Genome-Wide Association Study Identified Candidate Genes for Alkalinity Tolerance in Rice
Abstract
1. Introduction
2. Results
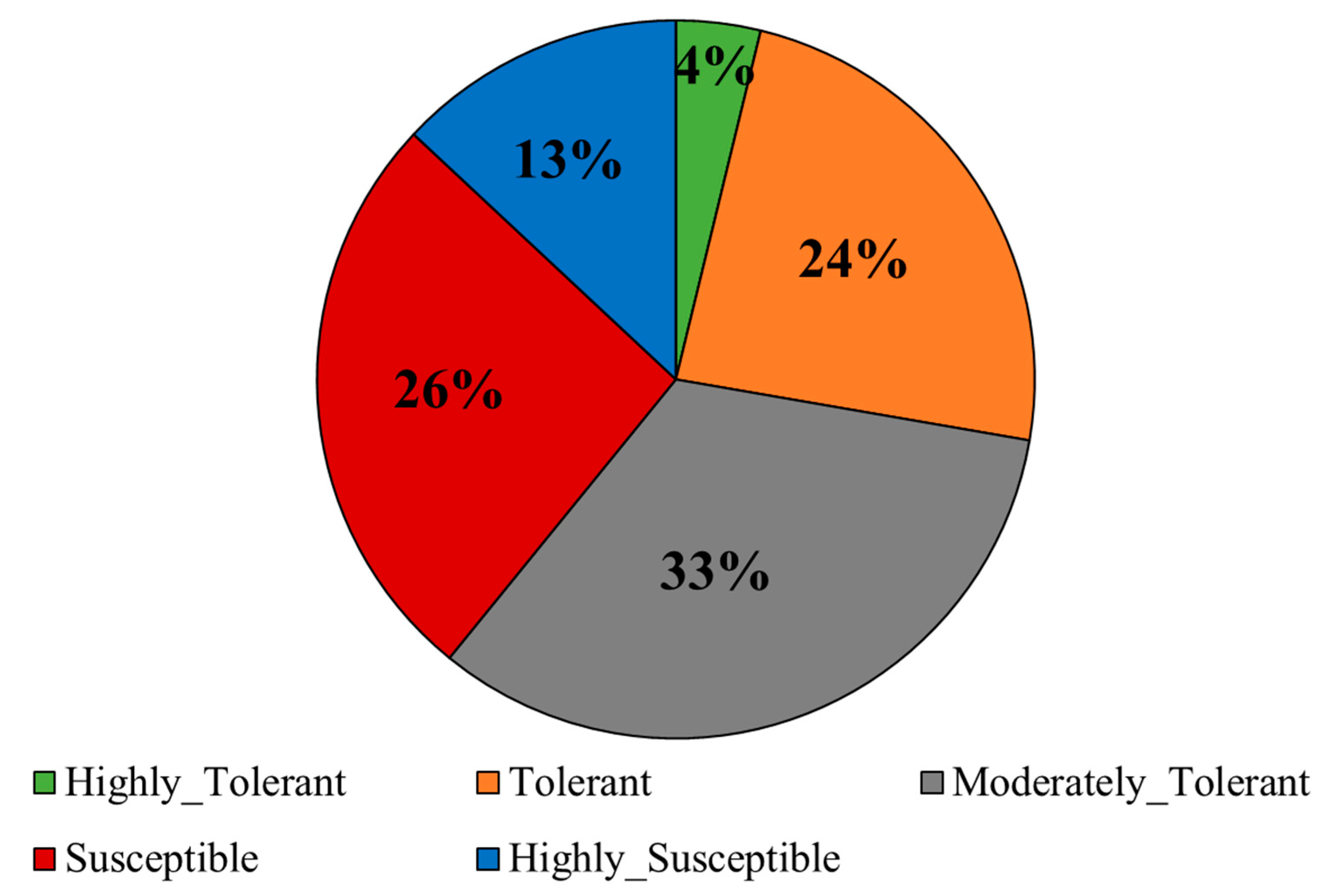
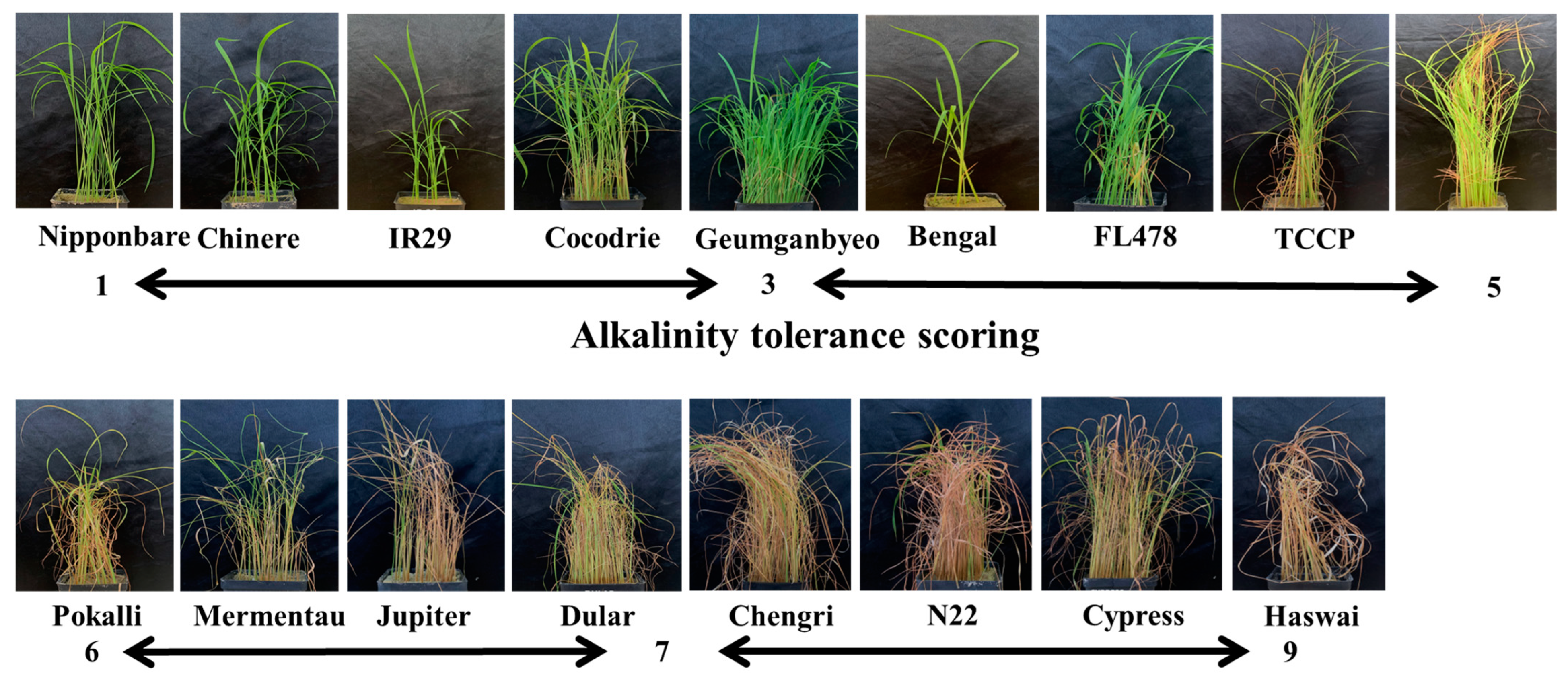
2.1. Phenotypic Evaluation under Alkaline Stress
2.2. Correlation Analysis
2.3. Principal Component Analysis (PCA)
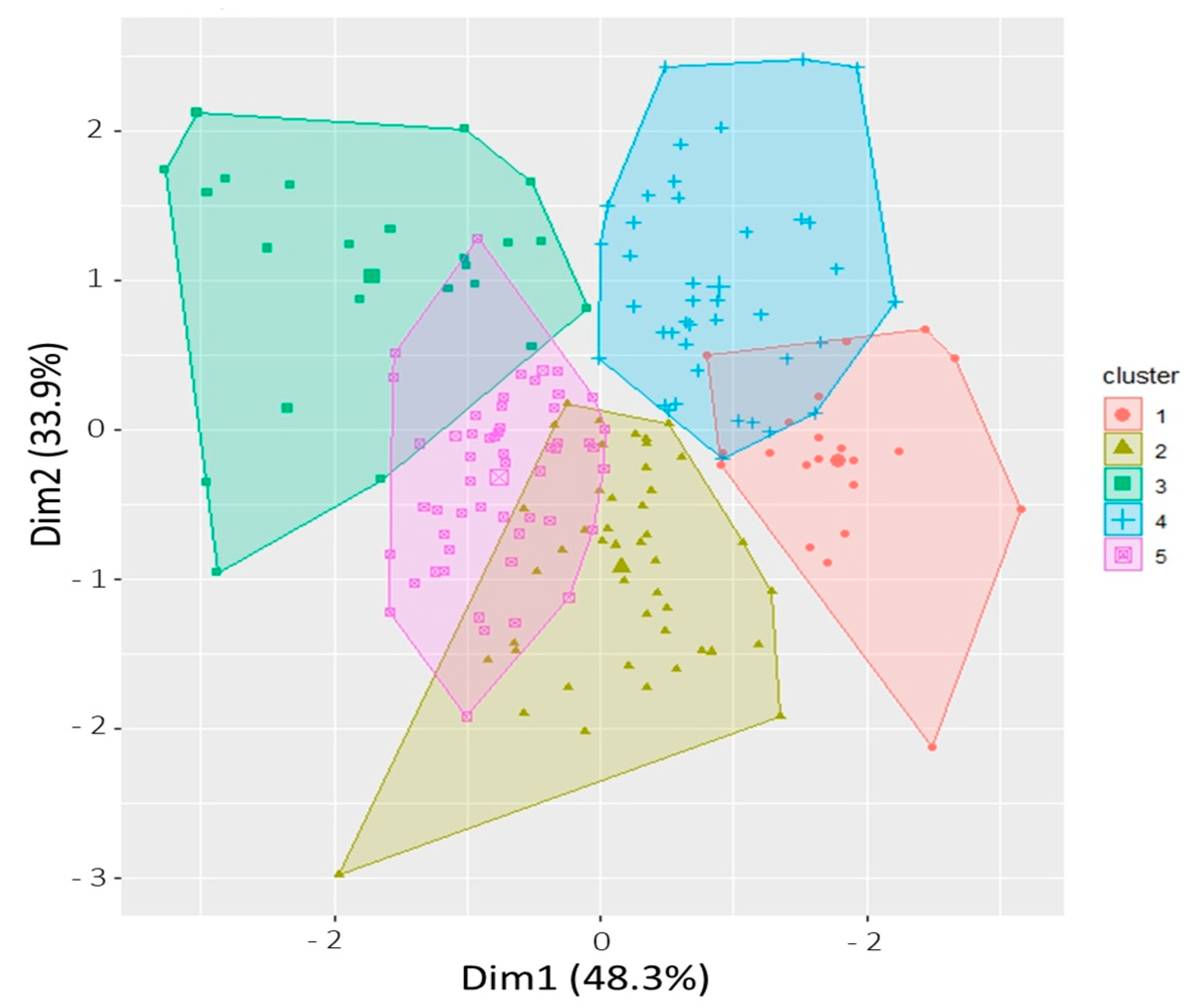
2.4. Phenotypic Clustering
2.5. Population Structure
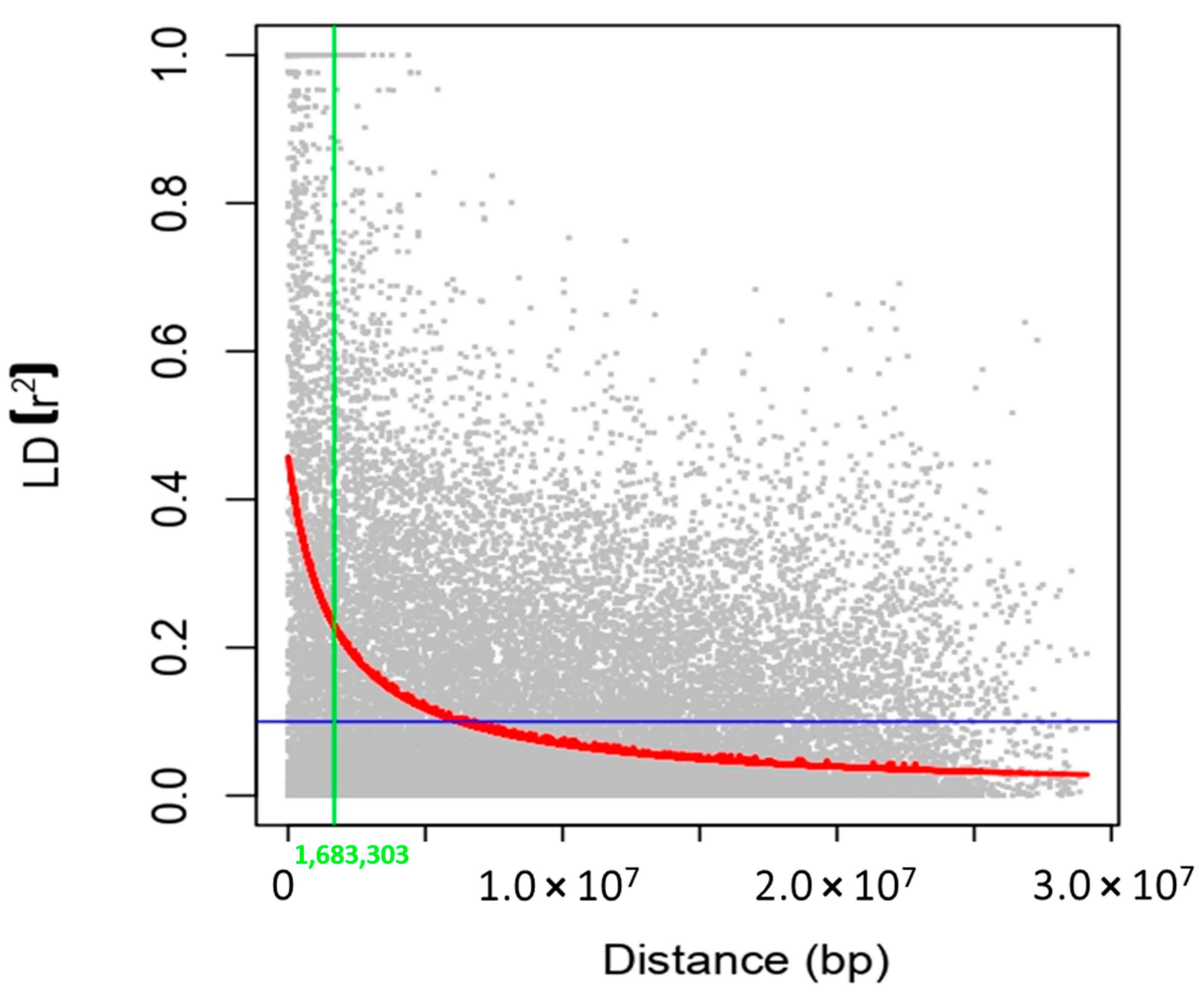
2.6. Linkage Disequilibrium (LD)
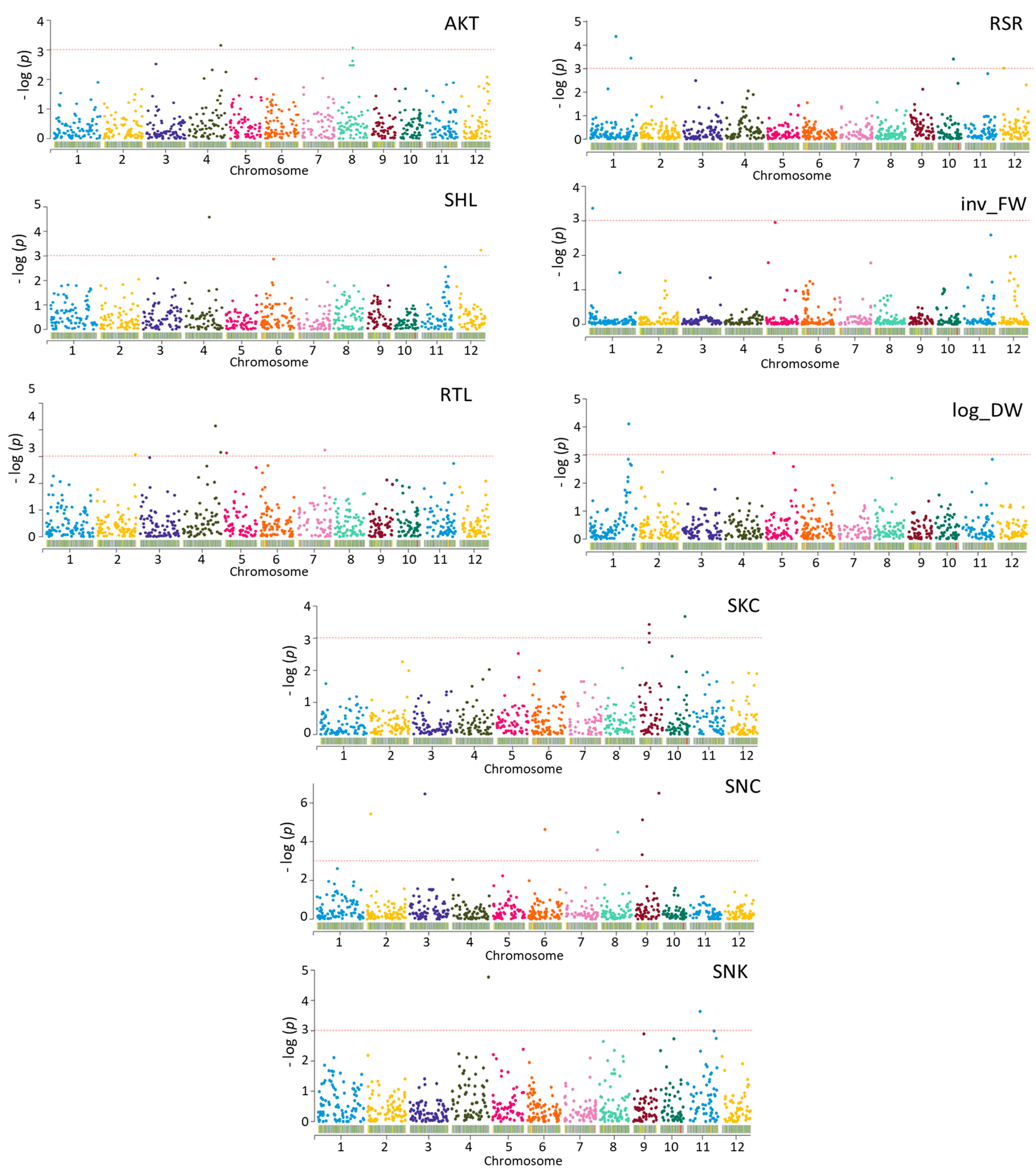
2.7. GWAS Analysis
2.8. Candidate Genes/QTLs for Alkalinity Tolerance
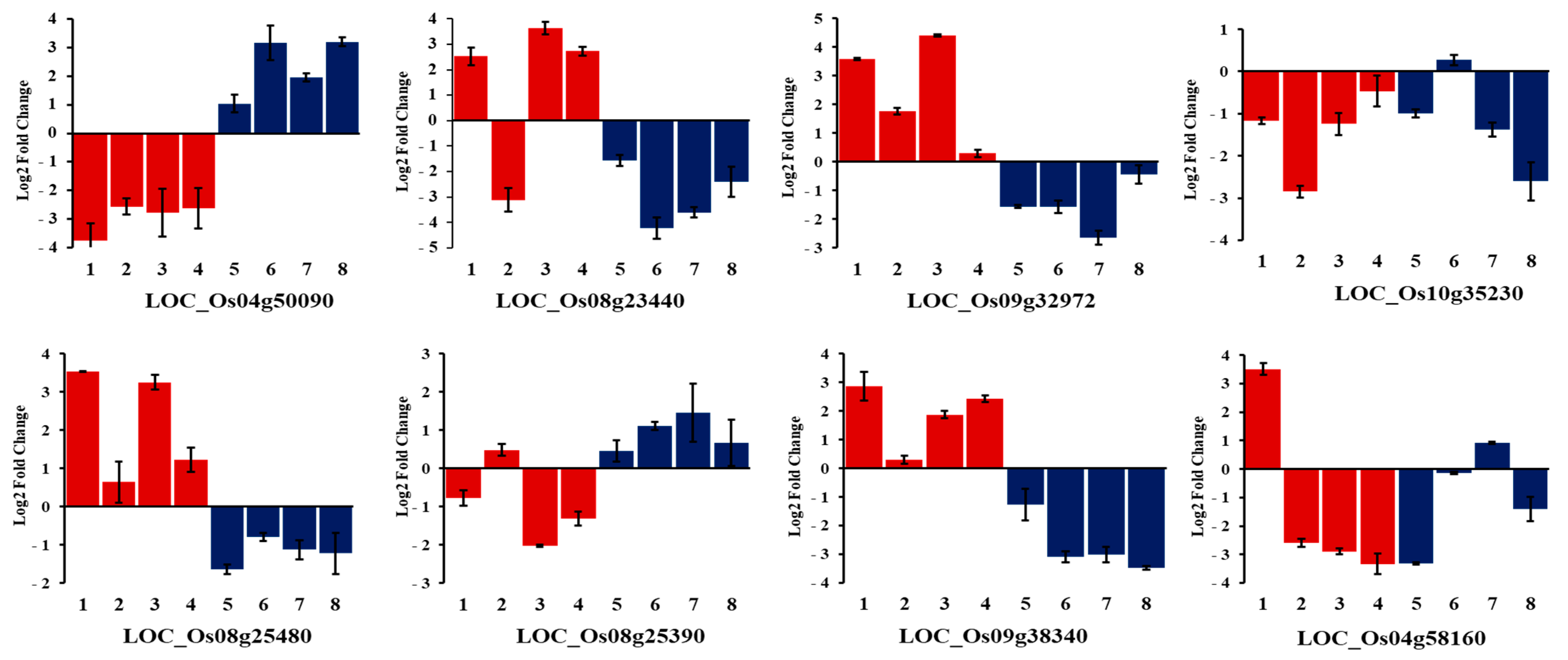
2.9. Expression Profiling of Selected Candidate Genes under Alkalinity Stress
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Materials
5.2. Statistical Analysis
5.3. SNP Genotyping and Quality Control
5.4. Structure Analysis and Linkage Disequilibrium
5.5. Association Mapping
5.6. Candidate Gene Analysis
5.7. Expression Profiling of Selected Genes by Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jin, H.; Plaha, P.; Park, J.Y.; Hong, C.P.; Lee, I.S.; Yang, Z.H.; Jiang, G.B.; Kwak, S.S.; Liu, S.K.; Lee, J.S.; et al. Comparative EST profiles of leaf and root of Leymus chinensis, a xerophilous grass adapted to high pH sodic soil. Plant Sci. 2006, 170, 1081–1086. [Google Scholar] [CrossRef]
- Singh, L.; Coronejo, S.; Pruthi, R.; Chapagain, S.; Subudhi, P.K. Integration of QTL mapping and whole genome sequencing identifies candidate genes for alkalinity tolerance in rice (Oryza sativa). Int. J. Mol. Sci. 2022, 23, 11791. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, L.; Cheng, Z.; Han, B.; Huang, X.; Wu, C.; Xiao, J.; Zhang, Q. Rice functional genomics research: Past decade and future. Mol. Plant 2018, 11, 359–380. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef]
- Lv, B.S.; Li, X.W.; Ma, H.Y.; Sun, Y.; Wei, L.X.; Jiang, C.J.; Liang, Z.-W. Differences in growth and physiology of rice in response to different saline-alkaline stress factors. Agron. J. 2013, 105, 1119–1128. [Google Scholar] [CrossRef]
- Singh, L.; Coronejo, S.; Pruthi, R.; Chapagain, S.; Bhattarai, U.; Subudhi, P.K. Genetic dissection of alkalinity tolerance at the seedling stage in rice (Oryza sativa) using a high-resolution linkage map. Plants 2022, 11, 3347. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X. Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef]
- Chen, W.; Cui, P.; Sun, H.; Guo, W.; Yang, C.; Jin, H.; Fang, B.; Shi, D. Comparative effects of salt and alkali stresses on organic acid accumulation and ionic balance of seabuckthorn (Hippophae rhamnoides L.). Ind. Crops Prod. 2009, 30, 351–358. [Google Scholar] [CrossRef]
- Tian, Z.J.; Li, J.P.; Jia, X.Y.; Yang, F.; Wang, Z.C. Assimilation and translocation of dry matter and phosphorus in rice genotypes affected by salt-alkaline stress. Sustainability 2016, 8, 568. [Google Scholar] [CrossRef]
- Chuamnakthong, S.; Nampei, M.; Ueda, A. Characterization of Na+ exclusion mechanism in rice under saline-alkaline stress conditions. Plant Sci. 2019, 287, 110171. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zheng, H.; Cui, J.; Wang, J.; Liu, H.; Sun, J.; Liu, T.; Zhao, H.; Lai, Y.; Zou, D. Genome-wide association study and candidate gene analysis of alkalinity tolerance in japonica rice germplasm at the seedling stage. Rice 2019, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, Z.; Chen, C.; Yang, C.; Shi, D. Effects of salt and alkali stresses on growth and ion balance in rice (Oryza sativa L.). Plant Soil Environ. 2011, 57, 286–294. [Google Scholar] [CrossRef]
- Guo, M.; Wang, R.; Wang, J.; Hua, K.; Wang, Y.; Liu, X.; Yao, S. ALT1, a Snf2 family chromatin remodeling ATPase, negatively regulates alkaline tolerance through enhanced defense against oxidative stress in rice. PLoS ONE 2014, 9, e112515. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Sun, J.; Wang, J.; Liu, H.; Zheng, H.; Yang, L.; Liang, Y.; Li, X.; Zou, D. QTL analysis for alkaline tolerance of rice and verification of a major QTL. Plant Breed. 2017, 136, 881–891. [Google Scholar] [CrossRef]
- Li, N.; Liu, H.; Sun, J.; Zheng, H.; Wang, J.; Yang, L.; Zhao, H.; Zou, D. Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci. Rep. 2018, 8, 9586. [Google Scholar] [CrossRef]
- El-Mahi, H.; Pérez-Hormaeche, J.; de Luca, A.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D.; et al. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef]
- Xiang, G.Q.; Ma, W.Y.; Gao, S.W.; Jin, Z.X.; Yue, Q.Y.; Yao, Y.X. Transcriptomic and phosphoproteomic profiling and metabolite analyses reveal the mechanism of NaHCO3-induced organic acid secretion in grapevine roots. BMC Plant Biol. 2019, 19, 383. [Google Scholar] [CrossRef]
- Li, X.Y.; Li, S.X.; Wang, J.H.; Lin, J.X. Exogenous abscisic acid alleviates harmful effect of salt and alkali stresses on wheat seedlings. Int. J. Environ. Res. Public Health 2020, 17, 3770. [Google Scholar] [CrossRef]
- Chen, T.X.; Shabala, S.; Niu, Y.N.; Chen, Z.H.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.L.; Zhou, M.X. Molecular mechanisms of salinity tolerance in rice. Crop J. 2021, 9, 506–520. [Google Scholar] [CrossRef]
- Galvan-Ampudia, C.S.; Julkowska, M.M.; Darwish, E.; Gandullo, J.; Korver, R.; Brunoud, G.; Haring, M.A.; Munnik, T.; Vernoux, T.; Testerink, C. Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 2013, 23, 2044–2055. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.L.; Zhang, R.X.; Yuan, H.Y.; Wang, M.M.; Yang, H.Y.; Ma, H.Y.; Liu, D.; Jiang, C.J.; Liang, Z.W. Root damage under alkaline stress is associated with reactive oxygen species accumulation in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yang, A.; Zhang, W.H. Efficient acquisition of iron confers greater tolerance to saline-alkaline stress in rice (Oryza sativa L.). J. Exp. Bot. 2016, 67, 6431–6444. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Qu, Y.; Yang, C.; Ma, X.; Cao, G.; Zhao, Z.; Zhang, S.; Zhang, T.; Han, L. Identification of QTLs associated with salt or alkaline tolerance at the seedling stage in rice under salt or alkaline stress. Euphytica 2015, 201, 441–452. [Google Scholar] [CrossRef]
- Cheng, H.T.; Jiang, H.; Xue, D.W.; Guo, L.B.; Zeng, D.L.; Zhang, G.H.; Qian, Q. Mapping of QTL underlying tolerance to alkali at germination and early seedling stages in rice. Acta Agron. Sin. 2008, 34, 1719–1727. [Google Scholar] [CrossRef]
- Qi, D.L.; Guo, G.Z.; Lee, M.C.; Zhang, J.G.; Cao, G.L.; Zhang, S.; Suh, S.; Zhou, Q.; Han, L. Identification of quantitative trait loci for the dead leaf rate and the seedling dead rate under alkaline stress in rice. J. Genet. Genom. 2008, 35, 299–305. [Google Scholar] [CrossRef]
- Luo, X.; Deng, H.; Wang, P.; Zhang, X.; Li, C.; Li, C.; Tan, J.; Wu, G.; Wang, Y.; Cheng, Q.; et al. Genetic analysis of germinating ability under alkaline and neutral salt stress by a high-density bin genetic map in rice. Euphytica 2020, 216, 107. [Google Scholar] [CrossRef]
- Obata, T.; Kitamoto, H.K.; Nakamura, A.; Fukuda, A.; Tanaka, Y. Rice shaker potassium channel OsKAT1 confers tolerance to salinity stress on yeast and rice cells. Plant Physiol. 2007, 144, 1978–1985. [Google Scholar] [CrossRef]
- Guo, J.; Sun, W.; Liu, H.; Chi, J.; Odiba, A.S.; Li, G.; Li-Ping, J.; Xin, C. Aldehyde dehydrogenase plays crucial roles in response to lower temperature stress in Solanum tuberosum and Nicotiana benthamiana. Plant Sci. 2020, 297, 110525. [Google Scholar] [CrossRef]
- Guan, Q.; Ma, H.; Wang, Z.J.; Wang, Z.Y.; Liu, S.K. A rice LSD1-like-type ZFP gene OsLOL5 enhances saline-alkaline tolerance in transgenic Arabidopsis thaliana, yeast and rice. BMC Genom. 2016, 17, 142. [Google Scholar] [CrossRef]
- Wang, B.; Xie, G.; Liu, Z.; He, R.; Han, J.; Huang, S.; Liu, L.; Cheng, X. Mutagenesis reveals that the OsPPa6 gene is required for enhancing the alkaline tolerance in rice. Front. Plant Sci. 2019, 10, 759. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Wang, S.; Shen, T.; Wang, Q.; Chen, C.; Xia, J.; Jiang, M. Calcium/calmodulin-dependent protein kinase OsDMI3 positively regulates saline-alkaline tolerance in rice roots. Plant Signal. Behav. 2020, 15, 1813999. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Kim, Y.A.; Shin, R.; Park, C.J. Nucleus-encoded thylakoid protein, OsY3IP1, confers enhanced tolerance to saline and alkaline stresses in rice. Rice Sci. 2022, 29, 225–236. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408. [Google Scholar] [CrossRef]
- Huang, X.; Yang, S.; Gong, J.; Zhao, Q.; Feng, Q.; Zhan, Q.; Zhao, Y.; Li, W.; Cheng, B.; Xia, J.; et al. Genomic architecture of heterosis for yield traits in rice. Nature 2016, 537, 629. [Google Scholar] [CrossRef]
- Du, X.; Xu, W.; Peng, C.; Li, C.; Zhang, Y.; Hu, L. Identification and validation of a novel locus, Qpm-3BL, for adult plant resistance to powdery mildew in wheat using multilocus GWAS. BMC Plant Biol. 2021, 21, 357. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef]
- Li, X.; Zheng, H.; Wu, W.; Liu, H.; Wang, J.; Jia, Y.; Li, J.; Yang, L.; Lei, L.; Zou, D.; et al. QTL Mapping and candidate gene analysis for alkali tolerance in japonica rice at the bud stage based on linkage mapping and genome-wide association study. Rice 2020, 13, 48. [Google Scholar] [CrossRef]
- Mei, S.; Zhang, G.; Jiang, J.; Lu, J.; Zhang, F. Combining genome-wide association study and gene-based haplotype analysis to identify candidate genes for alkali tolerance at the germination stage in rice. Front. Plant Sci. 2022, 13, 887239. [Google Scholar] [CrossRef]
- Wei, L.X.; Lv, B.S.; Wang, M.M.; Ma, H.Y.; Yang, H.Y.; Liu, X.L.; Jiang, C.J.; Liang, Z.W. Priming effect of abscisic acid on alkaline stress tolerance in rice (Oryza sativa L.) seedlings. Plant Physiol. Biochem. 2015, 90, 50–57. [Google Scholar] [CrossRef]
- De Leon, T.B.; Linscombe, S.; Gregorio, G.; Subudhi, P.K. Genetic variation in Southern USA rice genotypes for seedling salinity tolerance. Front. Plant Sci. 2015, 6, 374. [Google Scholar] [CrossRef]
- Chunthaburee, S.; Dongsansuk, A.; Sanitchon, J.; Pattanagul, W.; Theerakulpisut, P. Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage. Saudi J. Biol. Sci. 2016, 23, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Haque, T.; Elias, S.M.; Razzaque, S.; Biswas, S.; Khan, S.F.; Azad Jewel, G.M.N.; Rahman, M.S.; Juenger, T.E.; Seraj, Z.I. Salt tolerance QTLs of an endemic rice landrace, Horkuch at seedling and reproductive stages. Sci. Rep. 2022, 12, 17306. [Google Scholar] [CrossRef] [PubMed]
- Puram, V.R.R.; Ontoy, J.; Subudhi, P.K. Identification of QTLs for salt tolerance traits and prebreeding lines with enhanced salt tolerance in an introgression line population of rice. Plant Mol. Biol. Rep. 2018, 36, 695–709. [Google Scholar] [CrossRef]
- Rahman, M.A.; Bimpong, I.K.; Bizimana, J.B.; Pascual, E.D.; Arceta, M.; Swamy, B.P.M.; Diaw, F.; Rahman, M.S.; Singh, R.K. Mapping QTLs using a novel source of salinity tolerance from Hasawi and their interaction with environments in rice. Rice 2017, 10, 47. [Google Scholar] [CrossRef]
- Lu, H.; Redus, M.A.; Coburn, J.R.; Rutger, N.; McCouch, S.R.; Tai, T.H. Population structure and breeding patterns of 145 U.S. rice cultivars based on SSR marker analysis. Crop Sci. 2005, 45, 66–76. [Google Scholar] [CrossRef]
- Shi, D.; Sheng, Y. Effect of various salt-alkaline mixed stress conditions on sunflower seedlings and analysis of their stress factors. Environ. Exp. Bot. 2005, 54, 8–21. [Google Scholar] [CrossRef]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Pan, X.; Li, Y.; Cai, H.; Ji, W.; Chen, Q.; Zhu, Y. Osa-MIR393: A salinity- and alkaline stress-related microRNA gene. Mol. Biol. Rep. 2011, 38, 237–242. [Google Scholar] [CrossRef]
- Wang, N.; Fan, X.; Lin, Y.; Li, Z.; Wang, Y.; Zhou, Y.; Meng, W.; Peng, Z.; Zhang, C.; Ma, J. Alkaline stress induces different physiological, hormonal and gene expression responses in diploid and autotetraploid rice. Int. J. Mol. Sci. 2022, 23, 5561. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Ann. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Sahni, S.; Prasad, B.D.; Liu, Q.; Grbic, V.; Sharpe, A.; Singh, S.P.; Krishna, P. Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 2016, 6, 28298. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef]
- Khan, I.; Mohamed, S.; Regnault, T.; Mieulet, D.; Guiderdoni, E.; Sentenac, H.; Véry, A.A. Constitutive contribution by the rice OsHKT1;4 Na+ transporter to xylem sap desalinization and low Na+ accumulation in young leaves under low as high external Na+ conditions. Front. Plant Sci. 2020, 11, 1130. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Qiu, X.; Wang, L.; Xie, W.; Zhang, C.; Xiong, L.; Lian, X.; Zhang, Q. KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol. Genet. Genom. 2008, 280, 437–452. [Google Scholar] [CrossRef]
- Cotsaftis, O.; Plett, D.; Shirley, N.; Tester, M.; Hrmova, M. A two-staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS ONE 2012, 7, e39865. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Singh, B.; Panda, K.; Singh, B.P.; Singh, N.; Misra, P.; Rai, V.; Singh, N.K. Association of SNP haplotypes of HKT family genes with salt tolerance in Indian wild rice germplasm. Rice 2016, 9, 15. [Google Scholar] [CrossRef]
- Shohan, M.U.S.; Sinha, S.; Nabila, F.H.; Dastidar, S.G.; Seraj, Z.I. HKT1;5 transporter gene expression and association of amino acid substitutions with salt tolerance across rice genotypes. Front. Plant Sci. 2019, 10, 1420. [Google Scholar] [CrossRef]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W.-X. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef]
- Huang, X.; Li, K.; Jin, C.; Zhang, S. ICE1 of Pyrus ussuriensis functions in cold tolerance by enhancing PuDREBa transcriptional levels through interacting with PuHHP1. Sci. Rep. 2015, 5, 17620. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Man, L.; Xiang, D.; Wang, L.; Zhang, W.; Wang, X.; Qi, G. Stress-responsive gene RsICE1 from Raphanus sativus increases cold tolerance in rice. Protoplasma 2017, 254, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Ma, X.; Wang, Q.; Gao, Y.; Xue, Y.; Niu, X.; Yu, G.; Liu, Y.S. Significant improvement of stress tolerance in tobacco plants by overexpressing a stress-responsive aldehyde dehydrogenase gene from maize (Zea mays). Plant Mol. Biol. 2008, 68, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.S.; Chen, X.Y.; Yang, K.; Sun, Z.X.; Fu, Y.P.; Zhang, Y.M.; Fang, R.X. Overexpression of an F-box protein gene reduces abiotic stress tolerance and promotes root growth in rice. Mol. Plant 2011, 4, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, H.; Hong, Y.; Huang, L.; Li, X.; Zhang, Y.; Ouyang, Z.; Song, F. Genome-wide identification, biochemical characterization, and expression analyses of the YTH domain-containing RNA-binding protein family in Arabidopsis and rice. Plant Mol. Biol. Rep. 2014, 32, 1169–1186. [Google Scholar] [CrossRef]
- Dametto, A.; Buffon, G.; Dos Reis Blasi, E.A.; Sperotto, R.A. Ubiquitination pathway as a target to develop abiotic stress tolerance in rice. Plant Signal. Behav. 2015, 10, e1057369. [Google Scholar] [CrossRef] [PubMed]
- Razzaque, S.; Elias, S.M.; Haque, T.; Biswas, S.; Jewel, N.A.; Rahman, S.; Weng, X.; Ismail, A.M.; Walia, H.; Juenger, T.E.; et al. Gene expression analysis associated with salt stress in a reciprocally crossed rice population. Sci. Rep. 2019, 9, 8249. [Google Scholar] [CrossRef]
- Gautam, R.; Meena, R.K.; Woch, N.; Kirti, P. Ectopic expression of BrALDH7B2 gene encoding an antiquitin from Brassica rapa confers tolerance to abiotic stresses and improves photosynthetic performance under salt stress in tobacco. Environ. Exp. Bot. 2020, 180, 104223. [Google Scholar] [CrossRef]
- Lin, R.; Zheng, J.; Pu, L.; Wang, Z.; Mei, Q.; Zhang, M.; Jian, S. Genome-wide identification and expression analysis of aquaporin family in Canavalia rosea and their roles in the adaptation to saline-alkaline soils and drought stress. BMC Plant Biol. 2021, 21, 333. [Google Scholar] [CrossRef]
- Shim, J.S.; Park, S.H.; Lee, D.K.; Kim, Y.S.; Park, S.C.; Redillas, M.R.; Seo, J.S.; Kim, J.K. The rice GLYCINE-RICH PROTEIN 3 confers drought tolerance by regulating mRNA stability of ROS scavenging-related genes. Rice 2021, 14, 31. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef] [PubMed]
- De Leon, T.B.; Linscombe, S.; Subudhi, P.K. Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice 2016, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, U.; Subudhi, P.K. Genetic analysis of yield and agronomic traits under reproductive-stage drought stress in rice using a high-resolution linkage map. Gene 2018, 669, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef]
- Kim, J.M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef]
- Singroha, G.; Kumar, S.; Gupta, O.P.; Singh, G.P.; Sharma, P. Uncovering the epigenetic marks involved in mediating salt stress tolerance in plants. Front. Genet. 2022, 13, 811732. [Google Scholar] [CrossRef]
- Wang, T.; Chen, Y.; Zhang, M.; Chen, J.; Liu, J.; Han, H.; Hua, X. Arabidopsis AMINO ACID PERMEASE1 contributes to salt stress-induced proline uptake from exogenous sources. Front. Plant Sci. 2017, 8, 2182. [Google Scholar] [CrossRef]
- Rai, A.; Singh, R.; Shirke, P.A.; Tripathi, R.D.; Trivedi, P.K.; Chakrabarty, D. Expression of rice CYP450-like gene (Os08g01480) in Arabidopsis modulates regulatory network leading to heavy metal and other abiotic stress tolerance. PLoS ONE 2015, 10, e0138574. [Google Scholar] [CrossRef]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.V.; Jugulam, M. Role of cytochrome P450 enzymes in plant stress response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 Zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef]
- Meng, B.; Wang, T.; Luo, Y.; Guo, Y.; Xu, D.; Liu, C.; Zou, J.; Li, L.; Diao, Y.; Gao, Z.; et al. Identification and allele combination analysis of rice grain shape-related genes by genome-wide association study. Int. J. Mol. Sci. 2022, 23, 1065. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.N.U.; Jewel, G.A.; Azim, T.; Seraj, Z.I. Novel QTLs for salinity tolerance revealed by genome-wide association studies of biomass, chlorophyll and tissue ion content in 176 rice landraces from Bangladesh. PLoS ONE 2021, 16, e0259456. [Google Scholar] [CrossRef]
- Descalsota, G.I.L.; Swamy, B.P.M.; Zaw, H.; Inabangan-Asilo, M.A.; Amparado, A.; Mauleon, R.; Chadha-Mohanty, P.; Arocena, E.C.; Raghavan, C.; Leung, H.; et al. Genome-wide association mapping in a rice MAGIC plus population detects QTLs and genes useful for biofortification. Front. Plant Sci. 2018, 9, 1347. [Google Scholar] [CrossRef]
- Alkahtani, J. Genome-wide association study of grain quality traits in rice detected genomic regions of high-quality rice for increasing rice consumption. Biosci. Biotechnol. Res. Asia 2022, 19, 333–346. [Google Scholar] [CrossRef]
- Jones, J.B.; Case, V.W. Sampling, handling, and analyzing plant tissue samples. In Soil Testing and Plant Analysis, 3rd ed.; Book Series No. 3; Westerman, R.L., Ed.; Soil Science Society of America: Madison, WI, USA, 1990; pp. 389–427. [Google Scholar]
- SAS Institute Inc. SAS®9.4 System Options: Reference, 2nd ed.; SAS Institute Inc.: Cary, NC, USA, 2012; p. 69. [Google Scholar]
- R Foundation. R: A Language and Environment for Statistical Computing; Reference Index Version 2.2.1; R Foundation: Vienna, Austria, 2005; Volume 68. [Google Scholar]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martínez, C.T. Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev. 2003, 22, 9–111. [Google Scholar]
- Money, D.; Gardner, K.; Migicovsky, Z.; Schwaninger, H.; Zhong, G.Y.; Myles, S. LinkImpute: Fast and accurate genotype imputation for nonmodel organisms. G3 2015, 5, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Hill, W.G.; Weir, B.S. Variances and covariance of squared linkage disequilibria in finite populations. Theor. Popul. Biol. 1988, 33, 54–78. [Google Scholar] [CrossRef]
- Price, A.; Patterson, N.; Plenge, R.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Larsson, S.J.; Lipka, A.E.; Buckler, E.S. Lessons from Dwarf8 on the strengths and weaknesses of structured association mapping. PLoS Genet. 2013, 9, e1003246. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Pressoir, G.; Briggs, W.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Garcia, R.S.; Coronejo, S.; Tapia, R. Comparative transcriptional profiling of root tissues in two rice genotypes reveals differential expressed genes associated with root architecture under nitrogen stress. Int. J. Mol. Sci. 2020, 21, 5759. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Trait a | Min | Max | Mean | Standard Deviation | RIL Pr > Fc b | Heritability |
|---|---|---|---|---|---|---|
| AKT | 1.0 | 9.0 | 4.75 | 2.05 | 0.002 ** | 0.87 |
| SHL | 21.3 | 63.0 | 37.7 | 6.6 | 0.047 * | 0.64 |
| RTL | 7.0 | 22.7 | 16.4 | 3.2 | 0.029 * | 0.52 |
| RSR | 0.16 | 0.76 | 0.46 | 0.21 | 0.029 * | 0.69 |
| Inv_FW | 0.01 | 0.30 | 0.16 | 0.07 | 0.0003 ** | 0.61 |
| log_DW | −2.84 | −0.60 | −1.50 | 0.43 | 0.029 * | 0.53 |
| SNC | 661.6 | 3508.1 | 1730.1 | 440.3 | 0.032 * | 0.77 |
| SKC | 326.3 | 1077.9 | 657.7 | 138.3 | 0.048 * | 0.84 |
| SNK | 0.88 | 5.95 | 2.79 | 1.11 | 0.041 * | 0.81 |
| Trait a | AKT | SHL | RTL | RSR | Inv_FW | log_DW | SNC | SKC | SNK |
|---|---|---|---|---|---|---|---|---|---|
| AKT | 1.000 | ||||||||
| SHL | −0.123 * | 1.000 | |||||||
| RTL | −0.147 * | 0.02 | 1.000 | ||||||
| RSR | −0.154 * | −0.757 ** | −0.533 ** | 1.000 | |||||
| Inv_FW | −0.909 ** | 0.127 * | 0.174 * | −0.173 * | 1.000 | ||||
| log_DW | −0.954 ** | 0.139 * | 0.170 * | −0.181 * | 0.961 ** | 1.000 | |||
| SNC | 0.467 ** | −0.028 | −0.074 | −0.027 | −0.36 ** | −0.423 ** | 1.000 | ||
| SKC | −0.053 * | 0.004 | 0.009 | 0.002 | 0.155 * | 0.094 | −0.87 ** | 1.000 | |
| SNK | 0.321 ** | −0.028 | −0.002 | −0.015 | −0.18 ** | −0.258 ** | 0.742 ** | −0.674 ** | 1.000 |
| Clusters | Genotypes |
|---|---|
| Cluster 1 (Highly Susceptible) | Hasawi, Roy J, Djogolan, Dular, Cypress, Vegold, ChN1264, Toro-2, Belle Patna, N22, Magnolia, Glutinous Zenith, Jazzman-2, Toro, Chengri, Azucena, Chambal, Bluebonnet, Orion, Adair, Pratao Tipo Guedes, Dholamon 560, Hill medium, KN-1-B-361-1-8-67 |
| Cluster 2 (Tolerant) | PSBRC-50, CL111, Caloro, Cheriviruppu, CL131, Trenasse, Pirogue, LA0802140, Jupiter, LA0702085, Rexona, FL478, Geumgangbyeo, Neptune, CL261, FL318, Caffey, Lacassine, CLPK873, Cocodrie, Lacrosse, Sunbonnet, Lafitte, Dellmati, Carolina Gold, Bengal, Century Patna, CL152, Nato, MS-1996-9, Glutinous Selection, Saturn Rogue, Langmanbi, Milagrosa, Zhenshan 97, R-50, Mars, Kasalath, Sarioo50, IR 8, M202, Zenith, IR 64, Arkansas Fortuna, Texmont, Kranti, TP 49, Millie, Kirak, Chung yuen, IRGC1244, Newrex, RD, IRGC32567, Kitaake, Brazos, M-204, Delitus, Italica Livorno, CT-329 |
| Cluster 3 (Highly Tolerant) | Saturn, Della, JN100, Moroberekan, JN349, Nipponbare, Mercury, BHA1115, IR 29, Dellrose, Lotus, Agami, Neches, Epagri, Cheniere, CSR11, Vandana, Gu Ze, IR 50, Panidhan II, Koshihikari, Teqing, Taichung 65, Daido, Lemont, Quilloa 66304, Dellmont, Kanchan, Swarna, W149, Perum karuppan, Taipe 309, Hayamasari |
| Cluster 4 (Moderately Tolerant) | LA110, CL142, Century Rogue, Pokkali, Pecos, Nona Bokra, Wells, Gold Zenith, Skybonnet, Tebonnet, Nira, Vista, TCCP, Templeton, Nova 66, IRRI147, Taggert, Bluebelle, Arang, Ecrevisse, Smooth Zenith, Damodar, Kalia, MS-1995-15, SLO16, Rexark, V20B, Ning Yang Keng, Stormproof, Starbonnet, B573-A4-20-6, R-27, Gold Nato, Naylamp, Azaurel, Melrose, Jinheung, Arkrose, Dixiebelle, Nerretto, PSRR-1, Bala, Co39, San Tou Thou, IR4432-52-6-4, Hill LongGrain, Bharathy, H4, IARI 5823, Early Prolific, Fatehpur 3, Prelude, WC10380 |
| Cluster 5 (Susceptible) | Pinkaeo, LAH10, Mermentau, Evangeline, CR5272, R609, Jes, Della-2, CL162, R-54, Radin Ebos 33, Kokubelle, LaGrue, Jackson |
| Source of Variation | DF a | Sum of Squares | Mean Sum of Squares | Variance (%) | p-Value b |
|---|---|---|---|---|---|
| Among population | 4 | 495.8 | 123.9 | 61 | <0.0001 |
| Within population | 163 | 1129.8 | 6.9 | 39 | <0.001 |
| Total | 167 | 1625.6 | 100 |
| Chr. | No. of SNPs | Chr. Size (bp) † | SNP Density (bp/SNP) | LD $ Distance (bp) | |
|---|---|---|---|---|---|
| 1 | 93 | 43,270,923 | 465,279 | 15,193,454 | |
| 2 | 81 | 35,937,250 | 443,670 | 13,604,743 | |
| 3 | 78 | 36,413,819 | 466,844 | 12,513,847 | |
| 4 | 66 | 35,502,694 | 537,920 | 11,534,383 | |
| 5 | 64 | 29,958,434 | 468,101 | 10,451,492 | |
| 6 | 85 | 31,248,787 | 367,633 | 11,193,100 | |
| 7 | 59 | 29,697,621 | 503,350 | 11,473,349 | |
| 8 | 72 | 28,443,022 | 395,042 | 9,654,739 | |
| 9 | 58 | 23,012,720 | 396,772 | 7,434,300 | |
| 10 | 54 | 23,207,287 | 429,765 | 7,334,850 | |
| 11 | 59 | 29,021,106 | 491,884 | 9,261,105 | |
| 12 | 62 | 27,531,856 | 444,063 | 8,644,012 | |
| Total | 830 | 373,245,519 | Mean | 450,861 | 10,691,115 |
| Trait a | SNP | Locus | Annotation | QTLs/Genes in Previous Studies |
|---|---|---|---|---|
| AKT | S04_29881066 | Os04g50090 | Helix–loop–helix DNA-binding protein | qSNK4-2 [12] |
| S08_14184612 | Os08g23440 | amino acid permease family protein | LOC_Os08g23440 [16] | |
| SHL | S04_22808095 | Os04g38340 | ER-Golgi intermediate-compartment protein 3 | qDLR4 [26] |
| S12_23066809 | Os12g37570 | protein kinase family protein | ||
| S12_23108164 | Os12g37640 | xaa-Pro aminopeptidase | ||
| RTL | S02_35216781 | Os02g58139 | OsSigP1-Type I Signal Peptidase homolog | |
| S04_29715617 | Os04g49850 | Expressed protein | qSNK4-2 [12] | |
| S04_34925111 | Os04g58730 | AT-hook-motif-domain-containing protein | qSNK4-2 [12] | |
| S05_1487229 | Os05g03510 | Expressed protein | ||
| S07_28409912 | Os07g47500 | Histone-arginine methyltransferase CARM1 | qRGR7 [25] | |
| RSR | S01_23656773 | Os01g41790 | Expressed protein | |
| S01_37680628 | Os01g64910 | Anthocyanidin 5,3-O-glucosyltransferase | ||
| S05_24090514 | Os05g41130 | OsFBX168-F-box-domain-containing protein | qRRN5 [25] | |
| S10_18098744 | Os10g35570 | Expressed protein | qSKC10.18 [2,7] | |
| S12_3544726 | Os12g07210 | Expressed protein | ||
| log_DW | S01_36150523 | Os01g62450 | Expressed protein | |
| S05_7195992 | Os05g12510 | Expressed protein | qDLRa5-3 [24] | |
| inv_FW | S01_3236648 | Os01g06820 | hcr2-0B, putative | |
| SKC | S09_19322095 | Os09g32350 | Expressed protein | qSNC9.19 [2] |
| S09_19683788 | Os09g32972 | MYB protein | qSNC9.19 [2] | |
| S10_18834021 | Os10g35230 | Rf1, mitochondrial precursor | qSKC10.18 [2,7] | |
| SNC | S02_3477202 | Os02g06890 | OTU-like cysteine protease family protein | |
| S03_14554651 | Os03g25480 | Cytochrome P450 | Os03g25480 [16] | |
| S06_15335573 | Os06g39580 | Hypothetical protein | qARL6 [38] | |
| S07_29627590 | Os07g49470 | Protein kinase APK1B, chloroplast precursor | ||
| S08_15439243 | Os08g25390 | Bifunctional homoserine dehydrogenase | Os08g25390 [16] | |
| S09_22076185 | Os09g38340 | ZOS9-17-C2H2 zinc finger protein | ||
| SNK | S04_34643455 | Os04g58160 | Fiber protein Fb34, putative | qSNK4-2 [12] |
| Trait a | QTLs | Lead SNP | Position | p-Value | R2 (%) | QTLs in a Previous Study |
|---|---|---|---|---|---|---|
| SHL | qSHL12 | S12_23108164 | 23,108,164 | 0.00058 | 11 | - |
| log_DW | qlog_DW1 | S01_36150523 | 36,150,523 | 0.00008 | 14 | qSHL1.38 [2,7] |
| SNC | qSNC7 | S07_29627590 | 29,627,590 | 0.00027 | 11 | - |
| SKC | qSKC9 | S09_19322095 | 19,322,095 | 0.00037 | 22 | qSNC9.19 [2] |
| SKC | qSKC10 | S10_18834021 | 18,834,021 | 0.00021 | 18 | qSKC10.18 [2,7] |
| SNK | qSNK4 | S04_34643455 | 34,643,455 | 0.00002 | 16 | qSNK4-2 [12] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, L.; Pruthi, R.; Chapagain, S.; Subudhi, P.K. Genome-Wide Association Study Identified Candidate Genes for Alkalinity Tolerance in Rice. Plants 2023, 12, 2206. https://doi.org/10.3390/plants12112206
Singh L, Pruthi R, Chapagain S, Subudhi PK. Genome-Wide Association Study Identified Candidate Genes for Alkalinity Tolerance in Rice. Plants. 2023; 12(11):2206. https://doi.org/10.3390/plants12112206
Chicago/Turabian StyleSingh, Lovepreet, Rajat Pruthi, Sandeep Chapagain, and Prasanta K. Subudhi. 2023. "Genome-Wide Association Study Identified Candidate Genes for Alkalinity Tolerance in Rice" Plants 12, no. 11: 2206. https://doi.org/10.3390/plants12112206
APA StyleSingh, L., Pruthi, R., Chapagain, S., & Subudhi, P. K. (2023). Genome-Wide Association Study Identified Candidate Genes for Alkalinity Tolerance in Rice. Plants, 12(11), 2206. https://doi.org/10.3390/plants12112206








