Transcription Profile of Auxin Related Genes during Positively Gravitropic Hypocotyl Curvature of Brassica rapa
Abstract
1. Introduction
2. Results
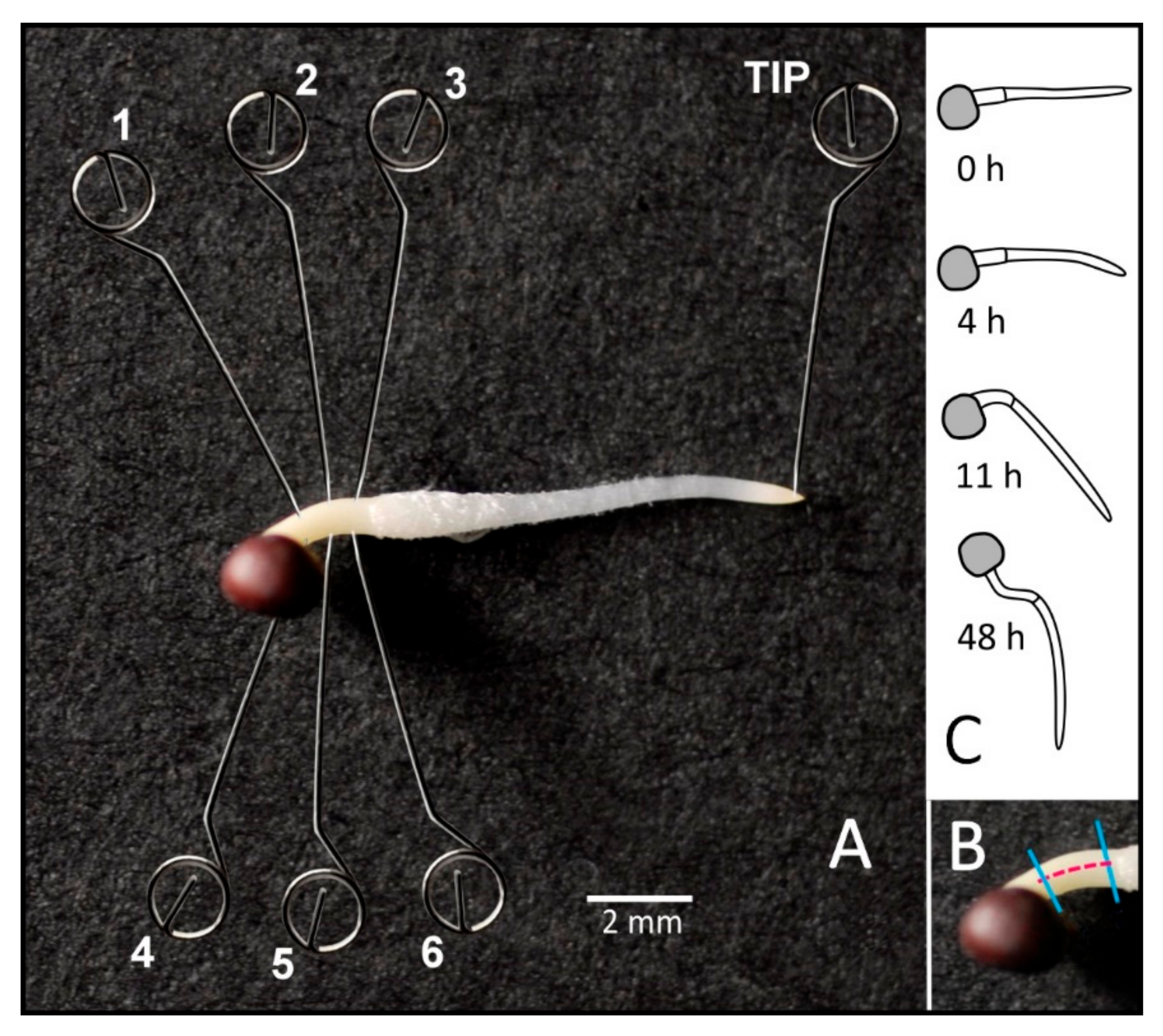
2.1. Time Course of Curvature
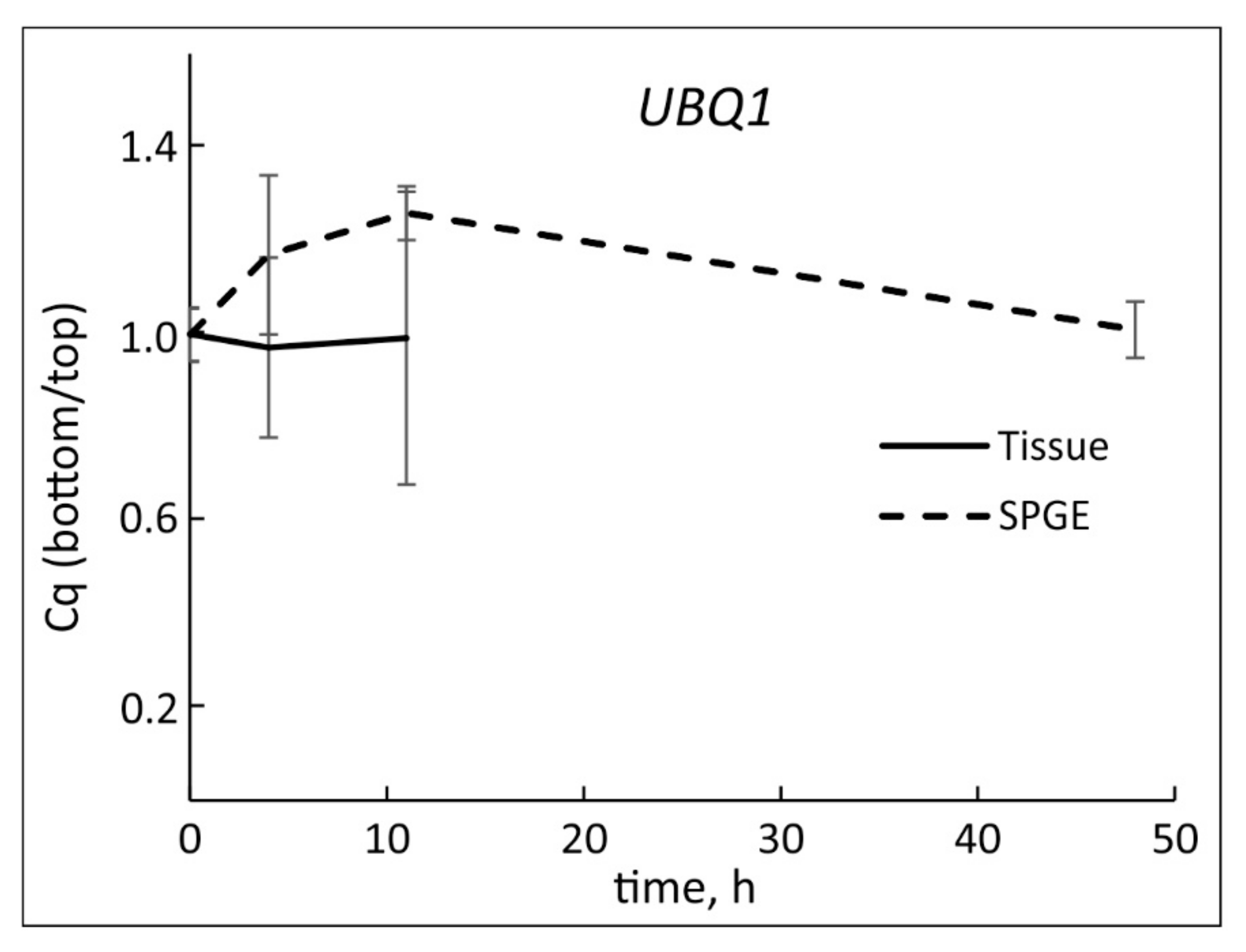
2.2. Nucleic Acid Extraction, Yield, and Reference
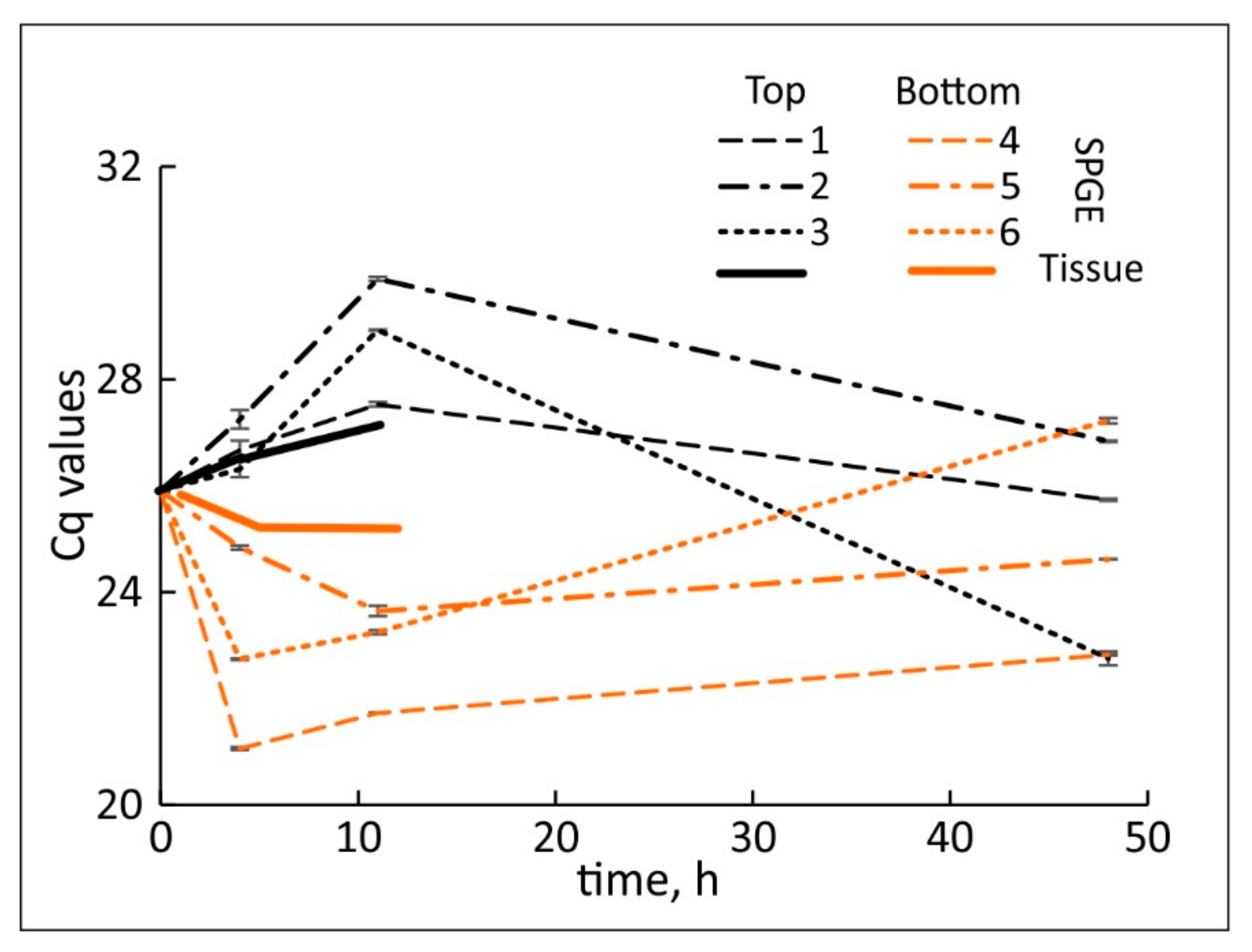
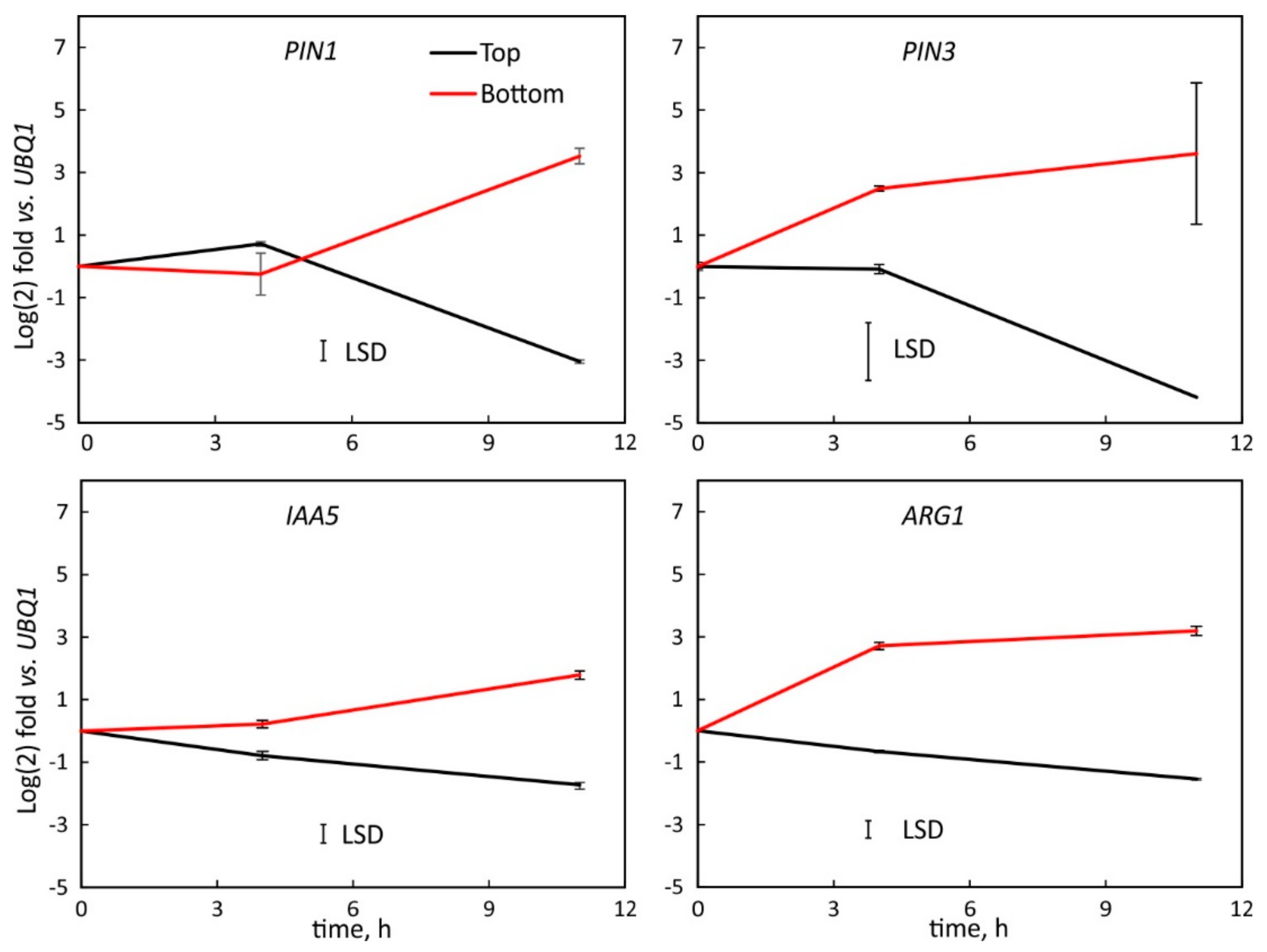
2.3. Transcription in the Hypocotyl Base Tissue
2.4. Tissue and SPGE Extraction
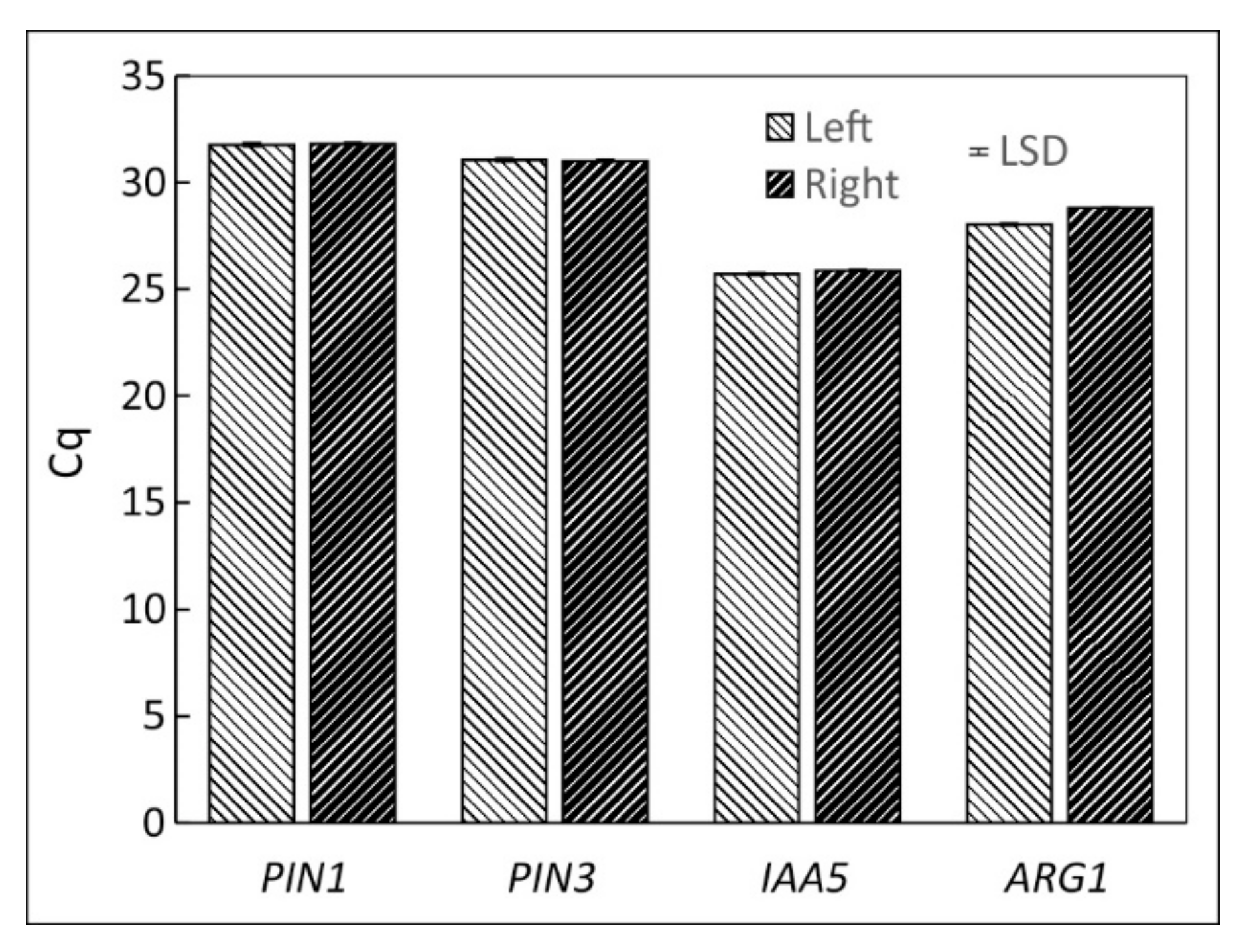
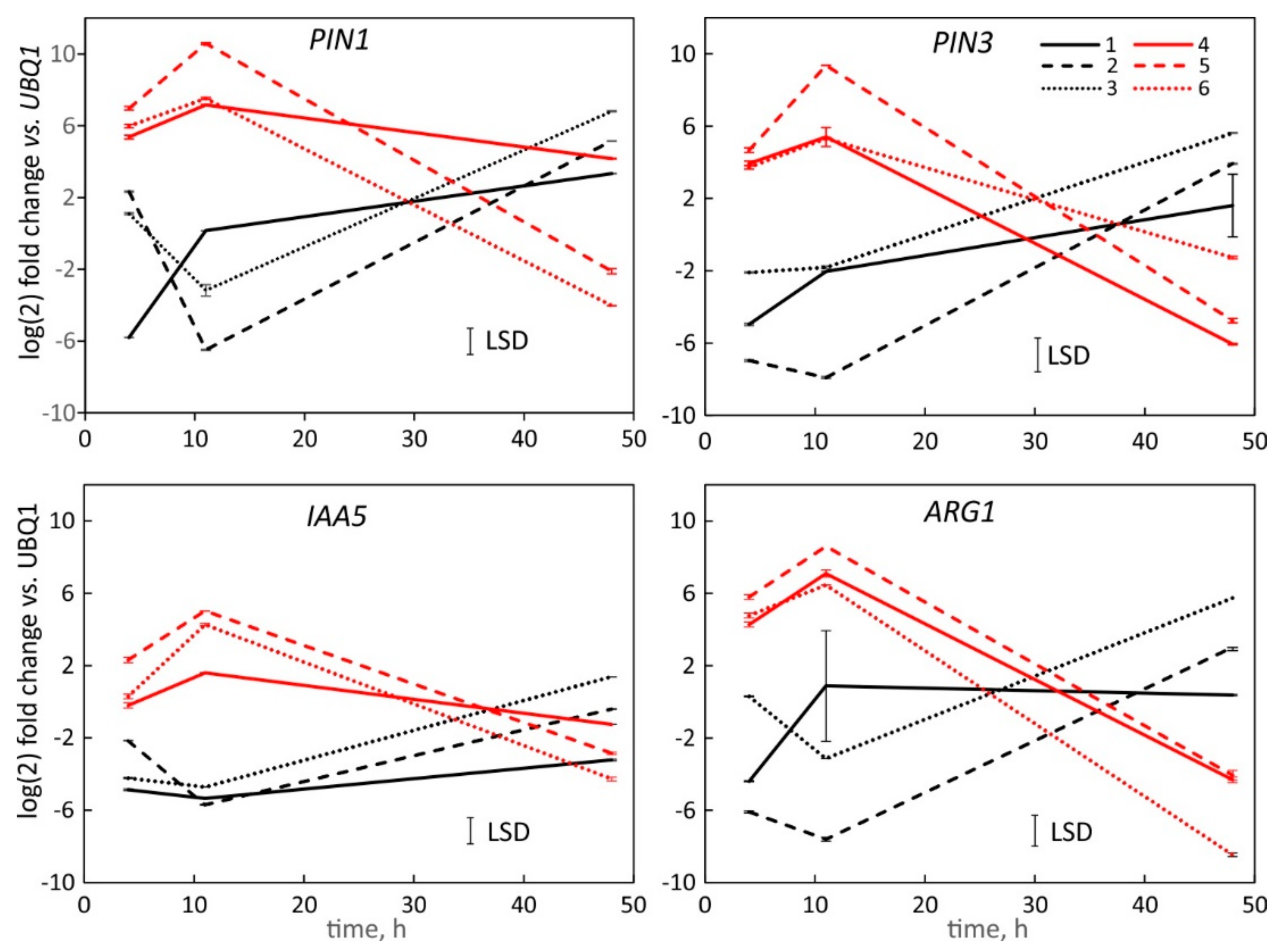
2.5. Comparison of the Hypocotyl and Root Tip
3. Discussion
3.1. UBQ1 as Reference
3.2. Transcription of Auxin Related Genes
3.3. Spatio-Temporal Changes during Hypocotyl Curvature
3.4. Comparison of Root Tip and Hypocotyl
3.5. Comparison of RNA Extraction Techniques
4. Materials and Methods
4.1. Plant Material
4.2. Gravistimulation and Tissue Sampling
4.3. RNA Extraction
4.4. SPGE Probe Preparation mRNA Extraction
4.5. Reverse Transcription
4.6. Transcript Quantification Using qPCR
4.7. Analyses
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ajala, C.; Hasenstein, K.H. Augmentation of root gravitropism by hypocotyl curvature in Brassica rapa seedlings. Plant Sci. 2019, 285, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Hasenstein, K.H.; Evans, M.L. Effects of cations on hormone transport in primary roots of Zea mays. Plant Physiol. 1988, 86, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Band, L.R.; Wells, D.M.; Larrieu, A.; Sun, J.Y.; Middleton, A.M.; French, A.P.; Brunoud, G.; Sato, E.M.; Wilson, M.H.; Peret, B.; et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 4668–4673. [Google Scholar] [CrossRef] [PubMed]
- Pernisova, M.; Prat, T.; Grones, P.; Harustiakova, D.; Matonohova, M.; Spichal, L.; Nodzynski, T.; Friml, J.; Hejatko, J. Cytokinins influence root gravitropism via differential regulation of auxin transporter expression and localization in Arabidopsis. New Phytol. 2016, 212, 497–509. [Google Scholar] [CrossRef]
- Lofke, C.; Zwiewka, M.; Heilmann, I.; Van Montagu, M.C.; Teichmann, T.; Friml, J. Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc. Natl. Acad. Sci. USA 2013, 110, 3627–3632. [Google Scholar] [CrossRef]
- Rood, S.; Kaufman, P.; Abe, H.; Pharis, R. Gibberellins and Gravitopism in maize shoots. Endogenous gibberellin-like substances and movementand metabolism of [3H]gibberellin A20. Plant Physiol. 1987, 83, 645–651. [Google Scholar] [CrossRef]
- Sukumar, P.; Edwards, K.S.; Rahman, A.; Delong, A.; Muday, G.K. PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol. 2009, 150, 722–735. [Google Scholar] [CrossRef]
- Hou, G.C.; Mohamalawari, D.R.; Blancaflor, E.B. Enhanced gravitropism of roots with a disrupted cap actin cytoskeleton. Plant Physiol. 2003, 131, 1360–1373. [Google Scholar] [CrossRef][Green Version]
- Blancaflor, E.B. The Cytoskeleton and Gravitropism in Higher Plants. J. Plant Growth Regul. 2002, 21, 120–136. [Google Scholar] [CrossRef]
- Roux, S.; Biro, R.; Hale, C. Calcium movements and the cellular basis of gravitropism. Cospar 1983, 3, 221–227. [Google Scholar] [CrossRef]
- Huang, L.K.; Liao, Y.Y.; Lin, W.H.; Lin, S.M.; Liu, T.Y.; Lee, C.H.; Pan, R.L. Potassium Stimulation of IAA Transport Mediated by the Arabidopsis Importer AUX1 Investigated in a Heterologous Yeast System. J. Membr. Biol. 2019, 252, 183–194. [Google Scholar] [CrossRef]
- Zhang, B.; Karnik, R.; Wang, Y.Z.; Wallmeroth, N.; Blatt, M.R.; Grefen, C. The Arabidopsis R-SNARE VAMP721 Interacts with KAT1 and KC1 K+ Channels to Moderate K+ Current at the Plasma Membrane. Plant Cell 2015, 27, 1697–1717. [Google Scholar] [CrossRef]
- Hasenstein, K.H.; Evans, M.L.; Stinemetz, C.L.; Moore, R.; Fondren, W.M.; Koon, E.C.; Higby, M.A.; Smucker, A.J.M. Comparative effectiveness of metal-ions in inducing curvature of primary roots of Zea mays. Plant Physiol. 1988, 86, 885–889. [Google Scholar] [CrossRef]
- Hornitschek, P.; Kohnen, M.V.; Lorrain, S.; Rougemont, J.; Ljung, K.; Lopez-Vidriero, I.; Franco-Zorrilla, J.M.; Solano, R.; Trevisan, M.; Pradervand, S.; et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012, 71, 699–711. [Google Scholar] [CrossRef]
- Fuchs, I.; Philippar, K.; Ljung, K.; Sandberg, G.; Hedrich, R. Blue light regulates an auxin-induced K+-channel gene in the maize coleoptile. Proc. Natl. Acad. Sci. USA 2003, 100, 11795–11800. [Google Scholar] [CrossRef]
- Yang, X.X.; Choi, H.W.; Li, N. A UV-light activated cinnamic acid isomer regulates plant growth and gravitropism via an ethylene receptor-independent pathway. Aust. J. Plant Physiol. 1999, 26, 325. [Google Scholar] [CrossRef]
- Vandenbrink, J.P.; Kiss, J.Z.; Herranz, R.; Medina, F.J. Light and gravity signals synergize in modulating plant development. Front. Plant Sci. 2014, 5, 563. [Google Scholar] [CrossRef]
- Sindelar, T.J.; Millar, K.D.L.; Kiss, J.Z. Red light effects on blue light-based phototropism in roots of Arabidopsis thaliana. Int. J. Plant Sci. 2014, 175, 731–740. [Google Scholar] [CrossRef]
- Zhang, N.G.; Hasenstein, K.H. Initiation and elongation of lateral roots in Lactuca sativa. Int. J. Plant Sci. 1999, 160, 511–519. [Google Scholar] [CrossRef]
- Rosquete, M.R.; von Wangenheim, D.; Marhavy, P.; Barbez, E.; Stelzer, E.H.K.; Benkova, E.; Maizel, A.; Kleine-Vehn, J. An Auxin Transport Mechanism Restricts Positive Orthogravitropism in Lateral Roots. Curr. Biol. 2013, 23, 817–822. [Google Scholar] [CrossRef]
- Leyser, O. Regulation of shoot branching by auxin. Trends Plant Sci. 2003, 8, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Friml, J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2010, 107, 12046–12051. [Google Scholar] [CrossRef]
- Ljung, K.; Hull, A.K.; Celenza, J.; Yamada, M.; Estelle, M.; Nonmanly, J.; Sandberg, G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 2005, 17, 1090–1104. [Google Scholar] [CrossRef]
- Bialek, K.; Michalczuk, L.; Cohen, J.D. Auxin biosynthesis during seed germination in Phaseolus vulgaris. Plant Physiol. 1992, 100, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Aloni, R.; Schwalm, K.; Langhans, M.; Ullrich, C.I. Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 2003, 216, 841–853. [Google Scholar] [CrossRef]
- Wisniewska, J.; Xu, J.; Seifertova, D.; Brewer, P.B.; Ruzicka, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN Localization Directs Auxin Flow in Plants. Science 2006, 312, 883. [Google Scholar] [CrossRef]
- Michniewicz, M.; Brewer, P.B.; Friml, J.I. Polar auxin transport and asymmetric auxin distribution. Arab. Book/Am. Soc. Plant Biol. 2007, 5, e0108. [Google Scholar] [CrossRef]
- Friml, J.; Palme, K. Polar Auxin Transport—Old Questions and New Concepts? Plant Mol. Biol. 2002, 49, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Ueda, J.; Komaki, M.; Bell, C.; Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Strohm, A.K.; Baldwin, K.L.; Masson, P.H. Multiple roles for membrane-associated protein trafficking and signaling in gravitropisnn. Front. Plant Sci. 2012, 3, 274. [Google Scholar] [CrossRef] [PubMed]
- Moore, I. Gravitropism: Lateral Thinking in Auxin Transport. Curr. Biol. 2002, 12, R452–R454. [Google Scholar] [CrossRef]
- Blancaflor, E.B.; Masson, P.H. Plant gravitropism. Unraveling the ups and downs of a complex process. Plant Physiol. 2003, 133, 1677–1690. [Google Scholar] [CrossRef]
- Friml, J.; Benkova, E.; Blilou, I.; Wisniewska, J.; Hamann, T.; Ljung, K.; Woody, S.; Sandberg, G.; Scheres, B.; Jurgens, G.; et al. Atpin4 Mediates Sink-Driven Auxin Gradients and Root Patterning in Arabidopsis. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef]
- Ottenschlager, I.; Wolff, P.; Wolverton, C.; Bhalerao, R.P.; Sandberg, G.; Ishikawa, H.; Evans, M.; Palme, K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 2003, 100, 2987–2991. [Google Scholar] [CrossRef]
- Rahman, A.; Takahashi, M.; Shibasaki, K.; Wu, S.; Inaba, T.; Tsurumi, S.; Baskin, T.I. Gravitropism of Arabidopsis thaliana Roots Requires the Polarization of PIN2 toward the Root Tip in Meristematic Cortical Cells. Plant Cell 2010, 22, 1762–1776. [Google Scholar] [CrossRef]
- Chen, R.; Rosen, E.; Masson, P.H. Gravitropism in Higher Plants. Plant Physiol. 1999, 120, 343. [Google Scholar] [CrossRef]
- Sack, F. The structure of the stem endodermis in etiolated pea seedlings. Can. J. Bot. 1987, 65, 1514–1519. [Google Scholar] [CrossRef]
- Sack, F.D. Plant gravity sensing. Int. Rev. Cytol. Surv. Cell Biol. 1991, 127, 193–252. [Google Scholar]
- Fukaki, H.; Wysocka-Diller, J.; Kato, T.; Fujisawa, H.; Benfey, P.N.; Tasaka, M. Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 1998, 14, 425–430. [Google Scholar] [CrossRef]
- Morita, M.T.; Tasaka, M. Gravity sensing and signaling. Curr. Opin. Plant Biol. 2004, 7, 712–718. [Google Scholar] [CrossRef]
- Tasaka, M.; Kato, T.; Fukaki, H. The endodermis and shoot gravitropism. Trends Plant Sci. 1999, 4, 103–107. [Google Scholar] [CrossRef]
- Friml, J.; Wisniewska, J.; Benkova, E.; Mendgen, K.; Palme, K. Lateral Relocation of Auxin Efflux Regulator Pin3 Mediates Tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef]
- Rakusova, H.; Gallego-Bartolome, J.; Vanstraelen, M.; Robert, H.S.; Alabadi, D.; Blazquez, M.A.; Benkova, E.; Friml, J. Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J. 2011, 67, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.R.; Masson, P.H. ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. Plant J. 2008, 53, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Sedbrook, J.C.; Chen, R.J.; Masson, P.H. Arg1 (Altered Response to Gravity) Encodes a Dnaj-Like Protein That Potentially Interacts With the Cytoskeleton. Proc. Natl. Acad. Sci. USA 1999, 96, 1140–1145. [Google Scholar] [CrossRef]
- Boonsirichai, K.; Sedbrook, J.C.; Chen, R.; Gilroy, S.; Masson, P.H. ALTERED RESPONSE TO GRAVITY is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes. Plant Cell 2003, 15, 2612–2625. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, J.; Bandyopadhyay, A.; Peer, W.; Makam, S.; Murphy, A. Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 2004, 134, 28–31. [Google Scholar] [CrossRef]
- Noh, B.; Bandyopadhyay, A.; Peer, W.A.; Spalding, E.P.; Murphy, A.S. Enhanced Gravi- and Phototropism in Plant Mdr Mutants Mislocalizing the Auxin Efflux Protein Pin1. Nature 2003, 423, 999–1002. [Google Scholar] [CrossRef]
- Gälweiler, L.; Guan, C.; Muller, A.; Wisman, E.; Mendgen, K.; Yephremov, A.; Palme, K. Regulation of Polar Auxin Transport by AtPIN1 in Arabidopsis Vascular Tissue. Science 1998, 282, 2226–2230. [Google Scholar] [CrossRef]
- Abel, S.; Nguyen, M.D.; Chow, W.; Theologis, A. ASC4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. J. Biol. Chem. 1995, 270, 19093–19099. [Google Scholar] [CrossRef]
- Nakamura, A.; Higuchi, K.; Goda, H.; Fujiwara, M.T.; Sawa, S.; Koshiba, T.; Shimada, Y.; Yoshida, S. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 2003, 133, 1843–1853. [Google Scholar] [CrossRef]
- Abel, S.; Theologis, A. Early Genes and Auxin Action. Plant Physiol. 1996, 111, 9–17. [Google Scholar] [CrossRef]
- Park, M.R.; Wang, Y.-H.; Hasenstein, K.H. Profiling Gene Expression in Germinating Brassica Roots. Plant Mol. Biol. Report. 2014, 32, 541–548. [Google Scholar] [CrossRef][Green Version]
- Khanlou, K.M.; Van Bockstaele, E. A critique of widely used normalization software tools and an alternative method to identify reliable reference genes in red clover (Trifolium pratense L.). Planta 2012, 236, 1381–1393. [Google Scholar] [CrossRef]
- Migocka, M.; Papierniak, A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 2011, 28, 343–357. [Google Scholar] [CrossRef]
- Xiao, D.; Zhang, N.-W.; Zhao, J.-J.; Bonnema, G.; Hou, X.-L. Validation of reference genes for real-time quantitative PCR normalisation in non-heading Chinese cabbage. Funct. Plant Biol. 2012, 39, 342–350. [Google Scholar] [CrossRef]
- Hobbie, L.J. Auxin: Molecular Genetic Approaches in Arabidopsis. Plant Physiol. Biochem. 1998, 36, 91–102. [Google Scholar] [CrossRef]
- Oono, Y.; Chen, Q.G.; Theologis, A. age Mutants of Arabidopsis Exhibit Altered Auxin-Regulated Gene Expression. Plant Cell 1998, 10, 1649–1662. [Google Scholar] [CrossRef]
- Park, M.R.; Hasenstein, K.H. Hormone-Induced Gene Expression During Gravicurvature of Brassica Roots. J. Plant Growth Regul. 2016, 35, 190–201. [Google Scholar] [CrossRef]
- Barcelo, P.; Lazzeri, P.; Martin, A.; Lorz, H. Competence of cereal leaf cells. 2. Influence of auxin, ammonium and explant age on regeneration. J. Plant Physiol. 1992, 139, 448–454. [Google Scholar] [CrossRef]
- Scherp, P.; Hasenstein, K.H. Solid phase gene extraction isolates mRNA at high spatial and temporal resolution. BioTechniques 2008, 45, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Liu, J.; Huang, S.M.; Guo, T.T.; Deng, L.B.; Hua, W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 2014, 538, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Fendrych, M.; Akhmanova, M.; Merrin, J.; Glanc, M.; Hagihara, S.; Takahashi, K.; Uchida, N.; Torii, K.U.; Friml, J. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat. Plants 2018, 4, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Rakusova, H.; Abbas, M.; Han, H.B.; Song, S.Y.; Robert, H.S.; Friml, J. Termination of Shoot Gravitropic Responses by Auxin Feedback on PIN3 Polarity. Curr. Biol. 2016, 26, 3026–3032. [Google Scholar] [CrossRef] [PubMed]
- Rakusova, H.; Han, H.B.; Valosek, P.; Friml, J. Genetic screen for factors mediating PIN polarization in gravistimulated Arabidopsis thaliana hypocotyls. Plant J. 2019, 98, 1048–1059. [Google Scholar] [CrossRef]
- Van den Berg, T.; Tusscher, K.H. Auxin Information Processing; Partners and Interactions beyond the Usual Suspects. Int. J. Mol. Sci. 2017, 18, 2585. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, T.; Palme, K.; Paponov, I.A. Glutathione Enhances Auxin Sensitivity in Arabidopsis Roots. Biomolecules 2020, 10, 1550. [Google Scholar] [CrossRef]
- Taniguchi, M.; Nakamura, M.; Tasaka, M.; Morita, M.T. Identification of gravitropic response indicator genes in Arabidopsis inflorescence stems. Plant Signal Behav. 2014, 17, e29570. [Google Scholar] [CrossRef]
- Kubes, M.; Yang, H.; Richter, G.L.; Cheng, Y.; Mlodzinska, E.; Wang, X.; Blakeslee, J.J.; Carraro, N.; Petrasek, J.; Zazimalova, E.; et al. The Arabidopsis concentration-dependent influx/efflux transporter ABCB4 regulates cellular auxin levels in the root epidermis. Plant J. 2012, 69, 640–654. [Google Scholar] [CrossRef]
- Tanimoto, E. Tall or short? Slender or thick? A plant strategy for regulating elongation growth of roots by low concentrations of gibberellin. Ann. Bot. 2012, 110, 373–381. [Google Scholar] [CrossRef]
- Zhao, Y.; Hasenstein, K.H. Primary Root Growth Regulation: The Role of Auxin and Ethylene Antagonists. J. Plant Growth Regul. 2009, 28, 309–320. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Kerr, M.K.; Churchill, G.A. Experimental design for gene expression microarrays. Biostatistics 2001, 2, 183–201. [Google Scholar] [CrossRef]


| Gene (Accession #) | Sequence (5′–3′) | Tm (°C) | Amplicon (bp) | Efficiency (%) * |
|---|---|---|---|---|
| UBQ1 (Z24738.1) | F: GGAGAGCAGTGACACCATCGA | 58 | 120 | 103.1 |
| R: GCCAAGGTACGACCATCTTCA | ||||
| PIN1 (AJ132363.1) | F: ATCTTCACACCGACGGTAAGTC | 54 | 155 | 111.5 |
| R: GTCAGATCTTCCACCCTGTTCA | ||||
| PIN3 (AJ249298.1) | F: TCTTAACGTTTCCGATGGAGCC | 58 | 167 | 103.2 |
| R: CTCTTCCAGCGAAACTAAACCG | ||||
| IAA5 (NM101427.4) | F: GCATGGATGGAGCTCCTTATCT | 58 | 171 | 96.5 |
| R: GCATCCAATCTCCATCTTTGTC | ||||
| ARG1 (AT1G68370.1) | F: TTTGGCGCGGTTTCAAGAAG | 54 | 120 | 95.3 |
| R: CCGCTTGGTGTCTTCACAAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ajala, C.; Hasenstein, K.H. Transcription Profile of Auxin Related Genes during Positively Gravitropic Hypocotyl Curvature of Brassica rapa. Plants 2022, 11, 1191. https://doi.org/10.3390/plants11091191
Ajala C, Hasenstein KH. Transcription Profile of Auxin Related Genes during Positively Gravitropic Hypocotyl Curvature of Brassica rapa. Plants. 2022; 11(9):1191. https://doi.org/10.3390/plants11091191
Chicago/Turabian StyleAjala, Chitra, and Karl H. Hasenstein. 2022. "Transcription Profile of Auxin Related Genes during Positively Gravitropic Hypocotyl Curvature of Brassica rapa" Plants 11, no. 9: 1191. https://doi.org/10.3390/plants11091191
APA StyleAjala, C., & Hasenstein, K. H. (2022). Transcription Profile of Auxin Related Genes during Positively Gravitropic Hypocotyl Curvature of Brassica rapa. Plants, 11(9), 1191. https://doi.org/10.3390/plants11091191







