Aluminum-Immobilizing Rhizobacteria Modulate Root Exudation and Nutrient Uptake and Increase Aluminum Tolerance of Pea Mutant E107 (brz)
Abstract
1. Introduction
2. Results
2.1. Plant Growth
2.2. Properties of Cupriavidus sp. D39
2.3. Distribution of Al in Hydroponic System
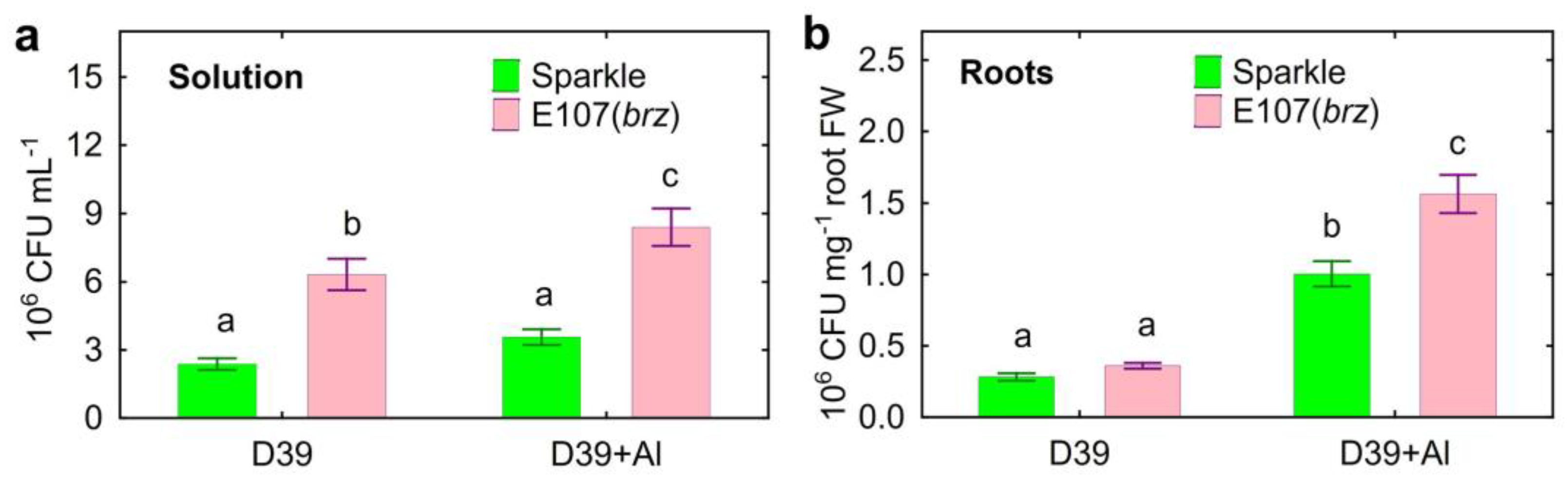
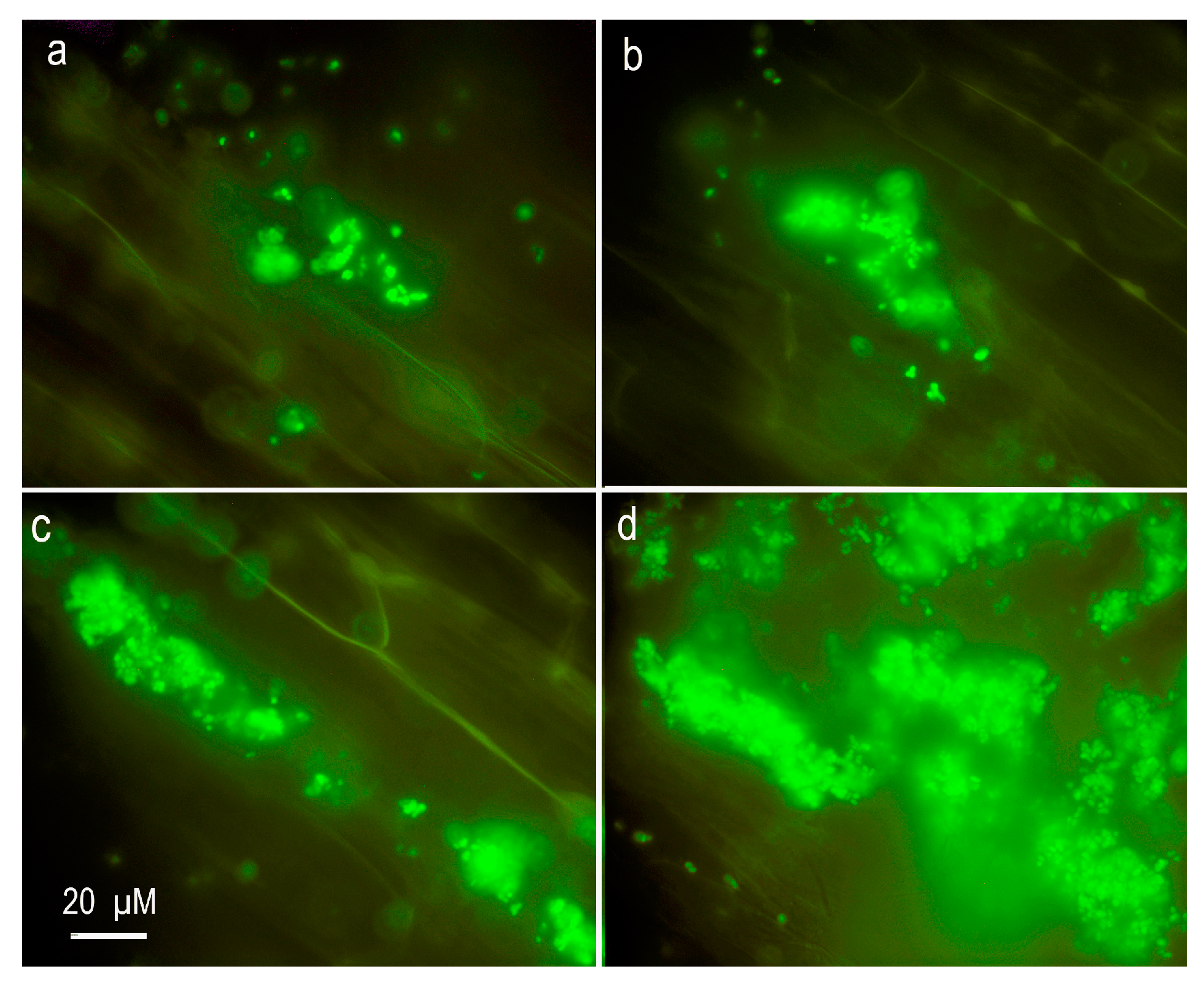
2.4. Presence of Cupriavidus sp. D39 in Hydroponic System
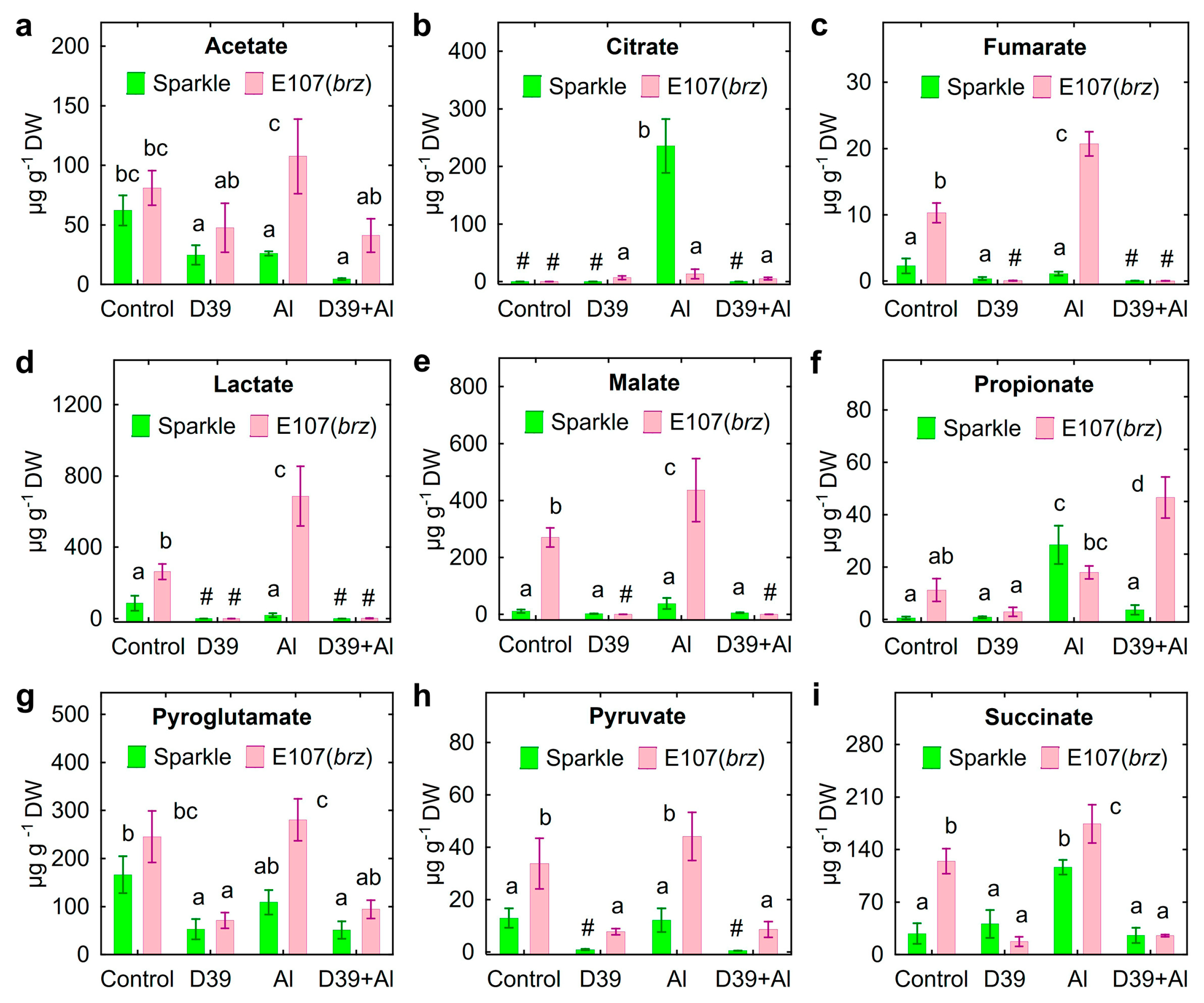
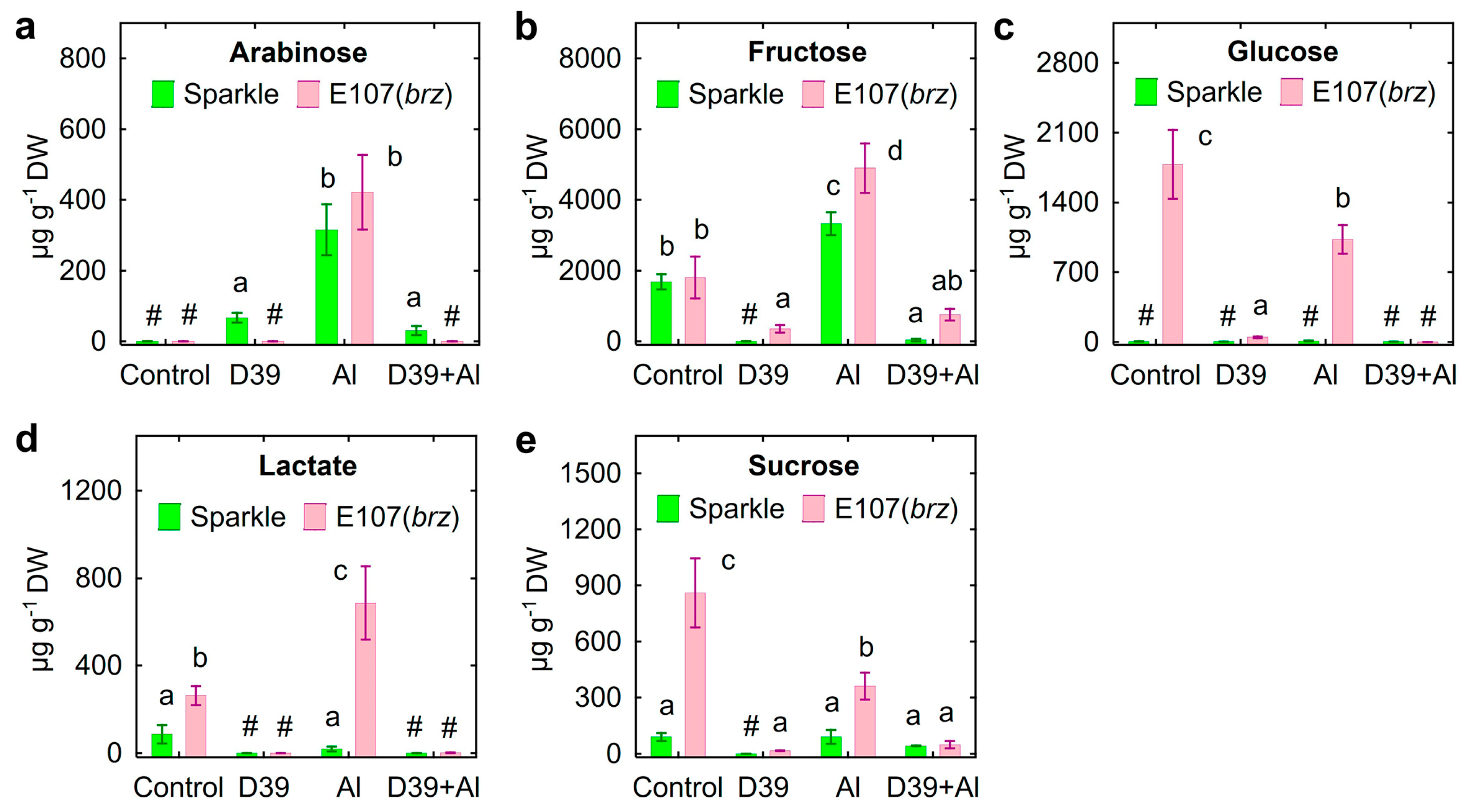
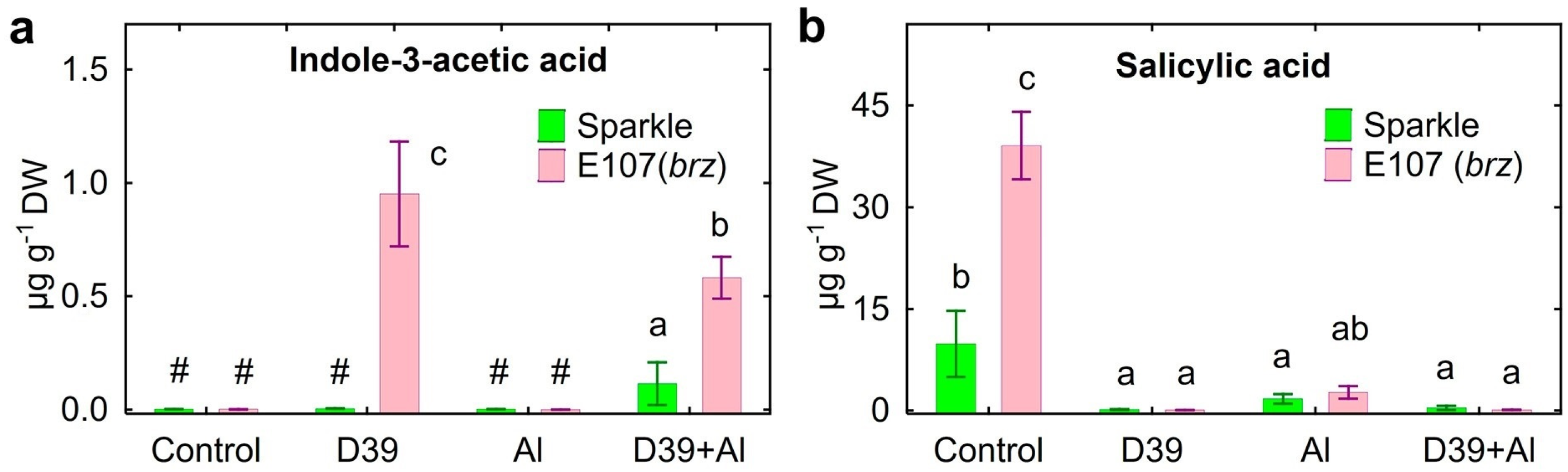
2.5. Root Exudation
2.6. Nutrient Uptake by Plants
3. Discussion
3.1. Comparison of Al-Immobilizing Strains for Plant Growth Promotion
3.2. Plant Biomass and Al Uptake
3.3. Root Exudation
3.4. Nutrient Uptake by Plants
4. Materials and Methods
4.1. Plants
4.2. Microorganisms
4.3. Plant Growth
4.4. Presence of Cupriavidus sp. D39 in Hydroponic System
4.5. Determination of Root Exudation
4.6. Elemental Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kochian, L.V.; Hoekenga, O.A.; Piñeros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef] [PubMed]
- Ofoe, R.; Thomas, R.H.; Asiedu, S.K.; Wang-Pruski, G.; Fofana, B.; Abbey, L. Aluminum in plant: Benefits, toxicity and tolerance mechanisms. Front. Plant Sci. 2023, 13, 1085998. [Google Scholar] [CrossRef]
- Kochian, L.V.; Piñeros, M.A.; Liu, J.; Magalhaes, J.V. Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 2015, 66, 571–598. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Gupta, N.; Gaurav, S.S.; Kumar, A. Molecular basis of aluminium toxicity in plants: A review. Am. J. Plant Sci. 2013, 4, 21–37. [Google Scholar] [CrossRef]
- Paulino, V.T.; Olivares, J.; Bedmar, E.J. Nodulation and nitrogenase activity of pea nodules as affected by low pH and aluminium. Plant Soil 1987, 101, 299–302. [Google Scholar] [CrossRef]
- Lazof, D.B.; Holland, M.J. Evaluation of the aluminium-induced root growth inhibition in isolation from low pH effects in Glycine max, Pisum sativum and Phaseolus vulgaris. Aust. J. Plant Physiol. 1999, 26, 147–157. [Google Scholar] [CrossRef]
- Arunakumara, K.K.I.U.; Walpola, B.C.; Yoon, M. Aluminum toxicity and tolerance mechanism in cereals and legumes—A review. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 1–9. [Google Scholar] [CrossRef]
- Singh, N.B.; Yadav, K.; Amist, N. Phytotoxic effects of aluminum on growth and metabolism of Pisum sativum L. Int. J. Innov. Biol. Chem. Sci. 2011, 2, 10–21. [Google Scholar]
- Motoda, H.; Kano, Y.; Hiragami, F.; Kawamura, K.; Matsumoto, H. Morphological changes in the apex of pea roots during and after recovery from aluminium treatment. Plant Soil 2010, 333, 49–58. [Google Scholar] [CrossRef]
- Matsumoto, H.; Motoda, H. Aluminum toxicity recovery processes in root apices. Possible association with oxidative stress. Plant Sci. 2012, 185–186, 1–8. [Google Scholar] [CrossRef]
- Matsumoto, H.; Motoda, H. Oxidative stress is associated with aluminum toxicity recovery in apex of pea root. Plant Soil 2013, 363, 399–410. [Google Scholar] [CrossRef]
- Kichigina, N.E.; Puhalsky, J.V.; Shaposhnikov, A.I.; Azarova, T.S.; Makarova, N.M.; Loskutov, S.I.; Safronova, V.I.; Tikhonovich, I.A.; Vishnyakova, M.A.; Semenova, E.V.; et al. Aluminum exclusion from root zone and maintenance of nutrient uptake are principal mechanisms of Al tolerance in Pisum sativum L. Physiol. Mol. Biol. Plants 2017, 23, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Rai, A.; Panyang, O. Hematoxylin staining as a potential screening technique for aluminium tolerance in pea (Pisum sativum L.). Cur. Sci. 2009, 96, 1029–1030. [Google Scholar]
- Yu, M.; Shen, R.; Liu, J.; Chen, R.; Xu, M.; Yang, Y.; Xiao, H.; Wang, H.; Wang, H.; Wang, C. The role of root border cells in aluminum resistance of pea (Pisum sativum) grown in mist culture. J. Plant Nutr. Soil Sci. 2009, 172, 528–534. [Google Scholar] [CrossRef]
- Sujkowska-Rybkowska, M.; Borucki, W. Pectins esterification in the apoplast of aluminum-treated pea root nodules. J. Plant Physiol. 2015, 184, 1–7. [Google Scholar] [CrossRef]
- Khan, A.G. Role of soil microbes in the rhizospheres of plants growing on trace metal contaminated soils in phytoremediation. J. Trace Elem. Med. Biol. 2005, 18, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Bukhat, S.; Imran, A.; Javaid, S.; Shahid, M.; Majeed, A.; Naqqash, T. Communication of plants with microbial world: Exploring the regulatory networks for PGPR mediated defense signaling. Microbiol. Res. 2020, 238, 126486. [Google Scholar] [CrossRef]
- Ha-Tran, D.M.; Nguyen, T.T.M.; Hung, S.-H.; Huang, E.; Huang, C.-C. Roles of plant growth-promoting rhizobacteria (PGPR) in stimulating salinity stress defense in plants: A review. Int. J. Mol. Sci. 2021, 22, 3154. [Google Scholar] [CrossRef]
- Kumar, M.; Giri, V.P.; Pandey, S.; Gupta, A.; Patel, M.K.; Bajpai, A.B.; Jenkins, S.; Siddique, K.H.M. Plant-growth-promoting rhizobacteria emerging as an effective bioinoculant to improve the growth, production, and stress tolerance of vegetable crops. Int. J. Mol. Sci. 2021, 22, 12245. [Google Scholar] [CrossRef]
- Kumawat, K.C.; Sharma, B.; Nagpal, S.; Kumar, A.; Tiwari, S.; Nair, R.M. Plant growth-promoting rhizobacteria: Salt stress alleviators to improve crop productivity for sustainable agriculture development. Front. Plant Sci. 2023, 13, 1101862. [Google Scholar] [CrossRef] [PubMed]
- Rai, R. Manganese-mediated resistance to aluminium and antibiotics in strains of Azospirillum brasilense and their interaction with rice genotypes in acid soil. J. Agric. Sci. 1986, 106, 279–285. [Google Scholar] [CrossRef]
- Rai, R. Aluminium-tolerant strains of Azospirillum brasilense and their associative nitrogen fixation with finger miller (Eleusine coracana) genotypes in an acid soil. J. Gen. Appl. Microbiol. 1991, 37, 9–24. [Google Scholar] [CrossRef]
- Belimov, A.A.; Kunakova, A.M.; Gruzdeva, E.V. Influence of soil pH on the interaction of associative bacteria with barley. Microbiologiya 1998, 67, 463–469. [Google Scholar]
- Thakur, R.; Sharma, K.C.; Gulati, A.; Sud, R.K.; Gulati, A. Stress-tolerant Viridibacillus arenosi strain IHB B 7171 from tea rhizosphere as a potential broad-spectrum microbial inoculant. Indian J. Microbiol. 2017, 57, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Singh, G.; Tiwari, A. Effect of microbial inoculation in combating the aluminium toxicity effect on growth of Zea mays. Cell Mol. Biol. 2017, 63, 79–82. [Google Scholar] [CrossRef]
- Shaposhnikov, A.I.; Vishnevskaya, N.A.; Shakhnazarova, V.Y.; Belimov, A.A.; Strunnikova, O.K. The role of barley root exudates as a food source in the relationship between Fusarium culmorum and Pseudomonas fluorescens. Mycol. Phytopathol. 2019, 53, 311–331. [Google Scholar] [CrossRef]
- Belimov, A.A.; Shaposhnikov, A.I.; Azarova, T.S.; Syrova, D.S.; Kitaeva, A.B.; Ulyanich, P.S.; Yuzikhin, O.S.; Sekste, E.A.; Safronova, V.I.; Vishnyakova, M.A.; et al. Rhizobacteria mitigate the negative effect of aluminum on pea growth by immobilizing the toxicant and modulating root exudation. Plants 2022, 11, 2416. [Google Scholar] [CrossRef] [PubMed]
- Piña, R.G.; Cervantes, C. Microbial interactions with aluminium. Biometals 1996, 9, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Wood, M. A mechanism of aluminium toxicity to soil bacteria and possible ecological implications. Plant Soil 1995, 171, 63–69. [Google Scholar] [CrossRef]
- Rogers, N.J.; Carson, K.C.; Glenn, A.R.; Dilworth, M.J.; Hughes, M.N.; Poole, R.K. Alleviation of aluminum toxicity to Rhizobium leguminosarum bv. viciae by the hydroxamate siderophore vicibactin. Biometals 2001, 14, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Kawai, F.; Zhang, D.; Sugimoto, M. Isolation and characterization of acid- and Al-tolerant microorganisms. FEMS Microbiol. Lett. 2000, 189, 143–147. [Google Scholar] [CrossRef]
- Wakao, N.; Yasuda, T.; Jojima, Y.; Yamanaka, S.; Hiraishi, A. Enhanced growth of acidocella facilis and related acidophilic bacteria at high concentrations of aluminum. Microbes Environ. 2002, 17, 98–104. [Google Scholar] [CrossRef]
- Shaposhnikov, A.; Yuzikhin, O.; Syrova, D.; Karlov, D.; Sazanova, A.; Azarova, T.; Sekste, E.; Safronova, V.; Belimov, A. Beneficial aluminum immobilizing microorganisms inhabiting the rhizosphere of pea. Biol. Commun. 2023, in press. [Google Scholar]
- Belimov, A.A.; Shaposhnikov, A.I.; Azarova, T.S.; Makarova, N.M.; Safronova, V.I.; Litvinskiy, V.A.; Nosikov, V.V.; Zavalin, A.A.; Tikhonovich, I.A. Microbial consortium of PGPR, rhizobia and arbuscular mycorrhizal fungus makes pea mutant SGECdt comparable with Indian mustard in cadmium tolerance and accumulation. Plants 2020, 9, 975. [Google Scholar] [CrossRef]
- Kichko, A.A.; Gladkov, G.V.; Ulianich, P.S.; Safronova, V.I.; Pinaev, A.G.; Sekste, E.A.; Belimov, A.A.; Andronov, E.E. Water stress, cadmium, and plant genotype modulate the rhizosphere microbiome of Pisum sativum L. Plants 2022, 11, 3013. [Google Scholar] [CrossRef]
- Guinel, F.C.; LaRue, T.A. Excessive aluminium accumulation in the pea mutant E107 (brz). Plant Soil 1993, 157, 75–82. [Google Scholar] [CrossRef]
- Kneen, B.E.; Larue, T.A.; Welch, R.M.; Weeden, N.F. Pleiotropic effects of brz: A mutation in Pisum sativum (L.) cv ‘Sparkle’ conditioning decreased nodulation and increased iron uptake and leaf necrosis. Plant Physiol. 1990, 93, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Mora, M.L.; Demanet, R.; Acuna, J.J.; Viscardi, S.; Jorquera, M.; Rengel, Z.; Duran, P. Aluminum-tolerant bacteria improve the plant growth and phosphorus content in ryegrass grown in a volcanic soil amended with cattle dung manure. Appl. Soil Ecol. 2017, 115, 19–26. [Google Scholar] [CrossRef]
- Silambarasan, S.; Logeswari, P.; Cornejo, P.; Kannan, V.R. Role of plant growth–promoting rhizobacterial consortium in improving the Vigna radiata growth and alleviation of aluminum and drought stresses. Environ. Sci. Pollut. Res. 2019, 26, 27647–27659. [Google Scholar] [CrossRef]
- Silambarasan, S.; Logeswari, P.; Valentine, A.; Cornejo, P. Role of Curtobacterium herbarum strain CAH5 on aluminum bioaccumulation and enhancement of Lactuca sativa growth under aluminum and drought stresses. Ecotoxicol. Environ. Saf. 2019, 183, 109573. [Google Scholar] [CrossRef] [PubMed]
- Silambarasan, S.; Logeswari, P.; Cornejo, P.; Abraham, J.; Valentine, A. Simultaneous mitigation of aluminum, salinity and drought stress in Lactuca sativa growth via formulated plant growth promoting Rhodotorula mucilaginosa CAM4. Ecotoxicol. Environ. Saf. 2019, 180, 63–72. [Google Scholar] [CrossRef]
- Guro, P.; Ulianich, P.; Shaposhnikov, A.; Yuzikhin, O.; Karlov, D.; Sazanova, A.; Safronova, V.; Belimov, A. Draft genome sequence of the bacterium Cupriavidus sp. strain D39, inhabiting the rhizosphere of pea plants (Pisum sativum L.). Microbiol. Resour. Announc. 2023, 12, e0135422. [Google Scholar] [CrossRef] [PubMed]
- Appanna, V.D.; Kepes, M.; Rochon, P. Aluminum tolerance in Pseudomonas fluorescens ATCC 13525: Involvement of a gelatinous lipid-rich residue. FEMS Microbiol. Lett. 1994, 119, 295–301. [Google Scholar] [CrossRef]
- Appanna, V.D.; St Pierre, M. Influence of phosphate on aluminum tolerance in Pseudomonas fluorescens. FEMS Microbiol. Lett. 1994, 124, 327–332. [Google Scholar] [CrossRef]
- Welch, R.M.; LaRue, T.A. Physiological characteristics of Fe accumulation in the ‘bronze’ mutant of Pisum sativum L., cv ‘Sparkle’ E107 (brz brz). Plant Physiol. 1990, 93, 723–729. [Google Scholar] [CrossRef]
- Pandey, P.; Srivastava, R.K.; Dubey, R.S. Salicylic acid alleviates aluminum toxicity in rice seedlings better than magnesium and calcium by reducing aluminum uptake, suppressing oxidative damage and increasing antioxidative defense. Ecotoxicology 2013, 22, 656–670. [Google Scholar] [CrossRef]
- Cheng, X.; Fang, T.; Zhao, E.; Zheng, B.; Huang, B.; An, Y.; Zhou, P. Protective roles of salicylic acid in maintaining integrity and functions of photosynthetic photosystems for alfalfa (Medicago sativa L.) tolerance to aluminum toxicity. Plant Physiol. Biochem. 2020, 155, 570–578. [Google Scholar] [CrossRef]
- Lan, T.; You, J.; Kong, L.; Yu, M.; Liu, M.; Yang, Z. The interaction of salicylic acid and Ca2+ alleviates aluminum toxicity in soybean (Glycine max L.). Plant Physiol. Biochem. 2016, 98, 146–154. [Google Scholar] [CrossRef]
- Yang, Z.-M.; Wang, J.; Wang, S.-H.; Xu, L.-L. Salicylic acid-induced aluminum tolerance by modulation of citrate efflux from roots of Cassia tora L. Planta 2003, 217, 168–174. [Google Scholar] [CrossRef]
- Liu, N.; Song, F.; Zhu, X.; You, J.; Yang, Z.; Li, X. Salicylic acid alleviates aluminum toxicity in soybean roots through modulation of reactive oxygen species metabolism. Front. Chem. 2017, 5, 96. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, Y.; Zhang, M.; Zhu, F.; Sun, M.; Lian, X.; Zhao, G.; Duan, D. Transcriptome and metabolome analysis of stress tolerance to aluminium in Vitis quinquangularis. Planta 2021, 254, 105. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Riaz, M.; Liu, Y.; Zeng, Y.; Jiang, C. Aluminum toxicity could be mitigated with boron by altering the metabolic patterns of amino acids and carbohydrates rather than organic acids in trifoliate orange. Tree Physiol. 2019, 39, 1572–1582. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, A.; AbdElgawad, H.; Fidalgo, F.; Teixeira, J.; Matos, M.; Hamed, B.A.; Selim, S.; Hozzein, W.N.; Beemster, G.T.S.; Asard, H. Al exposure increases proline levels by different pathways in an Al-sensitive and an Al-tolerant rye genotype. Sci. Rep. 2020, 10, 16401. [Google Scholar] [CrossRef]
- Wang, P.; Bi, S.; Wang, S.; Ding, Q. Variation of wheat root exudates under aluminum stress. J. Agric. Food Chem. 2006, 54, 10040–10046. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Jin, C.; Sun, C.; Wang, J.; Ye, Y.; Lu, L.; Lin, X. Elevation of arginine decarboxylase-dependent putrescine production enhances aluminum tolerance by decreasing aluminum retention in root cell walls of wheat. J. Hazard. Mater. 2015, 299, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wang, Y.; Zhang, J.; Shi, W.; Qian, C.; Peng, X. Identification of aluminum-responsive proteins in rice roots by a proteomic approach: Cysteine synthase as a key player in Al response. Proteomics 2007, 7, 737–749. [Google Scholar] [CrossRef]
- Song, H.; Xu, X.; Wang, H.; Wang, H.; Tao, Y. Exogenous γ-aminobutyric acid alleviates oxidative damage caused by aluminium and proton stresses on barley seedlings. J. Sci. Food Agric. 2010, 90, 1410–1416. [Google Scholar] [CrossRef]
- Wang, P.; Dong, Y.; Zhu, L.; Hao, Z.; Hu, L.; Hu, X.; Wang, G.; Cheng, T.; Shi, J.; Chen, J. The role of γ-aminobutyric acid in aluminum stress tolerance in a woody plant, Liriodendron chinense × tulipifera. Hortic. Res. 2021, 8, 80. [Google Scholar] [CrossRef]
- Sharma, P.; Dubey, R.S. Modulation of nitrate reductase activity in rice seedlings under aluminium toxicity and water stress: Role of osmolytes as enzyme protectant. J. Plant Physiol. 2005, 162, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Mangeon, A.; Pardal, R.; Menezes-Salgueiro, A.D.; Duarte, G.L.; de Seixas, R.; Cruz, F.P.; Cardeal, V.; Magioli, C.; Ricachenevsky, F.K.; Margis, R.; et al. AtGRP3 is implicated in root size and aluminum response pathways in arabidopsis. PLoS ONE 2016, 11, e0150583. [Google Scholar] [CrossRef]
- Zhou, P.; Su, L.; Lv, A.; Wang, S.; Huang, B.; An, Y. Gene Expression analysis of alfalfa seedlings response to acid-aluminum. Int. J. Genom. 2016, 2016, 2095195. [Google Scholar] [CrossRef]
- Su, L.; Lv, A.; Wen, W.; Fan, N.; Li, J.; Gao, L.; Zhou, P.; An, Y. MsMYB741 is involved in alfalfa resistance to aluminum stress by regulating flavonoid biosynthesis. Plant J. 2022, 112, 756–771. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Shen, H.; He, L.F.; Sasaki, T.; Yamamoto, Y.; Zheng, S.J.; Ligaba, A.; Yan, X.L.; Ahn, S.J.; Yamaguchi, M.; Sasakawa, H.; et al. Citrate secretion coupled with the modulation of soybean root tip under aluminum stress. up-regulation of transcription, translation, and threonine-oriented phosphorylation of plasma membrane H+-ATPase. Plant Physiol. 2005, 138, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Sinha, R.; Lal, S.K.; Bishi, S.K.; Singh, A.K. Phytohormone signalling and cross-talk to alleviate aluminium toxicity in plants. Plant Cell Rep. 2021, 40, 1331–1343. [Google Scholar] [CrossRef]
- Kamilova, F.; Kravchenko, L.V.; Shaposhnikov, A.I.; Makarova, N.M.; Azarova, T.S.; Lugtenberg, B. organic acids, sugars, and l-Tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant-Microbe Interact. 2006, 19, 250–256. [Google Scholar] [CrossRef]
- Wu, D.; Shen, H.; Yokawa, K.; Baluška, F. Alleviation of aluminium induced cell rigidity by overexpression of OsPIN2 in rice roots. J. Exp. Bot. 2014, 65, 5305–5315. [Google Scholar] [CrossRef]
- Kollmeier, M.; Felle, H.H.; Horst, W.J. Genotypical differences in aluminum resistance of maize are expressed in the distal part of the transition zone. is reduced basipetal auxin flow involved in inhibition of root elongation by aluminum? Plant Physiol. 2000, 122, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.-M.; Liu, W.-C.; Jin, Y.; Lu, Y.-T. Role of ROS and auxin in plant response to metal-mediated stress. Plant Signal. Behav. 2013, 8, e24671. [Google Scholar] [CrossRef] [PubMed]
- Kopittke, P.M. Role of phytohormones in aluminium rhizotoxicity. Plant Cell Environ. 2016, 39, 2319–2328. [Google Scholar] [CrossRef]
- Wang, S.; Ren, X.; Huang, B.; Wang, G.; Zhou, P.; An, Y. Aluminium-induced reduction of plant growth in alfalfa (Medicago sativa) is mediated by interrupting auxin transport and accumulation in roots. Sci. Rep. 2016, 6, 30079. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, G.; Liu, J.; Zhang, B.; Meng, W.; Müller, B.; Hayashi, K.; Zhang, X.; Zhao, Z.; De Smet, I.; et al. Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. EMBO Rep. 2017, 18, 1213–1230. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Gao, S.; Tian, H.; Wu, W.; Robert, H.S.; Ding, Z. local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. PLoS Genet. 2021, 17, e1009964. [Google Scholar] [CrossRef]
- Zerrouk, I.Z.; Benchabane, M.; Khelifi, L.; Yokawa, K.; Ludwig-Müller, J.; Baluska, F. A Pseudomonas strain isolated from date-palm rhizospheres improves root growth and promotes root formation in maize exposed to salt and aluminum stress. J. Plant Physiol. 2016, 191, 111–119. [Google Scholar] [CrossRef]
- Kuzmicheva, Y.V.; Shaposhnikov, A.I.; Petrova, S.N.; Makarova, N.M.; Tychinskaya, I.L.; Puhalsky, J.V.; Parahin, N.V.; Tikhonovich, I.A.; Belimov, A.A. Variety specific relationships between effects of rhizobacteria on root exudation, growth and nutrient uptake of soybean. Plant Soil 2017, 419, 83–96. [Google Scholar] [CrossRef]
- Matsumoto, H.; Yamaya, T. Inhibition of potassium uptake and regulation of membrane-associated Mg2+-ATPase activity of pea roots by aluminium. Soil Sci. Plant Nutr. 1986, 32, 179–188. [Google Scholar] [CrossRef]
- Belimov, A.A.; Shaposhnikov, A.I.; Syrova, D.S.; Kichko, A.A.; Guro, P.V.; Yuzikhin, O.S.; Azarova, T.S.; Sazanova, A.L.; Sekste, E.A.; Litvinskiy, V.A.; et al. The role of symbiotic microorganisms, nutrient uptake and rhizosphere bacterial community in response of pea (Pisum sativum L.) genotypes to elevated Al concentrations in soil. Plants 2020, 9, 1801. [Google Scholar] [CrossRef]
- Suarez, A.; Güttler, A.; Strätz, M.; Staendner, L.H.; Timmis, K.N.; Guzmán, C.A. Green fluorescent protein-based reporter systems for genetic analysis of bacteria including monocopy applications. Gene 1997, 196, 69–74. [Google Scholar] [CrossRef]
- Belimov, A.A.; Dodd, I.C.; Safronova, V.I.; Shaposhnikov, A.I.; Azarova, T.S.; Makarova, N.M.; Davies, W.J.; Tikhonovich, I.A. Rhizobacteria that produce auxins and contain ACC deaminase decrease amino acid concentrations in the rhizosphere and improve growth and yield of well-watered and water-limited potato (Solanum tuberosum). Ann. Appl. Biol. 2015, 167, 11–25. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Zvyagintsev, D.G. Methods for Soil Microbiology and Biochemistry; Moscow University Press: Moscow, Russia, 1991. (In Russian) [Google Scholar]
- Boykova, I.; Yuzikhin, O.; Novikova, I.; Ulianich, P.; Eliseev, I.; Shaposhnikov, A.; Yakimov, A.; Belimov, A. Strain Streptomyces sp. P-56 Produces Nonactin and Possesses Insecticidal, Acaricidal, Antimicrobial and Plant Growth-Promoting Traits. Microorganisms 2023, 11, 764. [Google Scholar] [CrossRef] [PubMed]





| Pea Genotype and Treatment | Root | Shoot | ||
|---|---|---|---|---|
| mg DW Plant−1 | Increase in Control, % | mg DW Plant−1 | Increase in Control, % | |
| Experiment 1 | ||||
| Sparkle | ||||
| Uninoculated control | 37.0 ± 5.4 | - | 43.8 ± 5.8 | - |
| Cupriavidus sp. D39 | 56.7 ± 4.1 * | +53 | 62.2 ± 4.0 * | +42 |
| Herbaspirillum sp. E52 | 39.4 ± 7.3 | +6 | 36.1 ± 7.7 | −18 |
| Paraburkholderia sediminicola C06 | 49.1 ± 6.1 | +33 | 47.8 ± 5.8 | +9 |
| Paraburkholderia sp. A35 | 50.2 ± 4.1 | +36 | 39.2 ± 3.5 | −11 |
| Rhodotorula sp. AL1 | 52.6 ± 4.1 * | +42 | 49.5 ± 4.8 | +13 |
| E107 (brz) | ||||
| Uninoculated control | 9.4 ± 1.2 | - | 15.7 ± 1.5 | - |
| Cupriavidus sp. D39 | 21.4 ± 2.3 * | +127 | 19.3 ± 1.2 | +23 |
| Herbaspirillum sp. E52 | 6.9 ± 1.1 | −27 | 10.3 ± 0.7 * | −35 |
| Paraburkholderia sediminicola C06 | 12.0 ± 0.7 | +28 | 21.0 ± 2.3 * | +34 |
| Paraburkholderia sp. A35 | 12.6 ± 1.3 | +34 | 23.2 ± 1.4 * | +48 |
| Rhodotorula sp. AL1 | 6.3 ± 0.4 | −33 | 11.8 ± 1.6 | −25 |
| Experiment 2 | ||||
| Sparkle | ||||
| Uninoculated control | 34.6 ± 2.8 | - | 39.5 ± 2.5 | - |
| Cupriavidus sp. D39 | 42.2 ± 1.6 * | +22 | 52.9 ± 2.8 * | +33 |
| Paraburkholderia sediminicola C06 | 44.0 ± 2.5 * | +27 | 43.3 ± 2.6 | +10 |
| Paraburkholderia sp. A35 | 41.3 ± 2.8 | +19 | 42.1 ± 2.3 | +7 |
| E107 (brz) | ||||
| Uninoculated control | 22.3 ± 3.0 | - | 29.1 ± 2.6 | - |
| Cupriavidus sp. D39 | 39.7 ± 7.0 * | +78 | 45.7 ± 8.1 * | +57 |
| Paraburkholderia sediminicola C06 | 30.0 ± 5.4 | +34 | 38.2 ± 4.9 | +31 |
| Paraburkholderia sp. A35 | 32.1 ± 4.0 * | +44 | 30.3 ± 4.5 | +4 |
| Treatments | Ca (µg g−1 DW) | Fe (µg g−1 DW) | K (mg g−1 DW) | Mg (mg g−1 DW) | Mn (µg g−1 DW) | P (mg g−1 DW) | S (mg g−1 DW) | Zn (µg g−1 DW) |
|---|---|---|---|---|---|---|---|---|
| Sparkle | ||||||||
| Control | 884 ± 37 a,b | 338 ± 38 a,b | 34 ± 1 b | 1.4 ± 0.1 a,b | 25 ± 1 a | 163 ± 6 a | 4.8 ± 0.3 a,b | 103 ± 14 a |
| Cupriavidus sp. D39 | 955 ± 116 a,b | 316 ± 52 a,b | 36 ± 1 b,c | 2.2 ± 0.2 c | 42 ± 5 b | 149 ± 2 a | 5.3 ± 0.4 b,c | 98 ± 21 a |
| AlCl3 | 898 ± 44 a,b | 439 ± 62 a,b | 35 ± 1 b,c | 1.2 ± 0.1 a | 24 ± 2 a | 194 ± 4 b | 5.4 ± 0.3 b,c | 120 ± 9 a |
| Cupriavidus sp. D39 + AlCl3 | 965 ± 26 b | 427 ± 31 a,b | 34 ± 2 b | 1.3 ± 0.1 a,b | 38 ± 4 b | 166 ± 6 a | 5.3 ± 0.3 b,c | 119 ± 9 a |
| E107 (brz) | ||||||||
| Control | 884 ± 15 a,b | 393 ± 121 a,b | 35 ± 3 b,c | 1.6 ± 0.1 b | 51 ± 8 b,c | 197 ± 7 b | 5.6 ± 0.1 b,c | 162 ± 18 b |
| Cupriavidus sp. D39 | 973 ± 73 a,b | 283 ± 89 a | 39 ± 1 c | 2.4 ± 0.3 c | 66 ± 3 c,d | 202 ± 2 b | 6.2 ± 0.3 c | 161 ± 16 b |
| AlCl3 | 867 ± 83 a,b | 701 ± 122 c | 20 ± 3 a | 1.2 ± 0.1 a | 61 ± 6 c | 229 ± 7 c | 4.1 ± 0.3 a | 169 ± 17 b |
| Cupriavidus sp. D39 + AlCl3 | 804 ± 58 a | 565 ± 91 b,c | 37 ± 3 b,c | 1.7 ± 0.2 b | 77 ± 7 d | 203 ± 4 b | 5.7 ± 0.2 b,c | 183 ± 7 b |
| Treatments | Ca (µg g−1 DW) | Fe (µg g−1 DW) | K (mg g−1 DW) | Mg (mg g−1 DW) | Mn (µg g−1 DW) | P (mg g−1 DW) | S (mg g−1 DW) | Zn (µg g−1 DW) |
|---|---|---|---|---|---|---|---|---|
| Sparkle | ||||||||
| Control | 808 ± 36 b,c | 81 ± 8 a | 4.0 ± 0.3 a | 2.3 ± 0.2 b | 21 ± 2 a | 58 ± 21 a | 1.8 ± 0.1 a,b | 76 ± 3 a |
| Cupriavidus sp. D39 | 1110 ± 69 d | 95 ± 10 a | 4.9 ± 0.6 a | 2.5 ± 0.3 b | 23 ± 2 a | 95 ± 4 a | 2.1 ± 0.2 b | 88 ± 5 a,b |
| AlCl3 | 744 ± 31 b | 93 ± 5 a | 4.6 ± 0.4 a | 2.2 ± 0.2 b | 21 ± 1 a | 87 ± 16 a | 1.8 ± 0.1 a,b | 84 ± 4 a,b |
| Cupriavidus sp. D39 + AlCl3 | 935 ± 29 c | 126 ± 18 b | 4.9 ± 0.2 a | 2.6 ± 0.2 b | 24 ± 1 a | 97 ± 14 a | 2.1 ± 0.1 b | 91 ± 2 b |
| E107 (brz) | ||||||||
| Control | 552 ± 24 a | 67 ± 1 a | 3.6 ± 0.4 a | 2.1 ± 0.1 b | 24 ± 2 a | 62 ± 23 a | 1.5 ± 0.1 a | 81 ± 5 a,b |
| Cupriavidus sp. D39 | 737 ± 47 b | 67 ± 10 a | 4.2 ± 0.2 a | 2.4 ± 0.1 b | 29 ± 7 a,b | 83 ± 23 a | 1.7 ± 0.1 a,b | 78 ± 5 a,b |
| AlCl3 | 424 ± 72 a | 88 ± 9 a | 4.7 ± 0.7 a | 1.7 ± 0.1 a | 28 ± 8 a,b | 78 ± 23 a | 1.3 ± 0.2 a | 84 ± 6 a,b |
| Cupriavidus sp. D39 + AlCl3 | 771 ± 79 b | 98 ± 2 a,b | 4.5 ± 1.0 a | 2.3 ± 0.2 b | 39 ± 3 b | 86 ± 26 a | 1.7 ± 0.2 a,b | 82 ± 6 a,b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belimov, A.A.; Shaposhnikov, A.I.; Azarova, T.S.; Yuzikhin, O.S.; Sekste, E.A.; Safronova, V.I.; Tikhonovich, I.A. Aluminum-Immobilizing Rhizobacteria Modulate Root Exudation and Nutrient Uptake and Increase Aluminum Tolerance of Pea Mutant E107 (brz). Plants 2023, 12, 2334. https://doi.org/10.3390/plants12122334
Belimov AA, Shaposhnikov AI, Azarova TS, Yuzikhin OS, Sekste EA, Safronova VI, Tikhonovich IA. Aluminum-Immobilizing Rhizobacteria Modulate Root Exudation and Nutrient Uptake and Increase Aluminum Tolerance of Pea Mutant E107 (brz). Plants. 2023; 12(12):2334. https://doi.org/10.3390/plants12122334
Chicago/Turabian StyleBelimov, Andrey A., Alexander I. Shaposhnikov, Tatiana S. Azarova, Oleg S. Yuzikhin, Edgar A. Sekste, Vera I. Safronova, and Igor A. Tikhonovich. 2023. "Aluminum-Immobilizing Rhizobacteria Modulate Root Exudation and Nutrient Uptake and Increase Aluminum Tolerance of Pea Mutant E107 (brz)" Plants 12, no. 12: 2334. https://doi.org/10.3390/plants12122334
APA StyleBelimov, A. A., Shaposhnikov, A. I., Azarova, T. S., Yuzikhin, O. S., Sekste, E. A., Safronova, V. I., & Tikhonovich, I. A. (2023). Aluminum-Immobilizing Rhizobacteria Modulate Root Exudation and Nutrient Uptake and Increase Aluminum Tolerance of Pea Mutant E107 (brz). Plants, 12(12), 2334. https://doi.org/10.3390/plants12122334






