize chitinases in the presence or absence of Trichoderma harzianum strain T22 and indications of a novel exo- endo-heterodimeric chitinase activity. BMC Plant Biol. 2010, 10, 136. [Google Scholar] [CrossRef]
- Howell, C.; Puckhaber, L. A study of the characteristics of “P” and “Q” strains of Trichoderma virens to account for differences in biological control efficacy against cotton seedling diseases. Biol. Control. 2005, 33, 217–222. [Google Scholar] [CrossRef]
- Reino, J.L.; Guerrero, R.F.; Hernandez-Galan, R.; Collado, I.G. Secondary metabolites from species of the biocontrol agent Trichoderma. Phytochem. Rev. 2008, 7, 89–123. [Google Scholar] [CrossRef]
- Sáenz-Mata, J.; Salazar-Badillo, F.B.; Jiménez-Bremont, J.F. Transcriptional regulation of Arabidopsis thaliana WRKY genes under interaction with beneficial fungus Trichoderma atroviride. Acta Physiol. Plant. 2014, 36, 1085–1093. [Google Scholar] [CrossRef]
- Rizhsky, L.; Davletova, S.; Liang, H.; Mittler, R. The Zinc Finger Protein Zat12 Is Required for Cytosolic Ascorbate Peroxidase 1 Expression during Oxidative Stress in Arabidopsis. J. Biol. Chem. 2004, 279, 11736–11743. [Google Scholar] [CrossRef]
- Scala, A.; Pazzagli, L.; Comparini, C.; Santini, A.; Tegli, S.; Cappugi, G. Cerato-platanin, an early-produced protein by Ceratocystis fimbriata f. sp. platani, elicits phytoalexin synthesis in host and non-host plants. J. Plant Pathol. 2004, 86, 27–33. [Google Scholar]
- Bennici, A.; Tani, C.; Pazzagli, L. Effects of cerato-platanin, a toxin from Ceratocystis fimbriataf. sp. platani, on Platanus acerifolialeaves: An ultrastructural study. Caryologia 2006, 59, 291–298. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis Thaliana-Pseudomonas Syringae Interaction. Arab. Book 2002, 1, e0039. [Google Scholar] [CrossRef]
- Isabel, C.M.A.; Ignacio, J.R.F.; Margarita, R.K.; Gill, S.S.; Alicia, B.F.; Francisco, J.B.J. Down-regulation of arginine decarboxylase gene-expression results in reactive oxygen species accumulation in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 506, 1071–1077. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas Moreno, M.M.; González-Pérez, E.; Rodríguez-Hernandez, A.A.; Ortega-Amaro, M.A.; Becerra-Flora, A.; Serrano, M.; Jiménez-Bremont, J.F. Expression of EPL1 from Trichoderma atroviride in Arabidopsis Confers Resistance to Bacterial and Fungal Pathogens. Plants 2023, 12, 2443. https://doi.org/10.3390/plants12132443
Rojas Moreno MM, González-Pérez E, Rodríguez-Hernandez AA, Ortega-Amaro MA, Becerra-Flora A, Serrano M, Jiménez-Bremont JF. Expression of EPL1 from Trichoderma atroviride in Arabidopsis Confers Resistance to Bacterial and Fungal Pathogens. Plants. 2023; 12(13):2443. https://doi.org/10.3390/plants12132443
Chicago/Turabian StyleRojas Moreno, Mónica Montserrat, Enrique González-Pérez, Aida Araceli Rodríguez-Hernandez, María Azucena Ortega-Amaro, Alicia Becerra-Flora, Mario Serrano, and Juan Francisco Jiménez-Bremont. 2023. "Expression of EPL1 from Trichoderma atroviride in Arabidopsis Confers Resistance to Bacterial and Fungal Pathogens" Plants 12, no. 13: 2443. https://doi.org/10.3390/plants12132443
APA StyleRojas Moreno, M. M., González-Pérez, E., Rodríguez-Hernandez, A. A., Ortega-Amaro, M. A., Becerra-Flora, A., Serrano, M., & Jiménez-Bremont, J. F. (2023). Expression of EPL1 from Trichoderma atroviride in Arabidopsis Confers Resistance to Bacterial and Fungal Pathogens. Plants, 12(13), 2443. https://doi.org/10.3390/plants12132443







