CRISPR/Cas9 Based Cell-Type Specific Gene Knock-Out in Arabidopsis Roots
Abstract
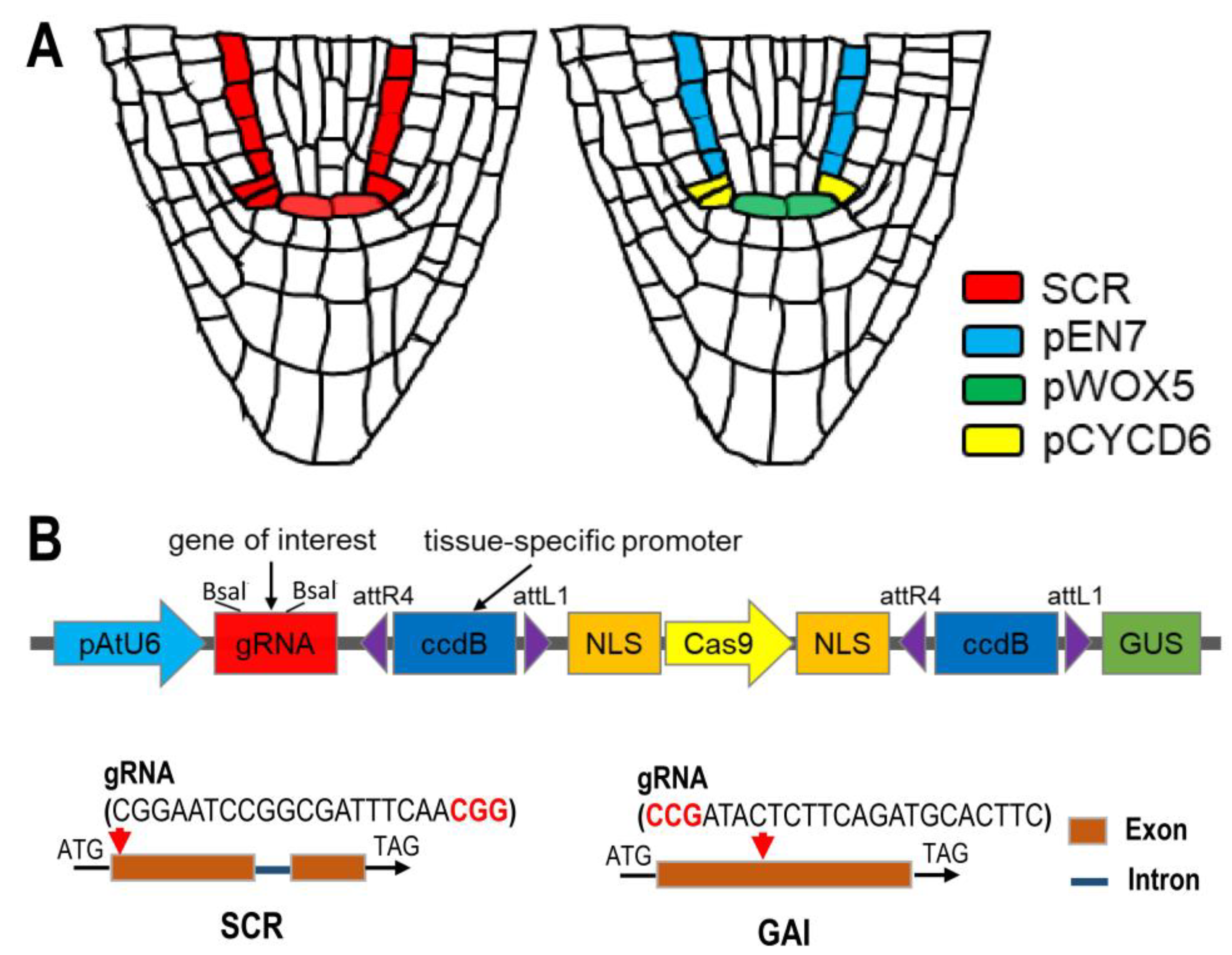
:1. Introduction
2. Results
2.1. Specific Knockout of SCR in QC Leads to Impaired QC Function and SCN Maintenance
2.2. Specific Knockout of SCR in CEI Allows Dissection of SCR Activity in Cell Division
2.3. Endodermis-Specific GAI Knockout Causes Stunted Root Phenotype
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction and Plant Transformation
4.3. Staining
4.4. Confocal Microscopy and Image Quantification
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Melnyk, C.W.; Molnar, A.; Baulcombe, D.C. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011, 30, 3553–3563. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelrahman, M.; Wei, Z.; Rohila, J.S.; Zhao, K. Multiplex genome-editing technologies for revolutionizing plant biology and crop improvement. Front. Plant Sci. 2021, 12, 721203. [Google Scholar] [CrossRef] [PubMed]
- Iliakis, G.; Wang, H.; Perrault, A.R.; Boecker, W.; Rosidi, B.; Windhofer, F.; Wu, W.; Guan, J.; Terzoudi, G.; Pantelias, G. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 2004, 104, 14–20. [Google Scholar] [CrossRef]
- Wyman, C.; Kanaar, R. DNA double-strand break repair: All’s well that ends well. Annu. Rev. Genet. 2006, 40, 363–383. [Google Scholar] [CrossRef]
- Ablain, J.; Durand, E.M.; Yang, S.; Zhou, Y.; Zon, L. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell 2015, 32, 756–764. [Google Scholar] [CrossRef] [Green Version]
- Port, F.; Chen, H.M.; Lee, T.; Lee, T.; Bullock, S. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E2967–E2976. [Google Scholar] [CrossRef] [Green Version]
- Carroll, K.J.; Makarewich, C.A.; McAnally, J.; Anderson, D.M.; Zentilin, L.; Liu, N.; Giacca, M.; Bassel-Duby, R.; Olson, E.N. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc. Natl. Acad. Sci. USA 2016, 113, 338–343. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wang, J.; Wilhelmsson, H.; Hansson, A.; Thoren, P.; Duffy, J.; Rustin, P.; Goran, N.G. Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc. Natl. Acad. Sci. USA 2000, 97, 3467–3472. [Google Scholar] [CrossRef] [Green Version]
- Elgadi, A.; Zemack, H.; Marcus, C.; Marcus, C.; Norgren, S. Tissue-specific knockout of TSHr in white adipose tissue increases adipocyte size and decreases TSH-induced lipolysis. Biochem. Biophys. Res. Commun. 2010, 393, 526–530. [Google Scholar] [CrossRef]
- Kim, G.B.; Pacheco, D.R.F.; Saxon, D.; Yang, A.; Sabet, S.; Dutra-Clarke, M.; Levy, R.; Watkins, A.; Park, H.; Akhtar, A.A.; et al. Rapid Generation of Somatic Mouse Mosaics with Locus-Specific, Stably Integrated Transgenic Elements. Cell 2019, 179, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Decaestecker, W.; Buono, R.A.; Pfeiffer, M.; Vangheluwe, N.; Jourquin, J.; Karimi, M.; Isterdael, G.V.; Beeckman, T.; Nowack, M.; Jacobs, T. CRISPR-TSKO: A Technique for Efficient Mutagenesis in Specific Cell Types, Tissues, or Organs in Arabidopsis. Plant Cell 2019, 31, 2868–2887. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.P.; Xing, H.L.; Dong, L.; Zhang, H.Y.; Han, C.Y.; Wang, X.C.; Chen, Q.J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.; Zhang, Z.; Feng, Z.; Wei, P.; Zhang, H.; Botella, J.R.; Zhu, J.K. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 2016, 14, 519–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Yamaji, N.; Huang, S.; Zhang, X.; Shi, M.; Fu, S.; Yang, G.; Ma, J.; Xia, J. OsCASP1 is required for Casparian strip formation at endodermal cells of rice roots for selective uptake of mineral elements. Plant Cell 2019, 31, 2636–2648. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ye, L.; Lyu, M.; Ursache, R.; Loytynoja, A.; Mahonen, A.P. An inducible genome editing system for plants. Nat. Plants 2020, 6, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Kim, J.; Cho, S.W.; Choi, Y.; Kim, J.S.; Coupland, G. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 2015, 241, 271–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, L.; Wei, S.; Wu, Y.; Hu, R.; Li, H.; Yang, W.; Xie, Q. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 2015, 8, 1820–1823. [Google Scholar] [CrossRef] [Green Version]
- Osakabe, Y.; Watanabe, T.; Sugano, S.S.; Ueta, R.; Ishihara, R.; Shinozaki, K.; Osakabe, K. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 2016, 6, 26685. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Zhang, Z.; Hua, K.; Gao, X.; Mao, Y.; Botella, J.R.; Zhu, J. A highly efficient cell division-specific CRISPR/Cas9 system generates homozygous mutants for multiple genes in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benfey, P.N.; Scheres, B. Root development. Curr. Biol. 2000, 10, R813–R815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolan, L.; Janmaat, K.; Willemsen, V.; Linstead, P.; Poethig, S.; Roberts, K.; Scheres, B. Cellular organisation of the Arabidopsis thaliana root. Development 1993, 119, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Scheres, B.; Wolkenfelt, H.; Willemsen, V.; Terlouw, M.; Lawson, E.; Dean, C.; Weisbeek, P. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 1994, 120, 2475–2487. [Google Scholar] [CrossRef]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef] [Green Version]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.S.; Amasino, R.; Scheres, B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xu, M.; Liang, N.; Zheng, Y.; Yu, Q.; Wu, S. Symplastic communication spatially directs local auxin biosynthesis to maintain root stem cell niche in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 4005–4010. [Google Scholar] [CrossRef] [Green Version]
- Di Laurenzio, L.; Wysocka-Diller, J.; Malamy, J.E.; Pysh, L.; Helariutta, Y.; Freshour, G.; Hahn, M.G.; Feldmann, K.A.; Benfey, P.N. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 1996, 86, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Paquette, A.J.; Benfey, P.N. Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol. 2005, 138, 636–640. [Google Scholar] [CrossRef] [Green Version]
- Shimotohno, A.; Heidstra, R.; Blilou, I.; Scheres, B. Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev. 2018, 32, 1085–1100. [Google Scholar] [CrossRef] [Green Version]
- Franke, R.; Schreiber, L. Suberin—A biopolyester forming apoplastic plant interfaces. Curr. Opin. Plant Biol. 2007, 10, 252–259. [Google Scholar] [CrossRef]
- Li, P.; Yu, Q.; Gu, X.; Qi, S.; Wang, H.; Zhong, F.; Baskin, T.; Rahman, A.; Wu, S. Construction of a functional casparian strip in non-endodermal lineages is orchestrated by two parallel signaling systems in Arabidopsis thaliana. Curr. Biol. 2018, 28, 2777–2786. [Google Scholar] [CrossRef] [Green Version]
- Bruno, L.; Pacenza, M.; Forgione, I.; Lamerton, L.R.; Greco, M.; Chiappetta, A.; Bironti, M.B. In Arabidopsis thaliana cadmium impact on the growth of primary root by altering SCR expression and auxin-cytokinin cross-talk. Front Plant Sci. 2017, 8, 1323. [Google Scholar] [CrossRef] [Green Version]
- Ubeda-Tomás, S.; Federici, F.; Casimiro, I.; Beemster, G.T.S.; Bhalerao, R.; Swarup, R.; Doerner, P.; Haseloff, J.; Bennett, M. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 2009, 19, 1194–1199. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Carol, P.; Richards, D.E.; Kingm, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockhart, J. Introducing CRISPR-TSKO: A Breakthrough in Precision Gene Editing. Plant Cell 2019, 31, 2831–2832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Eudes, A.; Yogiswara, S.; Jing, B.; Benites, V.T.; Yamanaka, R.; Cheng-Yue, C.; Baidoo, E.E.; Mortimer, J.C.; Scheller, H.V.; et al. A screening method to identify efficient sgRNAs in Arabidopsis, used in conjunction with cell-specific lignin reduction. Biotechnol. Biofuels 2019, 12, 130. [Google Scholar] [CrossRef] [Green Version]
- Wolter, F.; Klemm, J.; Puchta, H. Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J. 2018, 94, 735–746. [Google Scholar] [CrossRef] [Green Version]
- Arora, L.; Narula, A. Gene editing and crop improvement using CRISPR-Cas9 system. Front. Plant Sci. 2017, 8, 1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wu, S.; Xu, J.; Sui, C.; Wei, J. Application of CRISPR/Cas9 in plant biology. Acta Pharm. Sin. B 2017, 7, 292–302. [Google Scholar] [CrossRef]
- Sozzani, R.; Cui, H.; Moreno-Risueno, M.A.; Busch, W.; Van Norman, J.M.; Vernoux, T.; Brady, S.M.; Dewitte, W.; Murray, J.A.H.; Benfey, P.N. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 2010, 466, 128–132. [Google Scholar] [CrossRef] [Green Version]
- Forzani, C.; Aichinger, E.; Sornay, E.; Willemsen, V.; Laux, T.; Dewitte, W.; Murray, J. WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 2014, 24, 1939–1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nambara, E.; Kawaide, H.; Kamiya, Y.; Naito, S. Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol. 1998, 39, 853–858. [Google Scholar] [CrossRef]
- Che, P.; Wu, E.; Simon, M.K.; Anand, A.; Lowe, K.; Gao, H.; Sigmund, A.L.; Yang, M.; Albertsen, M.C.; Gordon-Kamm, W.; et al. Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. Commun. Biol. 2022, 5, 344. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Machin, F.; Wang, S.; Wang, S.; Saplaoura, E.; Kragler, F. Heritable transgene-free genome editing in plants by grafting of wild-type shoots to transgenic donor rootstocks. Nat. Biotechnol. 2023, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Franke-van Dijk, M.; Vencken, R.J.; Quint, A.; Hooykaas, P.; Offringa, R. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 2001, 128, 4289–4299. [Google Scholar] [CrossRef]
- Xu, M.; Gu, X.; Liang, N.; Bian, X.; Wang, H.; Qin, Y.; Pi, L.; Wu, S. Intersected functional zone of transcriptional regulators patterns stemness within stem cell niche of root apical meristem. J. Inter. Plant Biol. 2020, 62, 897–911. [Google Scholar] [CrossRef]



| Name | GUS | Root Number | Phenotypes | Editing Efficiency |
|---|---|---|---|---|
| pWOX5:Cas9-SCR | + | 30 | 3 | 10% |
| pCYCD6;1:Cas9-SCR | + | 30 | 5 | 16.1% |
| pEN7:Cas9-GAI | + | 31 | 7 | 22.6% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Niu, X.; Li, S.; Fu, S.; Li, Q.; Xu, M.; Wang, C.; Wu, S. CRISPR/Cas9 Based Cell-Type Specific Gene Knock-Out in Arabidopsis Roots. Plants 2023, 12, 2365. https://doi.org/10.3390/plants12122365
Li M, Niu X, Li S, Fu S, Li Q, Xu M, Wang C, Wu S. CRISPR/Cas9 Based Cell-Type Specific Gene Knock-Out in Arabidopsis Roots. Plants. 2023; 12(12):2365. https://doi.org/10.3390/plants12122365
Chicago/Turabian StyleLi, Meng, Xufang Niu, Shuang Li, Shasha Fu, Qianfang Li, Meizhi Xu, Chunhua Wang, and Shuang Wu. 2023. "CRISPR/Cas9 Based Cell-Type Specific Gene Knock-Out in Arabidopsis Roots" Plants 12, no. 12: 2365. https://doi.org/10.3390/plants12122365
APA StyleLi, M., Niu, X., Li, S., Fu, S., Li, Q., Xu, M., Wang, C., & Wu, S. (2023). CRISPR/Cas9 Based Cell-Type Specific Gene Knock-Out in Arabidopsis Roots. Plants, 12(12), 2365. https://doi.org/10.3390/plants12122365







