Specific Roles of Lipoxygenases in Development and Responses to Stress in Plants
Abstract
1. Introduction
2. Lipoxygenases
3. Mechanism of LOX-Mediated Reaction
4. Regulation of LOX Signaling Associated Scheme
5. LOX and Related Metabolites in Stress Mitigation via Phytohormonal Interaction
6. LOX in Seed Germination and Growth of Seedling
7. LOXs in Male Gametophyte
8. LOX in Pathogen Attack
9. LOX Activity as a Molecular Marker for Stress Tolerance/Stress and LOX Activity
10. LOX Activity and Their Products
11. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 10-OPEA | 10-oxo-11-phytoenoic acid |
| AA | amino acid |
| ABA | abscisic acid |
| AOC | allene oxide cyclase |
| AOS | allene oxide synthase |
| BR | brassinosteroid |
| DES | divinyl ether synthase |
| ET | ethylene |
| HFA | 9-hydro (peroxy) fatty acid |
| HPL | hydroperoxide lyase |
| HPO | hydroperoxy fatty acid |
| 9-HPODE | (9S,10E,12Z)-9-hydroperoxy-10,12-octadecadienoic acid |
| 13-HPODE | (13S,9Z,10E)-13-hydroperoxy-9,11-octadecadienoic acid |
| ICK1 | inhibitor/interactor of cyclin-dependent kinase 1 |
| JA | jasmonic acid |
| LA | α-linoleic acid |
| LOX | lipoxygenase |
| MeJA | methyl jasmonate |
| OPDA | cis-12-oxophytodienoic acid |
| PAL | phenylalanine ammonia-lyase |
| PSbMV | pea seed-borne mosaic virus |
| PUFA | polyunsaturated fatty acid |
| VSP | vegetative storage protein |
References
- Song, H.; Wang, P.; Li, C.; Han, S.; Lopez-Baltazar, J.; Zhang, X.; Wang, X. Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci. Rep. 2016, 6, 35245. [Google Scholar] [CrossRef] [PubMed]
- Hayward, S.; Cilliers, T.; Swart, P. Lipoxygenases: From isolation to application. Compr. Rev. Food Sci. Food Saf. 2016, 16, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Sumner, J.B.; Somers, G.F. Miscellaneous oxidizing enzymes. In Chemistry and Methods of Enzymes, 3rd ed.; Sumner, J.B., Somers, G.F., Eds.; Academic Press: New York, NY, USA, 1953; pp. 300–320. [Google Scholar] [CrossRef]
- Craig, F.N. A fat oxidation system in Lupinus albus. J. Biol. Chem. 1936, 114, 727–746. [Google Scholar] [CrossRef]
- Tauber, H. Unsaturated fat oxidase. J. Am. Chem. Soc. 1940, 62, 2251. [Google Scholar] [CrossRef]
- Sumner, R.J. Lipid oxidase studies: III. The relation between carotene oxidation and the enzymic peroxidation of unsaturated fats. J. Biol. Chem. 1942, 146, 215–218. [Google Scholar] [CrossRef]
- Theorell, H.; Holman, R.T.; Akeson, A. A note on the preparation of crystalline soy bean lipoxidase. Arch. Biochem. 1947, 14, 250–252. [Google Scholar]
- Viswanath, K.K.; Varakumar, P.; Pamuru, R.R.; Basha, S.J.; Mehta, S.; Rao, A.D. Plant lipoxygenases and their role in plant physiology. J. Plant Biol. 2020, 63, 83–95. [Google Scholar] [CrossRef]
- Shi, Y.; Mandal, R.; Singh, A.; Pratap Singh, A. Legume lipoxygenase: Strategies for application in food industry. Legume Sci. 2020, 2, e44. [Google Scholar] [CrossRef]
- Aanangi, R.; Kotapati, K.V.; Palaka, B.K.; Kedam, T.; Kanika, N.D.; Ampasala, D.R. Purification and characterization of lipoxygenase from mung bean (Vigna radiata L.) germinating seedlings. 3 Biotech 2016, 6, 113. [Google Scholar] [CrossRef]
- Mitsuda, H.; Yasumoto, K.; Yamamoto, A.; Kusano, T. Study on soybean lipoxygenase: Part I. Preparation of crystalline enzyme and assay by polarographic method. Agric. Biol. Chem. 1967, 31, 115–118. [Google Scholar] [CrossRef]
- Murphy, P.A. Soybean proteins. In Soybeans: Chemistry, Production, Processing, and Utilization; Johnson, L.A., White, P.J., Galloway, R., Eds.; AOCS Press: Urbana, IL, USA, 2008; pp. 229–267. [Google Scholar] [CrossRef]
- Offenbacher, A.R.; Holman, T.R. Fatty acid allosteric regulation of C-H activation in plant and animal lipoxygenases. Molecules 2020, 25, 3374. [Google Scholar] [CrossRef]
- Casey, R. Lipoxygenases. In Seed Proteins; Shewry, P.R., Casey, R., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 685–708. [Google Scholar] [CrossRef]
- Nemchenko, A.; Kunze, S.; Feussner, I.; Kolomiets, M. Duplicate maize 13-lipoxygenase genes are differentially regulated by circadian rhythm, cold stress, wounding, pathogen infection, and hormonal treatments. J. Exp. Bot. 2006, 57, 3767–3779. [Google Scholar] [CrossRef]
- Szymanowska, U.; Jakubczyk, A.; Baraniak, B.; Kur, A. Characterisation of lipoxygenase from pea seeds (Pisum sativum var. Telephone L.). Food Chem. 2009, 116, 906–910. [Google Scholar] [CrossRef]
- Liavonchanka, A.; Feussner, I. Lipoxygenases: Occurrence, functions and catalysis. J. Plant Physiol. 2006, 163, 348–357. [Google Scholar] [CrossRef]
- Leone, A.; Melillo, M.T.; Bleve-Zacheo, T. Lipoxygenase in pea roots subjected to biotic stress. Plant Sci. 2001, 161, 703–717. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Nita, M.; Grzybowski, A. The Role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxid. Med. Cell. Longev. 2016, 2016, 3164734. [Google Scholar] [CrossRef]
- Kurutas, E.B. The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: Current state. Nutr. J. 2016, 15, 71. [Google Scholar] [CrossRef]
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef]
- Gaschler, M.M.; Stockwell, B.R. Lipid peroxidation in cell death. Biochem. Biophys. Res. Commun. 2017, 482, 419–425. [Google Scholar] [CrossRef]
- Dennis, E.A.; Norris, P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015, 15, 511–523. [Google Scholar] [CrossRef]
- Porta, H.; Rueda-Benitez, P.; Campos, F.; Colmenero-Flores, J.M.; Colorado, J.M.; Carmona, M.J.; Covarrubias, A.A.; Rocha-Sosa, M. Analysis of lipoxygenase mRNA accumulation in the common bean (Phaseolus vulgaris L.) during development and under stress conditions. Plant Cell Physiol. 1999, 40, 850–858. [Google Scholar] [CrossRef]
- Babenko, L.M.; Shcherbatiuk, M.M.; Skaterna, T.D.; Kosakivska, I.V. Lipoxygenases and their metabolites in formation of plant stress tolerance. Ukr. Biochem. J. 2017, 89, 5–21. [Google Scholar] [CrossRef]
- Howe, G.A.; Lee, G.I.; Itoh, A.; Li, L.; DeRocher, A.E. Cytochrome P450-dependent metabolism of oxylipins in tomato. Cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol. 2000, 123, 711–724. [Google Scholar] [CrossRef]
- Sivasankar, S.; Sheldrick, B.; Rothstein, S.J. Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 2000, 122, 1335–1342. [Google Scholar] [CrossRef]
- Grechkin, A.N. Hydroperoxide lyase and divinyl ether synthase. Prostaglandins Other Lipid Mediat. 2002, 68-69, 457–470. [Google Scholar] [CrossRef]
- Froehlich, J.E.; Itoh, A.; Howe, G.A. Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 2001, 125, 306–317. [Google Scholar] [CrossRef]
- Laudert, D.; Weiler, E.W. Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 1998, 15, 675–684. [Google Scholar] [CrossRef]
- Arif, Y.; Sami, F.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salicylic acid in relation to other phytohormones in plant: A study towards physiology and signal transduction under challenging environment. Environ. Exp. Bot. 2020, 175, 104040. [Google Scholar] [CrossRef]
- Hughes, R.K.; De Domenico, S.; Santino, A. Plant cytochrome CYP74 family: Biochemical features, endocellular localisation, activation mechanism in plant defence and improvements for industrial applications. ChemBioChem 2009, 10, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Gullner, G.; Künstler, A.; Király, L.; Pogány, M.; Tóbiás, I. Up-regulated expression of lipoxygenase and divinyl ether synthase genes in pepper leaves inoculated with Tobamoviruses. Physiol. Mol. Plant Pathol. 2010, 74, 387–393. [Google Scholar] [CrossRef]
- Meng, Y.; Liang, Y.; Liao, B.; He, W.; Liu, Q.; Shen, X.; Xu, J.; Chen, S. Genome-wide identification, characterization and expression analysis of lipoxygenase gene family in Artemisia annua L. Plants (Basel) 2022, 11, 655. [Google Scholar] [CrossRef]
- Melan, M.A.; Dong, X.; Endara, M.E.; Davis, K.R.; Ausubel, F.M.; Peterman, T.K. An Arabidopsis thaliana lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate. Plant Physiol. 1993, 101, 441–450. [Google Scholar] [CrossRef]
- Bell, E.; Creelman, R.A.; Mullet, J.E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 1995, 92, 8675–8679. [Google Scholar] [CrossRef]
- Vellosillo, T.; Martínez, M.; López, M.A.; Vicente, J.; Cascón, T.; Dolan, L.; Hamberg, M.; Castresana, C. Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 2007, 19, 831–846. [Google Scholar] [CrossRef]
- Caldelari, D.; Wang, G.; Farmer, E.E.; Dong, X. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 2010, 75, 25–33. [Google Scholar] [CrossRef]
- Santomauro, F.; Donato, R.; Pini, G.; Sacco, C.; Ascrizzi, R.; Bilia, A. Liquid and vapor-phase activity of Artemisia annua essential oil against pathogenic Malassezia spp. Planta Med. 2017, 84, 160–167. [Google Scholar] [CrossRef]
- Umate, P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal. Behav. 2011, 6, 335–338. [Google Scholar] [CrossRef]
- Marla, S.S.; Singh, V.K. LOX genes in blast fungus (Magnaporthe grisea) resistance in rice. Funct. Integr. Genom. 2012, 12, 265–275. [Google Scholar] [CrossRef]
- Savchenko, T.V.; Zastrijnaja, O.M.; Klimov, V.V. Oxylipins and plant abiotic stress resistance. Biochemistry (Moscow) 2014, 79, 362–375. [Google Scholar] [CrossRef]
- Lorenzo, O.; Solano, R. Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 2005, 8, 532–540. [Google Scholar] [CrossRef]
- Fabbri, A.A.; Fanelli, C.; Reverberi, M.; Ricelli, A.; Camera, E.; Urbanelli, S.; Rossini, A.; Picardo, M.; Altamura, M.M. Early physiological and cytological events induced by wounding in potato tuber. J. Exp. Bot. 2000, 51, 1267–1275. [Google Scholar] [CrossRef]
- Maccarrone, M.; Melino, G.; Finazzi-Agro, A. Lipoxygenases and their involvement in programmed cell death. Cell Death Differ. 2001, 8, 776–784. [Google Scholar] [CrossRef]
- Chandra, S.; Heinstein, P.F.; Low, P.S. Activation of phospholipase A by plant defense elicitors. Plant Physiol. 1996, 110, 979–986. [Google Scholar] [CrossRef]
- Babenko, L.M.; Kosakivska, I.V.; Akimov, Y.A.; Klymchuk, D.O.; Skaternya, T.D. Effect of temperature stresses on pigment content, lipoxygenase activity and cell ultrastructure of winter wheat seedlings. Genet. Plant Physiol. 2014, 4, 117–125. [Google Scholar]
- Feussner, I.; Wasternack, C. The lipoxygenase pathway. Annu. Rev. Plant Biol. 2002, 53, 275–297. [Google Scholar] [CrossRef]
- Santino, A.; Iannacone, R.; Hughes, R.; Casey, R.; Mita, G. Cloning and characterisation of an almond 9-lipoxygenase expressed early during seed development. Plant Sci. 2005, 168, 699–706. [Google Scholar] [CrossRef]
- Nishiuchi, T.; Hamada, T.; Kodama, H.; Iba, K. Wounding changes the spatial expression pattern of the Arabidopsis plastid omega-3 fatty acid desaturase gene (FAD7) through different signal transduction pathways. Plant Cell 1997, 9, 1701–1712. [Google Scholar] [CrossRef]
- Kolupaev, Y.E.; Yastreb, T.O.; Lugova, G.A. Role of jasmonates in plant adaptation to abiotic stressors. Genet. Plant Physiol. 2016, 48, 95–111. [Google Scholar] [CrossRef]
- Cohen, Y. Local and systemic protection against Phytophthora infestans induced in potato and tomato plants by jasmonic acid and jasmonic methyl ester. Phytopathology 1993, 83, 1054. [Google Scholar] [CrossRef]
- Birkenmeier, G.F.; Ryan, C.A. Wound signaling in tomato plants. Evidence that aba is not a primary signal for defense gene activation. Plant Physiol. 1998, 117, 687–693. [Google Scholar] [CrossRef]
- Wang, H.; Qi, Q.; Schorr, P.; Cutler, A.J.; Crosby, W.L.; Fowke, L.C. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 1998, 15, 501–510. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, H.; Gilmer, S.; Whitwill, S.; Fowke, L.C. Effects of co-expressing the plant CDK inhibitor ICK1 and D-type cyclin genes on plant growth, cell size and ploidy in Arabidopsis thaliana. Planta 2003, 216, 604–613. [Google Scholar] [CrossRef]
- Ozeretskovskaya, O.L.; Vasyukova, N.I.; Chalenko, G.I.; Gerasimova, N.G.; Revina, T.A.; Valueva, T.A. Wound healing and induced resistance in potato tubers. Appl. Biochem. Microbiol. 2009, 45, 199–203. [Google Scholar] [CrossRef]
- Campos-Vargas, R.; Saltveit, M.E. Involvement of putative chemical wound signals in the induction of phenolic metabolism in wounded lettuce. Physiol. Plant. 2002, 114, 73–84. [Google Scholar] [CrossRef]
- Moore, J.P.; Paul, N.D.; Whittaker, J.B.; Taylor, J.E. Exogenous jasmonic acid mimics herbivore-induced systemic increase in cell wall bound peroxidase activity and reduction in leaf expansion. Funct. Ecol. 2003, 17, 549–554. [Google Scholar] [CrossRef]
- Park, J.-H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef]
- Koshio, K.; Takahashi, H.; Ota, Y. Induction of browning of male flowers of Cryptomeria japonica by treatment with fatty acids: Mechanism and the role of trans-2-hexenal. Plant Cell Physiol. 1995, 36, 1511–1517. [Google Scholar] [CrossRef]
- Vick, B.A.; Zimmerman, D.C. Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 1984, 75, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Creelman, R.A.; Mullet, J.E. Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 355–381. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef] [PubMed]
- Kućko, A.; Florkiewicz, A.B.; Wolska, M.; Miętki, J.; Kapusta, M.; Domagalski, K.; Wilmowicz, E. Jasmonate-dependent response of the flower abscission zone cells to drought in yellow lupine. Plants (Basel) 2022, 11, 527. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Ye, J.; Shen, L.; Luo, Y. Effect of lipoxygenase and jasmonic acid on ethylene biosynthesis during tomato fruit ripening. Acta Hortic. 2003, 119–125. [Google Scholar] [CrossRef]
- Kausch, K.D.; Handa, A.K. Molecular cloning of a ripening-specific lipoxygenase and its expression during wild-type and mutant tomato fruit development. Plant Physiol. 1997, 113, 1041–1050. [Google Scholar] [CrossRef]
- Pirrung, M.C. Mechanism of a lipoxygenase model for ethylene biosynthesis. Biochemistry 1986, 25, 114–119. [Google Scholar] [CrossRef]
- Yu, M.; Shen, L.; Fan, B.; Zhao, D.; Zheng, Y.; Sheng, J. The effect of MeJA on ethylene biosynthesis and induced disease resistance to Botrytis cinerea in tomato. Postharvest Biol. Technol. 2009, 54, 153–158. [Google Scholar] [CrossRef]
- Hou, S.; Lin, L.; Lv, Y.; Xu, N.; Sun, X. Responses of lipoxygenase, jasmonic acid, and salicylic acid to temperature and exogenous phytohormone treatments in Gracilariopsis lemaneiformis (Rhodophyta). J. Appl. Phycol. 2018, 30, 3387–3394. [Google Scholar] [CrossRef]
- Fukuda, A.; Nakamura, Y.; Ohigashi, H.; Osawa, T.; Uchida, K. Cellular response to the redox active lipid peroxidation products: Induction of glutathione S-transferase P by 4-hydroxy-2-nonenal. Biochem. Biophys. Res. Commun. 1997, 236, 505–509. [Google Scholar] [CrossRef]
- Thakur, M.; Udayashankar, A.C. Lipoxygenases and their function in plant innate mechanism. In Bioactive Molecules in Plant Defense: Signaling in Growth and Stress; Jogaiah, S., Abdelrahman, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 133–143. [Google Scholar] [CrossRef]
- Peña-Cortés, H.; Fisahn, J.; Willmitzer, L. Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc. Natl. Acad. Sci. USA 1995, 92, 4106–4113. [Google Scholar] [CrossRef]
- Royo, J.; Vancanneyt, G.; Pérez, A.G.; Sanz, C.; Störmann, K.; Rosahl, S.; Sánchez-Serrano, J.J. Characterization of three potato lipoxygenases with distinct enzymatic activities and different organ-specific and wound-regulated expression patterns. J. Biol. Chem. 1996, 271, 21012–21019. [Google Scholar] [CrossRef]
- Roychoudhury, A.; Basu, S.; Sengupta, D.N. Effects of exogenous abscisic acid on some physiological responses in a popular aromatic indica rice compared with those from two traditional non-aromatic indica rice cultivars. Acta Physiol. Plant. 2009, 31, 915–926. [Google Scholar] [CrossRef]
- Deluc, L.G.; Quilici, D.R.; Decendit, A.; Grimplet, J.; Wheatley, M.D.; Schlauch, K.A.; Mérillon, J.-M.; Cushman, J.C.; Cramer, G.R. Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC Genom. 2009, 10, 212. [Google Scholar] [CrossRef]
- Turner, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14 (Suppl. 1), S153–S164. [Google Scholar] [CrossRef]
- Zhang, K.; An, Y.; Hu, Z.; Yang, D.; Sheng, Y. Relationship between lipoxygenase and ABA and JA in wounded signal transduction of heathy populus seedlings. For. Res. 2005, 18, 300–304. [Google Scholar]
- Fedina, E.O.; Karimova, F.G.; Chechetkin, I.R.; Tarchevsky, I.A.; Khripach, V.A. Contribution of lipoxygenase metabolism to the brassinosteroid signaling pathway. Dokl. Biochem. Biophys. 2004, 395, 80–83. [Google Scholar] [CrossRef]
- Bajguz, A.; Chmur, M.; Gruszka, D. Comprehensive overview of the brassinosteroid biosynthesis pathways: Substrates, products, inhibitors, and connections. Front. Plant Sci. 2020, 11, 1034. [Google Scholar] [CrossRef]
- Weber, H.; Chételat, A.; Caldelari, D.; Farmer, E.E. Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell 1999, 11, 485–494. [Google Scholar] [CrossRef]
- Han, C.; Yin, X.; He, D.; Yang, P. Analysis of proteome profile in germinating soybean seed, and its comparison with rice showing the styles of reserves mobilization in different crops. PLoS ONE 2013, 8, e56947. [Google Scholar] [CrossRef]
- Siedow, J. Plant lipoxygenase: Structure and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 145–188. [Google Scholar] [CrossRef]
- Huang, J.; Cai, M.; Long, Q.; Liu, L.; Lin, Q.; Jiang, L.; Chen, S.; Wan, J. OsLOX2, a rice type I lipoxygenase, confers opposite effects on seed germination and longevity. Transgenic Res. 2014, 23, 643–655. [Google Scholar] [CrossRef]
- Xu, H.; Wei, Y.; Zhu, Y.; Lian, L.; Xie, H.; Cai, Q.; Chen, Q.; Lin, Z.; Wang, Z.; Xie, H.; et al. Antisense suppression of LOX3 gene expression in rice endosperm enhances seed longevity. Plant Biotechnol. J. 2014, 13, 526–539. [Google Scholar] [CrossRef]
- Leenhardt, F.; Lyan, B.; Rock, E.; Boussard, A.; Potus, J.; Chanliaud, E.; Remesy, C. Genetic variability of carotenoid concentration, and lipoxygenase and peroxidase activities among cultivated wheat species and bread wheat varieties. Eur. J. Agron. 2006, 25, 170–176. [Google Scholar] [CrossRef]
- Clemente, A.; Olías, R.; Olías, J.M. Purification and characterization of broad bean lipoxygenase isoenzymes. J. Agric. Food Chem. 2000, 48, 1070–1075. [Google Scholar] [CrossRef]
- Sanz, L.C.; Perez, A.G.; Rios, J.J.; Olias, J.M. Positional specificity of ketodienes from linoleic acid aerobically formed by lipoxygenase isozymes from kidney bean and pea. J. Agric. Food Chem. 1993, 41, 696–699. [Google Scholar] [CrossRef]
- Burow, G.B.; Gardner, H.W.; Keller, N.P. A peanut seed lipoxygenase responsive to Aspergillus colonization. Plant Mol. Biol. 2000, 42, 689–701. [Google Scholar] [CrossRef]
- Jensen, A.B.; Poca, E.; Rigaud, M.; Freyssinet, G.; Pagès, M. Molecular characterization of L2 lipoxygenase from maize embryos. Plant Mol. Biol. 1997, 33, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Weichert, H.; Kolbe, A.; Kraus, A.; Wasternack, C.; Feussner, I. Metabolic profiling of oxylipins in germinating cucumber seedlings—Lipoxygenase-dependent degradation of triacylglycerols and biosynthesis of volatile aldehydes. Planta 2002, 215, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Oenel, A.; Fekete, A.; Krischke, M.; Faul, S.C.; Gresser, G.; Havaux, M.; Mueller, M.J.; Berger, S. Enzymatic and non-enzymatic mechanisms contribute to lipid oxidation during seed aging. Plant Cell Physiol. 2017, 58, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Porta, H.; Rocha-Sosa, M. Plant lipoxygenases. Physiological and molecular features. Plant Physiol. 2002, 130, 15–21. [Google Scholar] [CrossRef]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 2012, 63, 695–709. [Google Scholar] [CrossRef]
- Vardar, F.; ÜNal, M. Immunolocalization of lipoxygenase in the anther wall cells of Lathyrus undulatus Boiss. during programmed cell death. Not. Bot. Horti Agrobot. Cluj-Napoca 2011, 39, 71. [Google Scholar] [CrossRef][Green Version]
- Kolomiets, M.V.; Hannapel, D.J.; Chen, H.; Tymeson, M.; Gladon, R.J. Lipoxygenase is involved in the control of potato tuber development. Plant Cell 2001, 13, 613–626. [Google Scholar] [CrossRef]
- Perla, V.; Jayanty, S.S.; Holm, D.G.; Davidson, R.D. Relationship between tuber storage proteins and tuber powdery scab resistance in potato. Am. J. Potato Res. 2014, 91, 233–245. [Google Scholar] [CrossRef]
- Pokotylo, I.V. Lipoxygenases and plant cell metabolism regulation. Ukr. Biochem. J. 2015, 87, 41–55. [Google Scholar] [CrossRef][Green Version]
- Kaur, K.D.; Jha, A.; Sabikhi, L.; Singh, A.K. Significance of coarse cereals in health and nutrition: A review. J. Food Sci. Technol. 2014, 51, 1429–1441. [Google Scholar] [CrossRef]
- Park, Y.-S.; Kunze, S.; Ni, X.; Feussner, I.; Kolomiets, M.V. Comparative molecular and biochemical characterization of segmentally duplicated 9-lipoxygenase genes ZmLOX4 and ZmLOX5 of maize. Planta 2010, 231, 1425–1437. [Google Scholar] [CrossRef]
- Vellosillo, T.; Aguilera, V.; Marcos, R.; Bartsch, M.; Vicente, J.; Cascón, T.; Hamberg, M.; Castresana, C. Defense activated by 9-lipoxygenase-derived oxylipins requires specific mitochondrial proteins. Plant Physiol. 2013, 161, 617–627. [Google Scholar] [CrossRef]
- Christensen, S.A.; Huffaker, A.; Kaplan, F.; Sims, J.; Ziemann, S.; Doehlemann, G.; Ji, L.; Schmitz, R.J.; Kolomiets, M.V.; Alborn, H.T.; et al. Maize death acids, 9-lipoxygenase-derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc. Natl. Acad. Sci. USA 2015, 112, 11407–11412. [Google Scholar] [CrossRef]
- López, M.A.; Vicente, J.; Kulasekaran, S.; Vellosillo, T.; Martínez, M.; Irigoyen, M.L.; Cascón, T.; Bannenberg, G.; Hamberg, M.; Castresana, C. Antagonistic role of 9-lipoxygenase-derived oxylipins and ethylene in the control of oxidative stress, lipid peroxidation and plant defence. Plant J. 2011, 67, 447–458. [Google Scholar] [CrossRef]
- Hwang, I.S.; Hwang, B.K. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 2010, 152, 948–967. [Google Scholar] [CrossRef]
- Hou, Y.; Ban, Q.; Meng, K.; He, Y.; Han, S.; Jin, M.; Rao, J. Overexpression of persimmon 9-lipoxygenase DkLOX3 confers resistance to Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea in Arabidopsis. Plant Growth Regul. 2017, 84, 179–189. [Google Scholar] [CrossRef]
- Montillet, J.-L.; Leonhardt, N.; Mondy, S.; Tranchimand, S.; Rumeau, D.; Boudsocq, M.; Garcia, A.V.; Douki, T.; Bigeard, J.; Laurière, C.; et al. An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 2013, 11, e1001513. [Google Scholar] [CrossRef]
- Woldemariam, M.G.; Ahern, K.; Jander, G.; Tzin, V. A role for 9-lipoxygenases in maize defense against insect herbivory. Plant Signal. Behav. 2018, 13, e1422462. [Google Scholar] [CrossRef]
- Gao, X.; Stumpe, M.; Feussner, I.; Kolomiets, M. A novel plastidial lipoxygenase of maize (Zea mays) ZmLOX6 encodes for a fatty acid hydroperoxide lyase and is uniquely regulated by phytohormones and pathogen infection. Planta 2007, 227, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Ozalvo, R.; Cabrera, J.; Escobar, C.; Christensen, S.A.; Borrego, E.J.; Kolomiets, M.V.; Castresana, C.; Iberkleid, I.; Brown Horowitz, S. Two closely related members of Arabidopsis 13-lipoxygenases (13-LOXs), LOX3 and LOX4, reveal distinct functions in response to plant-parasitic nematode infection. Mol. Plant. Pathol. 2014, 15, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Cerna, H.; Černý, M.; Habánová, H.; Šafářová, D.; Abushamsiya, K.; Navrátil, M.; Brzobohatý, B. Proteomics offers insight to the mechanism behind Pisum sativum L. response to pea seed-borne mosaic virus (PSbMV). J. Proteom. 2017, 153, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Han, B. Differential expression pattern of an acidic 9/13-lipoxygenase in flower opening and senescence and in leaf response to phloem feeders in the tea plant. BMC Plant Biol. 2010, 10, 228. [Google Scholar] [CrossRef]
- Sujatha, B.; Devi, P.U.M. Antifungal potential of papaya lipoxygenase metabolites against Phytophthora palmivora. J. Pure Appl. Microbiol. 2012, 6, 433–438. [Google Scholar]
- Stolterfoht, H.; Rinnofner, C.; Winkler, M.; Pichler, H. Recombinant lipoxygenases and hydroperoxide lyases for the synthesis of green leaf volatiles. J. Agric. Food Chem. 2019, 67, 13367–13392. [Google Scholar] [CrossRef]
- Saraiva, T.; Morais, K.; Pereira, V.; Azevedo, M.; Rocha, C.; Prosperi, C.; Gomes-Santos, A.; Bermudez-Humaran, L.; Faria, A.; Blottiere, H.; et al. Milk fermented with a 15-lipoxygenase-1-producing Lactococcus lactis alleviates symptoms of colitis in a murine model. Curr. Pharm. Biotechnol. 2015, 16, 424–429. [Google Scholar] [CrossRef]
- Ben-Hayyim, G.; Gueta-Dahan, Y.; Avsian-Kretchmer, O.; Weichert, H.; Feussner, I. Preferential induction of a 9-lipoxygenase by salt in salt-tolerant cells of Citrus sinensis L. Osbeck. Planta 2001, 212, 367–375. [Google Scholar] [CrossRef]
- Yang, X.-Y.; Jiang, W.-J.; Yu, H.-J. The expression profiling of the lipoxygenase (LOX) family genes during fruit development, abiotic stress and hormonal treatments in cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2012, 13, 2481–2500. [Google Scholar] [CrossRef]
- Mao, L.-C.; Wang, G.-Z.; Zhu, C.-G.; Pang, H.-Q. Involvement of phospholipase D and lipoxygenase in response to chilling stress in postharvest cucumber fruits. Plant Sci. 2007, 172, 400–405. [Google Scholar] [CrossRef]
- Skaterna, T.D.; Kharchenko, O.V. Effect of phosphatidic acid on the reaction of linoleic acid oxidation by 5-lipooxygenase from potatoes. Ukr. Biochem. J. 2008, 80, 21–30. [Google Scholar]
- Kopich, V.N.; Kretynin, S.V.; Kharchenko, O.V.; Litvinovskaya, R.P.; Chashina, N.M.; Khripach, V.A. Effect of 24-epibrassinolide on lipoxygenase activity in maize seedlings under cold stress. Biopolym. Cell 2010, 26, 218–224. [Google Scholar] [CrossRef]
- Kosakivska, I.; Babenko, L.; Ustinova, A.; Skaterna, T. The influence of temperature conditions on lipoxygenase activity in seedling of rape Brassica napus var. Oleifera. Rep. Natl. Acad. Sci. Ukr. 2012, 6, 134–137. [Google Scholar]
- Kosakivska, I.; Babenko, L.; Skaterna, T.; Ustinova, A. Influence of hypo- and hyperthermia on lipoxygenase activity, content of pigments and soluble proteins in Triticum aestivum L. cv. Yatran 60 seedlings. Genet. Plant Physiol. 2014, 46, 212–220. [Google Scholar]
- Andreou, A.; Feussner, I. Lipoxygenases—Structure and reaction mechanism. Phytochemistry 2009, 70, 1504–1510. [Google Scholar] [CrossRef]
- Schwarz, K.; Walther, M.; Anton, M.; Gerth, C.; Feussner, I.; Kuhn, H. Structural basis for lipoxygenase specificity. J. Biol. Chem. 2001, 276, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Delporte, A.; Lannoo, N.; Vandenborre, G.; Ongenaert, M.; Van Damme, E.J.M. Jasmonate response of the Nicotiana tabacum agglutinin promoter in Arabidopsis thaliana. Plant Physiol. Biochem. 2011, 49, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, A.; Bajguz, A. Conjugates of abscisic acid, brassinosteroids, ethylene, gibberellins, and jasmonates. Phytochemistry 2011, 72, 2097–2112. [Google Scholar] [CrossRef] [PubMed]
- Floerl, S.; Majcherczyk, A.; Possienke, M.; Feussner, K.; Tappe, H.; Gatz, C.; Feussner, I.; Kües, U.; Polle, A. Verticillium longisporum infection affects the leaf apoplastic proteome, metabolome, and cell wall properties in Arabidopsis thaliana. PLoS ONE 2012, 7, e31435. [Google Scholar] [CrossRef] [PubMed]
- Quaglia, M.; Fabrizi, M.; Zazzerini, A.; Zadra, C. Role of pathogen-induced volatiles in the Nicotiana tabacum–Golovinomyces cichoracearum interaction. Plant Physiol. Biochem. 2012, 52, 9–20. [Google Scholar] [CrossRef]
- Copolovici, L.; Kännaste, A.; Pazouki, L.; Niinemets, Ü. Emissions of green leaf volatiles and terpenoids from Solanum lycopersicum are quantitatively related to the severity of cold and heat shock treatments. J. Plant Physiol. 2012, 169, 664–672. [Google Scholar] [CrossRef]
- Stumpe, M.; Göbel, C.; Demchenko, K.; Hoffmann, M.; Klösgen, R.B.; Pawlowski, K.; Feussner, I. Identification of an allene oxide synthase (CYP74C) that leads to formation of α-ketols from 9-hydroperoxides of linoleic and linolenic acid in below-ground organs of potato. Plant J. 2006, 47, 883–896. [Google Scholar] [CrossRef]
- Huang, F.-C.; Schwab, W. Cloning and characterization of a 9-lipoxygenase gene induced by pathogen attack from Nicotiana benthamiana for biotechnological application. BMC Biotechnol. 2011, 11, 30. [Google Scholar] [CrossRef]
- Liu, F.; Li, H.; Wu, J.; Wang, B.; Tian, N.; Liu, J.; Sun, X.; Wu, H.; Huang, Y.; Lü, P.; et al. Genome-wide identification and expression pattern analysis of lipoxygenase gene family in banana. Sci. Rep. 2021, 11, 9948. [Google Scholar] [CrossRef]
- Chauvin, A.; Caldelari, D.; Wolfender, J.L.; Farmer, E.E. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: A role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2012, 197, 566–575. [Google Scholar] [CrossRef]
- Yan, L.; Zhai, Q.; Wei, J.; Li, S.; Wang, B.; Huang, T.; Du, M.; Sun, J.; Kang, L.; Li, C.-B.; et al. Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 2013, 9, e1003964. [Google Scholar] [CrossRef]
- Zhang, B.; Yin, X.-r.; Li, X.; Yang, S.-l.; Ferguson, I.B.; Chen, K.-s. Lipoxygenase gene expression in ripening kiwifruit in relation to ethylene and aroma production. J. Agric. Food Chem. 2009, 57, 2875–2881. [Google Scholar] [CrossRef]
- Sarde, S.J.; Bouwmeester, K.; Venegas-Molina, J.; David, A.; Boland, W.; Dicke, M. Involvement of sweet pepper CaLOX2 in jasmonate-dependent induced defence against Western flower thrips. J. Integr. Plant Biol. 2019, 61, 1085–1098. [Google Scholar] [CrossRef]
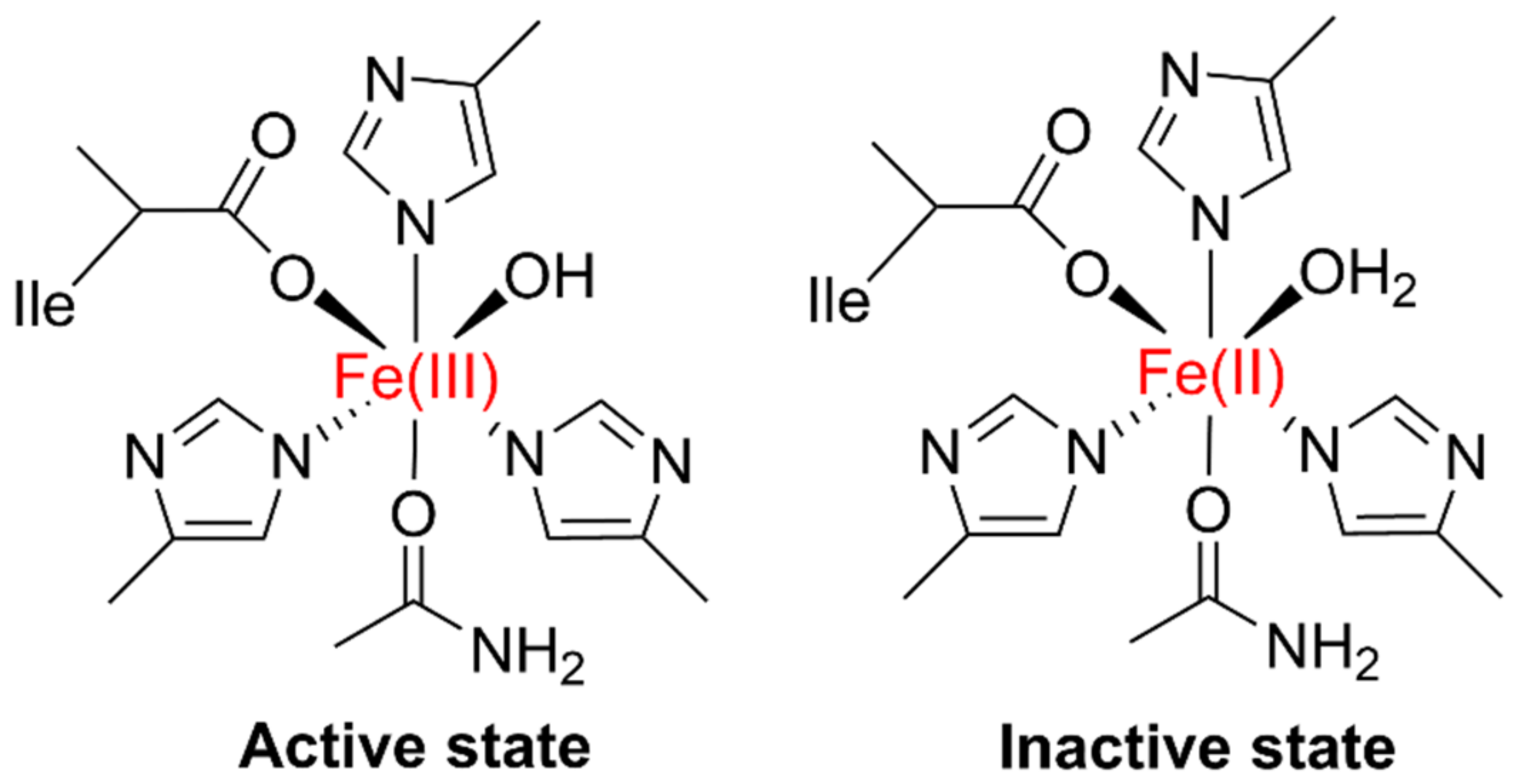
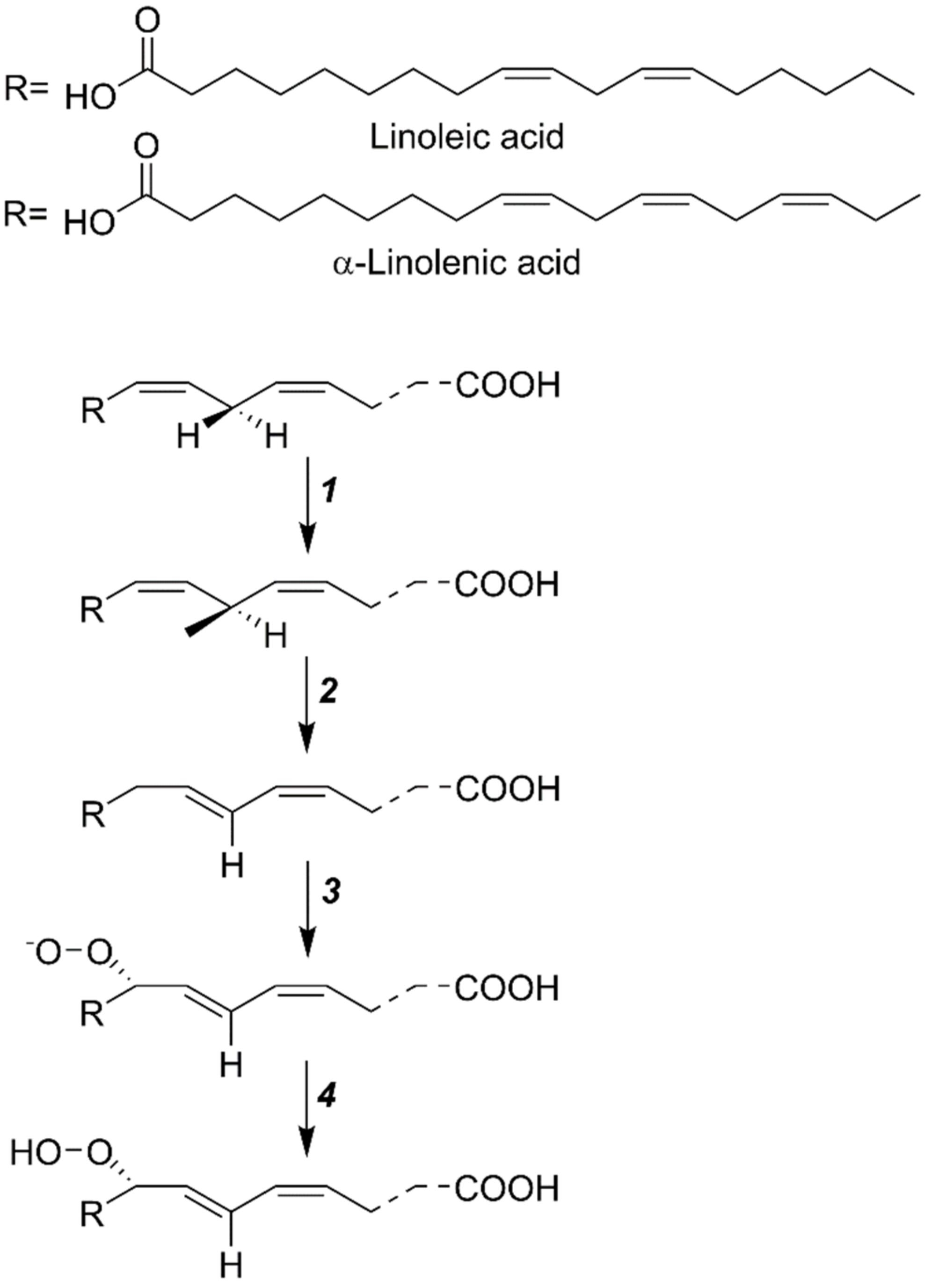
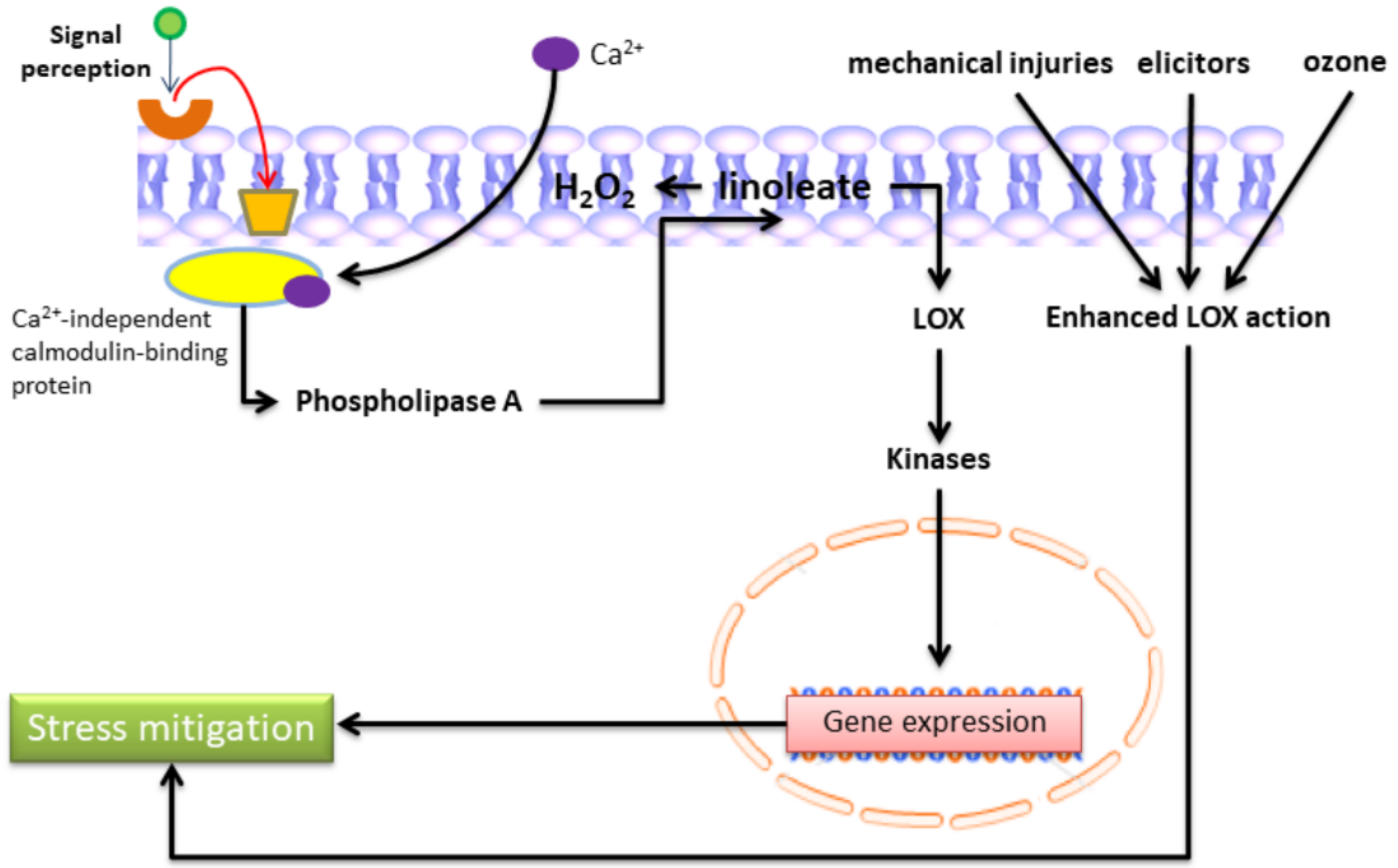
| Gene Locus | Gene Name | Gene Function | Total Amino Acids |
|---|---|---|---|
| AT1G55020 | lipoxygenase 1 (LOX1) | conferring resistance pathogens | 859 |
| AT3G45140 | lipoxygenase 2 (LOX2) | biosynthesis of JA | 896 |
| AT1G17420 | lipoxygenase 3 (LOX3) | flower development, catalyze the oxygenation of fatty acids | 919 |
| AT1G72520 | lipoxygenase, putative (LOX4) | flower development, 13-lipoxygenase induced by abiotic stresses, triggers defense responses | 926 |
| AT3G22400 | lipoxygenase 5 (LOX5) | defense response | 886 |
| AT1G67560 | lipoxygenase family protein (LOX6) | biosynthesis of JA, PLAT/LH2 domain-containing lipoxygenase family protein | 917 |
| Gene Locus | Gene Function | Total Amino Acids | Chromosome Number |
|---|---|---|---|
| LOC_Os11g36719 | lipoxygenase, putative, expressed | 869 | 11 |
| LOC_Os12g37260 | lipoxygenase 2.1, chloroplast precursor, putative, expressed | 923 | 12 |
| LOC_Os12g37320 | lipoxygenase 2.2, chloroplast precursor, putative, expressed | 359 | 12 |
| LOC_Os02g10120 | lipoxygenase, putative, expressed | 927 | 2 |
| LOC_Os02g19790 | lipoxygenase 4, putative, expressed | 297 | 2 |
| LOC_Os03g08220 | lipoxygenase protein, putative, expressed | 919 | 3 |
| LOC_Os03g49260 | lipoxygenase, putative, expressed | 868 | 3 |
| LOC_Os03g49380 | lipoxygenase, putative, expressed | 878 | 3 |
| LOC_Os03g52860 | lipoxygenase, putative, expressed | 871 | 3 |
| LOC_Os04g37430 | lipoxygenase protein, putative, expressed | 798 | 4 |
| LOC_Os05g23880 | lipoxygenase, putative, expressed | 848 | 5 |
| LOC_Os06g04420 | lipoxygenase 4, putative | 126 | 6 |
| LOC_Os08g39840 | lipoxygenase, chloroplast precursor, putative, expressed | 925 | 8 |
| LOC_Os08g39850 | lipoxygenase, chloroplast precursor, putative, expressed | 942 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, P.; Arif, Y.; Miszczuk, E.; Bajguz, A.; Hayat, S. Specific Roles of Lipoxygenases in Development and Responses to Stress in Plants. Plants 2022, 11, 979. https://doi.org/10.3390/plants11070979
Singh P, Arif Y, Miszczuk E, Bajguz A, Hayat S. Specific Roles of Lipoxygenases in Development and Responses to Stress in Plants. Plants. 2022; 11(7):979. https://doi.org/10.3390/plants11070979
Chicago/Turabian StyleSingh, Priyanka, Yamshi Arif, Edyta Miszczuk, Andrzej Bajguz, and Shamsul Hayat. 2022. "Specific Roles of Lipoxygenases in Development and Responses to Stress in Plants" Plants 11, no. 7: 979. https://doi.org/10.3390/plants11070979
APA StyleSingh, P., Arif, Y., Miszczuk, E., Bajguz, A., & Hayat, S. (2022). Specific Roles of Lipoxygenases in Development and Responses to Stress in Plants. Plants, 11(7), 979. https://doi.org/10.3390/plants11070979








