Development, Validation, and Application of Reverse Transcription Real-Time and Droplet Digital PCR Assays for the Detection of the Potyviruses Watermelon Mosaic Virus and Zucchini Yellow Mosaic Virus in Cucurbits
Abstract
:1. Introduction
2. Results
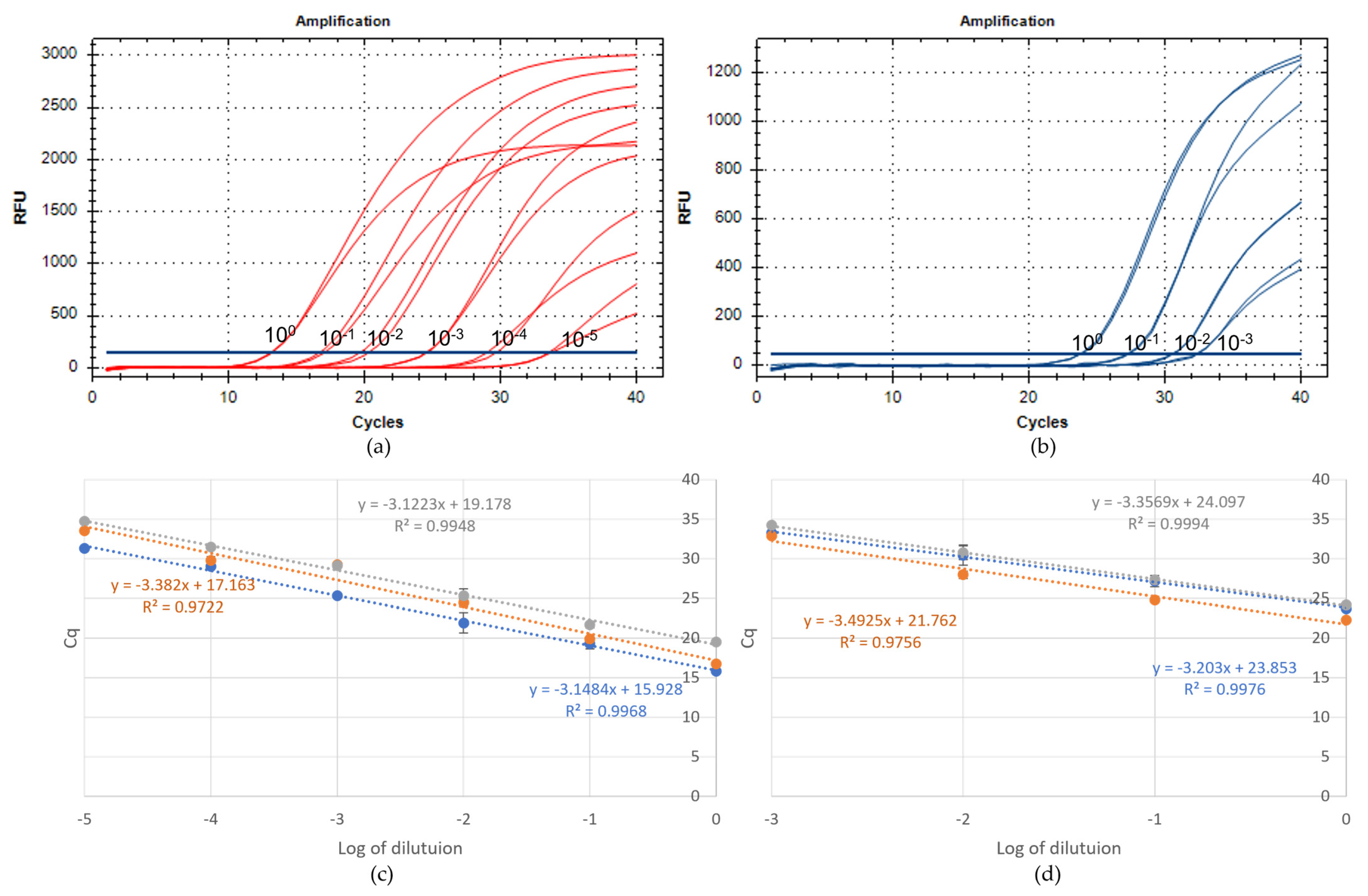
2.1. Set up of the Real-Time RT-PCR Assays
2.2. Validation of the Real-Time RT-PCR Assays
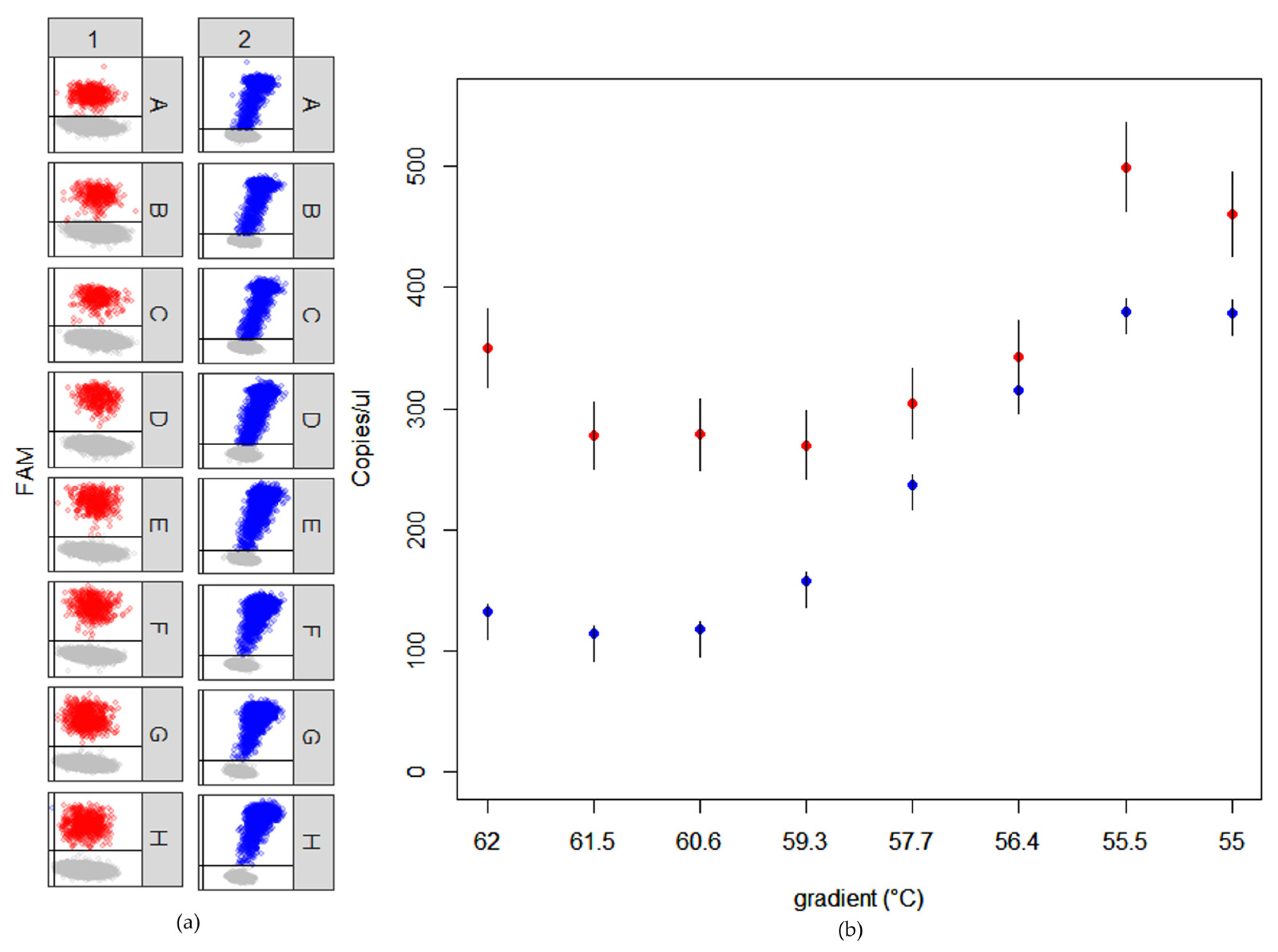
2.3. RT-ddPCR Assays
2.4. Test of Naturally Infected Samples
3. Discussion
4. Materials and Methods
4.1. Plant Material and RNA Extraction
4.2. Primer and Probe Design
4.3. Real-Time RT-PCR Set Up
4.4. Validation of the Real-Time RT-PCR Assays
4.4.1. Analytical Sensitivity
4.4.2. Analytical Specificity
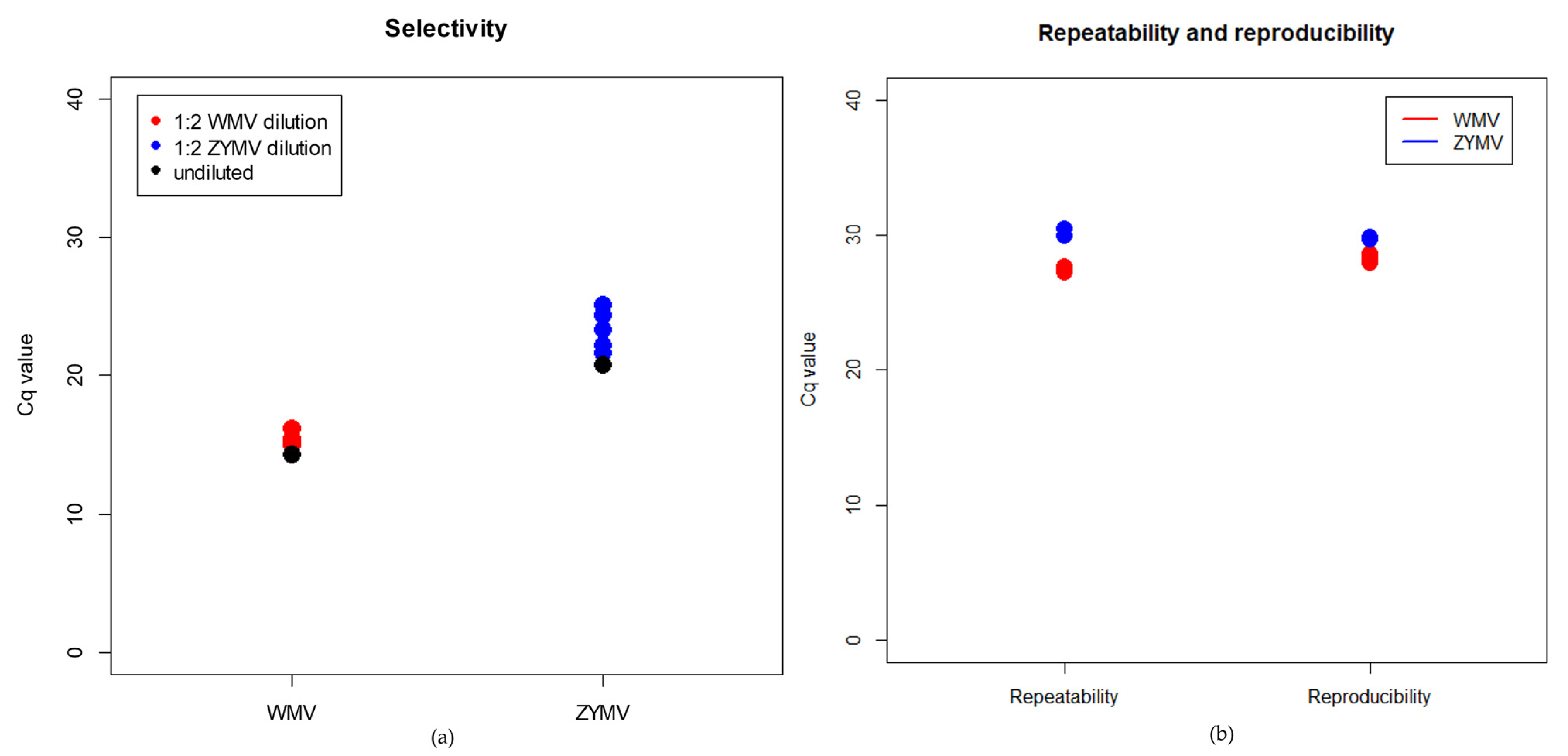
4.4.3. Selectivity
4.4.4. Repeatability and Reproducibility
4.5. RT-ddPCR Set Up
4.6. Test of Naturally Infected Samples
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grumet, R.; McCreight, J.D.; McGregor, C.; Weng, Y.; Mazourek, M.; Reitsma, K.; Labate, J.; Davis, A.; Fei, Z. Genetic Resources and Vulnerabilities of Major Cucurbit Crops. Genes 2021, 12, 1222. [Google Scholar] [CrossRef]
- Lecoq, H.; Desbiez, C. Viruses of Cucurbit Crops in the Mediterranean Region. An Ever-Changing Picture. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2012; Volume 84. [Google Scholar]
- Gadhave, K.R.; Gautam, S.; Rasmussen, D.A.; Srinivasan, R. Aphid Transmission of Potyvirus: The Largest Plant-Infecting RNA Virus Genus. Viruses 2020, 12, 773. [Google Scholar] [CrossRef]
- Ali, A. Epidemiology and Evolution of Poytviruses Infecting Cucurbits. In Applied Plant Virology: Advances, Detection, and Antiviral Strategies; Awasthi, L.P., Ed.; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- De Moya-Ruiz, C.; Gómez, P.; Juárez, M. Occurrence, Distribution, and Management of Aphid-Transmitted Viruses in Cucurbits in Spain. Pathogens 2023, 12, 422. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, E.; Perotto, M.C.; Bertin, S.; Manglli, A.; Luciani, C.; Conci, V.C.; Tomassoli, L. Molecular Variability of Watermelon Mosaic Virus Isolates from Argentina. Eur. J. Plant. Pathol. 2020, 156, 1091–1099. [Google Scholar] [CrossRef]
- Desbiez, C.; Wipf-Scheibel, C.; Millot, P.; Berthier, K.; Girardot, G.; Gognalons, P.; Hirsch, J.; Moury, B.; Nozeran, K.; Piry, S.; et al. Distribution and Evolution of the Major Viruses Infecting Cucurbitaceous and Solanaceous Crops in the French Mediterranean Area. Virus Res. 2020, 286, 198042. [Google Scholar] [CrossRef]
- Lecoq, H.; Fabre, F.; Joannon, B.; Wipf-Scheibel, C.; Chandeysson, C.; Schoeny, A.; Desbiez, C. Search for Factors Involved in the Rapid Shift in Watermelon mosaic virus (WMV) Populations in South-Eastern France. Virus Res. 2011, 159, 115–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juarez, M.; Legua, P.; Mengual, C.M.; Kassem, M.A.; Sempere, R.N.; Gómez, P.; Truniger, V.; Aranda, M.A. Relative Incidence, Spatial Distribution and Genetic Diversity of Cucurbit Viruses in Eastern Spain. Ann. Appl. Biol. 2013, 162, 362–370. [Google Scholar] [CrossRef]
- Bertin, S.; Manglli, A.; McLeish, M.; Tomassoli, L. Genetic Variability of Watermelon Mosaic Virus Isolates Infecting Cucurbit Crops in Italy. Arch. Virol. 2020, 165, 937–946. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, P.; Zhou, Q.; Zhou, X.; Guo, Z.; Cheng, L.; Zhu, L.; He, X.; Zhu, Y.; Hu, Y. Detection of Disease in Cucurbita maxima Duch. Ex Lam. Caused by a Mixed Infection of Zucchini yellow mosaic virus, Watermelon mosaic virus, and Cucumber mosaic virus in Southeast China Using a Novel Small RNA Sequencing Method. PeerJ 2019, 2019, e7930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-De-Castro, A.; Martınez de Alba, A.E.; Sáez, C.; Flores, A.; Sifres, A.; Gómez-Guillamón, M.L.; López, C.; Picó, B. Incidence and Genetic Diversity of Cucurbit Viruses in Spain. Acta Hortic. 2020, 1294, 203–210. [Google Scholar] [CrossRef]
- Khanal, V.; Wells, H.; Ali, A. High Prevalence of Three Potyviruses Infecting Cucurbits in Oklahoma and Phylogenetic Analysis of Cucurbit Aphid-Borne Yellows Virus Isolated from Pumpkins. Pathogens 2021, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Syller, J. Facilitative and Antagonistic Interactions between Plant Viruses in Mixed Infections. Mol. Plant. Pathol. 2012, 13, 204–216. [Google Scholar] [CrossRef]
- Mascia, T.; Gallitelli, D. Synergies and Antagonisms in Virus Interactions. Plant. Sci. 2016, 252, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Salvaudon, L.; De Moraes, C.M.; Mescher, M.C. Outcomes of Co-Infection by Two Potyviruses: Implications for the Evolution of Manipulative Strategies. Proc. R. Soc. B Biol. Sci. 2013, 280, 20122959. [Google Scholar] [CrossRef] [Green Version]
- Dietzgen, R.G.; Herrington, M.E. A Sensitive Semi-Quantitative Biotin-Streptavidin ELISA for the Detection of Potyviruses Infecting Cucurbits. Aust. J. Agric. Res. 1991, 42, 417–427. [Google Scholar] [CrossRef]
- Mohammed, H.S.; Zicca, S.; Manglli, A.; Mohamed, M.E.; El Siddig, M.A.; Tomassoli, L.; El Hussein, A.A. Identification and Phylogenetic Analysis of Common Pumpkin Viruses in Sudan. J. Plant Pathol. 2014, 96, 77–84. [Google Scholar] [CrossRef]
- Rajbanshi, N.; Ali, A. Simultaneous Detection of Three Common Potyviruses Infecting Cucurbits by Multiplex Reverse Transcription Polymerase Chain Reaction Assay. J. Virol. Methods 2019, 273, 113725. [Google Scholar] [CrossRef]
- Zhao, Z.; Xiang, J.; Tian, Q.; Zhao, W.; Zhou, T.; Zhao, L.; Zhang, Y. Development of One-Step Multiplex RT-PCR Assay for Rapid Simultaneous Detection of Five RNA Viruses and Acidovorax citrulli in Major Cucurbitaceous Crops in China. Arch. Microbiol. 2022, 204, 696. [Google Scholar] [CrossRef]
- Kuan, C.-P.; Deng, T.-C.; Huang, H.-C.; Chi, H.-H.; Lu, Y.-L. Use of Reverse Transcription Loop-Mediated Isothermal Amplification for the Detection of Zucchini yellow mosaic virus. J. Phytopathol. 2014, 162, 238–244. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, Y.; Wu, Y.; Hao, X. Rapid Detection of Watermelon Viruses by Reverse Transcription Loop-Mediated Isothermal Amplification. J. Phytopathol. 2016, 164, 330–336. [Google Scholar] [CrossRef]
- Rubio, L.; Giménez, K.; Romero, J.; Font-San-Ambrosio, M.I.; Alfaro-Fernández, A.; Galipienso, L. Detection and Absolute Quantitation of Watermelon Mosaic Virus by Real-Time RT-PCR with a TaqMan Probe. J. Virol. Methods 2022, 300, 114416. [Google Scholar] [CrossRef]
- Taglienti, A.; Donati, L.; Ferretti, L.; Tomassoli, L.; Sapienza, F.; Sabatino, M.; Di Massimo, G.; Fiorentino, S.; Vecchiarelli, V.; Nota, P.; et al. In Vivo Antiphytoviral Activity of Essential Oils and Hydrosols from Origanum vulgare, Thymus vulgaris, and Rosmarinus officinalis to Control Zucchini Yellow Mosaic Virus and Tomato Leaf Curl New Delhi Virus in Cucurbita pepo L. Front. Microbiol. 2022, 13, 840893. [Google Scholar] [CrossRef]
- Taglienti, A.; Donati, L.; Dragone, I.; Ferretti, L.; Gentili, A.; Araniti, F.; Sapienza, F.; Astolfi, R.; Fiorentino, S.; Vecchiarelli, V.; et al. In Vivo Antiphytoviral and Aphid Repellency Activity of Essential Oils and Hydrosols from Mentha suaveolens and Foeniculum vulgare to Control Zucchini Yellow Mosaic Virus and Its Vector Aphis gossypii. Plants 2023, 12, 1078. [Google Scholar] [CrossRef] [PubMed]
- Chinnadurai, C.; Kollam, M.; Ramsubhag, A.; Jayaraman, J. Genome Characterization of Zucchini Yellow Mosaic Virus Infecting Cucurbits Reveals the Presence of a New Genotype in Trinidad and Tobago in the Caribbean Region. Arch. Virol. 2021, 166, 1661–1669. [Google Scholar] [CrossRef] [PubMed]
- Mehle, N.; Dobnik, D.; Ravnikar, M.; Pompe Novak, M. Validated Reverse Transcription Droplet Digital PCR Serves as a Higher Order Method for Absolute Quantification of Potato virus Y Strains. Anal. Bioanal. Chem. 2018, 410, 3815–3825. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Wang, Q.; Zhang, Y.; Shen, W.; Li, R.; Cao, M.; Chen, L.; Li, X.; Zhou, C.; et al. Development of a Sensitive and Reliable Reverse Transcription Droplet Digital PCR Assay for the Detection of Citrus Yellow Vein Clearing Virus. Arch. Virol. 2019, 164, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.; Mallik, I.; Gudmestad, N.C. Development and Application of a Real-Time Reverse-Transcription PCR and Droplet Digital PCR Assays for the Direct Detection of Potato mop top virus in Soil. Phytopathology 2020, 110, 58–67. [Google Scholar] [CrossRef]
- Villamil, C.; Calderon, M.N.; Arias, M.M.; Leguizamon, J.E. Validation of Droplet Digital Polymerase Chain Reaction for Salmonella Spp. Quantification. Front. Microbiol. 2020, 11, 1512. [Google Scholar] [CrossRef]
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet Digital PCR of Viral DNA/RNA, Current Progress, Challenges, and Future Perspectives. J. Med. Virol. 2021, 93, 4182–4197. [Google Scholar] [CrossRef]
- EPPO Standard-Diagnostics. PM 7/98 (5) Specific Requirements for Laboratories Preparing Accreditation for a Plant Pest Diagnostic Activity. EPPO Bull. 2021, 51, 468–498. [Google Scholar] [CrossRef]
- Manglli, A.; Bertin, S.; Tomassoli, L. Preliminary Analysis of ZYMV and WMV Interaction in Mixed Infection by ΔΔCt Rt-QPCR. In Proceedings of the International Advances in Plant Virology 2019, Rome, Italy, 29–31 October 2019; 95. [Google Scholar]
- Coutts, B.A.; Kehoe, M.A.; Webster, C.G.; Wylie, S.J.; Jones, R.A.C. Zucchini Yellow Mosaic Virus: Biological Properties, Detection Procedures and Comparison of Coat Protein Gene Sequences. Arch. Virol. 2011, 156, 2119–2131. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, R.G. Mathematics of Quantitative Kinetic PCR and the Application of Standard Curves. Nucleic Acids Res. 2003, 31, e93. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Version 4.0.5; R Foundation for Statistical Computing: Vienna, Austria, 2021; p. 55. ISBN 3-900051-07-0. Available online: https://www.R-project.org/ (accessed on 5 May 2023).
- Bidshahri, R.; Attali, D.; Fakhfakh, K.; McNeil, K.; Karsan, A.; Won, J.R.; Wolber, R.; Bryan, J.; Hughesman, C.; Haynes, C. Quantitative Detection and Resolution of BRAF V600 Status in Colorectal Cancer Using Droplet Digital PCR and a Novel Wild-Type Negative Assay. J. Mol. Diagn. 2016, 18, 190–204. [Google Scholar] [CrossRef] [Green Version]
| Name | Sequence (5′-3′) | Position (Sequence ID) | Reference |
|---|---|---|---|
| WMV-CP Probe | FAM-CCAACAAAAGCTGGCACAGTCAGCAA-BH1 | 9061-9086 (MN_296125.1) | [33] |
| WMV-CP F | TGGGCAGGGTAGCAAGGA | 9042-9059 (MN_296125.1) | |
| WMV-CP R | CCTTTTGATCCAACGTTCACATC | 9088-9110 (MN_296125.1) | |
| ZYMV-CP Probe | FAM-AGCCAACTGTGGCAGATGCTGGAGCT-BH1 | 8552-8573 (NC_003224.1) | This study |
| ZYMV-CP F | CCTACAAGCCCTCCATCAAG | 8484-8503 (NC_003224.1) | |
| ZYMV-CP R | ACTGTTTTCTCACCTGAGCC | 8632-8651 (NC_003224.1) |
| Real-Time RT-PCR | RT-ddPCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phytosanitary Status | Sample ID | Host | Test Set-Up | Analytical Sensitivity | Analytical Specificity—Inclusivity | Analytical Specificity—Exclusivity | Selectivity | Repeatability/ Reproducibility | Test Set-Up | Analytical Sensitivity |
| WMV | 36/12 | C. pepo | W | W | Z | W | W | |||
| WMV | 75/20 | C. pepo | W | W | W | Z | W | W | W | W |
| WMV | 136/20 | C. pepo | W | W | W | Z | W | W | W | |
| WMV | 163/17 | C. pepo | W | Z | ||||||
| WMV + CABYV | 86/18 | C. melo | W | Z | ||||||
| WMV + ZYMV | 191/17 | C. melo | WZ | |||||||
| WMV + ZYMV | 202/17 | C. melo | WZ | |||||||
| ZYMV | 23/12 | C. pepo | Z | W | ||||||
| ZYMV | 32Syn/12 | C. pepo | Z | W | ||||||
| ZYMV | 1ZB | C. pepo | Z | Z | Z | W | Z | Z | Z | Z |
| ZYMV | ZK | C. pepo | Z | Z | Z | W | Z | Z | Z | |
| ZYMV | ZB10.2 | C. pepo | Z | Z | W | Z | Z | |||
| ZYMV | 31Syn/12 | C. pepo | Z | W | ||||||
| Healthy | - | C. pepo | WZ | WZ | WZ | WZ | WZ | WZ | WZ | |
| Healthy | - | C. lanatus | WZ | WZ | ||||||
| Healthy | - | C. sativus | WZ | WZ | ||||||
| Healthy | - | C. melo | WZ | WZ | ||||||
| Healthy | - | C. moschata | WZ | WZ | ||||||
| MNSV | MNSV | C. lanatus | WZ | |||||||
| CMV | 104/16 | C. pepo | WZ | |||||||
| MWMV + PRSV | 144/19 | C. pepo | WZ | |||||||
| SLCV | PC-1271 | unknown | WZ | |||||||
| WmCSV | PV-0830 | unknown | WZ | |||||||
| CABYV | 153/17 | C. melo | WZ | |||||||
| ToLCNDV | 198/18 | C. pepo | WZ | |||||||
| PRSV | 45Syn/12 | C. pepo | WZ | |||||||
| CYSDV | 178/17 | C. pepo | WZ | |||||||
| BYPV | 60/16 | C. melo | WZ | |||||||
| Primers | |||||
|---|---|---|---|---|---|
| WMV | ZYMV | ||||
| 450 nM | 900 nM | 450 nM | 900 nM | ||
| Probe | 125 nM | 317 copies/µL | NT | 178 copies/µL | NT |
| 250 nM | 241 copies/µL | 343 copies/µL | 219 copies/µL | 315 copies/µL | |
| Dilutions | ||||||
|---|---|---|---|---|---|---|
| 10−7 | 10−6 | 10−5 | 10−4 | 10−3 | ||
| Samples | WMV 1 | 49 | 120 | 450 | 3880 | 28,900 |
| WMV 2 | 9 | 29 | 54 | 1790 | 21,240 | |
| WMV 3 | - | 12.4 | 40 | 395 | 2880 | |
| ZYMV 1 | - | - | 8 | 35 | 559 | |
| ZYMV 2 | - | - | 22 | 131 | 610 | |
| ZYMV 3 | - | - | 47 | 229 | 2160 | |
| SAMPLES | WMV | ZYMV | ||||||
|---|---|---|---|---|---|---|---|---|
| ID/Year | Host | Region/Location | RT-PCR | Real-Time RT-PCR (Cq) | RT-ddPCR (Copies/µL) | RT-PCR | Real-Time RT-PCR (Cq) | RT-ddPCR (Copies/µL) |
| 102/2017 | C. melo | Sardinia/Arborea | + | 13.37 | NT | - | - | - |
| 105/2017 | C. melo | Sardinia/Serramanna | + | 12.13 | NT | - | - | - |
| 106/2017 | C. melo | Sardinia/Uta | - | - | - | - | - | - |
| 113/2017 | C. melo | Sardinia/Arborea | - | 32.82 | 64 | - | - | - |
| 130/2017 | C. melo | Sardinia/Arborea | - | 27.71 | NT | - | - | - |
| 136/2017 | C. melo | Sardinia/Arborea | + | 14.70 | NT | - | - | - |
| 137/2017 | C. melo | Sardinia/Arborea | + | 17.03 | NT | - | - | - |
| 139/2017 | C. melo | Sardinia/Arborea | - | 30.85 | 375 | - | - | - |
| 163/2017 | C. melo | Sardinia/Arborea | + | 15.98 | NT | - | - | - |
| 178/2017 | C. melo | Sardinia/Uta | - | - | - | - | - | - |
| 182/2017 | C. melo | Sardinia/Uta | - | - | 11 | - | - | - |
| 189/2017 | C. melo | Sardinia/Serramanna | + | 22.07 | NT | + | 23.98 | NT |
| 190/2017 | C. melo | Sardinia/Serramanna | + | 22.77 | NT | + | 25.49 | NT |
| 191/2017 | C. melo | Sardinia/Serramanna | - | 29.48 | 87 | - | - | - |
| 192/2017 | C. melo | Sardinia/Serramanna | + | 20.11 | NT | + | 27.05 | NT |
| 201/2017 | C. melo | Sardinia/Serramanna | + | 18.57 | NT | + | 25.30 | NT |
| 203/2017 | C. melo | Sardinia/Serramanna | - | 30.76 | 240 | - | - | 38 |
| 143/2019 | C.pepo | Latium/Fondi | - | - | - | - | - | - |
| 147/2019 | C. pepo | Latium/Fondi | - | - | - | - | - | - |
| 148/2019 | C. pepo | Latium/Fondi | - | - | - | - | - | - |
| 51/2020 | C. pepo | Calabria/L. Terme | + | 20.32 | NT | - | - | 9 |
| 52/2020 | C. pepo | Calabria/L. Terme | + | 14.46 | NT | - | - | - |
| 53/2020 | C. pepo | Calabria/Cosenza | - | 27.50 | NT | - | - | - |
| 60/2020 | C. pepo | E. Romagna/Cesena | - | 26.89 | NT | - | - | - |
| 61/2020 | C. pepo | E. Romagna/Cesena | + | 23.37 | NT | + | 24.02 | NT |
| 62/2020 | C. pepo | E. Romagna/Cesena | + | 20.58 | NT | - | - | - |
| 63/2020 | C. pepo | E. Romagna/Cesena | + | 24.87 | NT | + | 23.72 | NT |
| 64/2020 | C. pepo | E. Romagna/Cesena | - | - | - | + | 22.71 | NT |
| 364/2021 | C. pepo | Veneto/Verona | + | 18.04 | NT | - | - | - |
| 365/2021 | C. pepo | Veneto/Verona | + | 18.73 | NT | - | - | - |
| 366/2021 | C. pepo | Veneto/Verona | + | 18.07 | NT | - | - | - |
| 367/2021 | C. pepo | Veneto/Verona | + | 23.35 | NT | - | - | - |
| 368/2021 | C. pepo | Veneto/Verona | + | 26.12 | NT | - | - | - |
| 369/2021 | C. lanatus | Marche/Pesaro-Urbino | + | 16.13 | NT | - | - | - |
| 370/2021 | C. pepo | Veneto/Verona | + | 15.15 | NT | - | - | - |
| 407/2021 | C. pepo | Veneto/Rovigo | + | 20.33 | NT | - | - | - |
| 423/2021 | C. pepo | Piedmont/Torino | + | 18.74 | NT | + | 27.95 | NT |
| 424/2021 | C. pepo | Piedmont/Torino | + | 19.35 | NT | + | 30.53 | 400 |
| 7/2022 | C. maxima | Latium | - | - | - | - | - | - |
| 8/2022 | C. maxima | Latium | - | - | - | - | - | 9 |
| 37/2022 | C. pepo | Latium/Latina | + | 18.70 | NT | + | 23.50 | NT |
| 38/2022 | C. pepo | Latium/Latina | - | - | - | + | 24.61 | NT |
| 39/2022 | C. pepo | Latium/Latina | + | 13.76 | NT | + | 22.87 | NT |
| 40/2022 | C. pepo | Latium/Latina | + | 19.04 | NT | + | 25.71 | NT |
| 41/2022 | C. pepo | Latium/Latina | + | 13.78 | NT | + | 23.71 | NT |
| 42/2022 | C. pepo | Latium/Latina | + | 18.07 | NT | + | 24.06 | NT |
| 43/2022 | C. pepo | Latium/Latina | + | 13.33 | NT | + | 22.36 | NT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luigi, M.; Manglli, A.; Corrado, C.L.; Tiberini, A.; Costantini, E.; Ferretti, L.; Tomassoli, L.; Bertin, S. Development, Validation, and Application of Reverse Transcription Real-Time and Droplet Digital PCR Assays for the Detection of the Potyviruses Watermelon Mosaic Virus and Zucchini Yellow Mosaic Virus in Cucurbits. Plants 2023, 12, 2364. https://doi.org/10.3390/plants12122364
Luigi M, Manglli A, Corrado CL, Tiberini A, Costantini E, Ferretti L, Tomassoli L, Bertin S. Development, Validation, and Application of Reverse Transcription Real-Time and Droplet Digital PCR Assays for the Detection of the Potyviruses Watermelon Mosaic Virus and Zucchini Yellow Mosaic Virus in Cucurbits. Plants. 2023; 12(12):2364. https://doi.org/10.3390/plants12122364
Chicago/Turabian StyleLuigi, Marta, Ariana Manglli, Carla Libia Corrado, Antonio Tiberini, Elisa Costantini, Luca Ferretti, Laura Tomassoli, and Sabrina Bertin. 2023. "Development, Validation, and Application of Reverse Transcription Real-Time and Droplet Digital PCR Assays for the Detection of the Potyviruses Watermelon Mosaic Virus and Zucchini Yellow Mosaic Virus in Cucurbits" Plants 12, no. 12: 2364. https://doi.org/10.3390/plants12122364
APA StyleLuigi, M., Manglli, A., Corrado, C. L., Tiberini, A., Costantini, E., Ferretti, L., Tomassoli, L., & Bertin, S. (2023). Development, Validation, and Application of Reverse Transcription Real-Time and Droplet Digital PCR Assays for the Detection of the Potyviruses Watermelon Mosaic Virus and Zucchini Yellow Mosaic Virus in Cucurbits. Plants, 12(12), 2364. https://doi.org/10.3390/plants12122364








