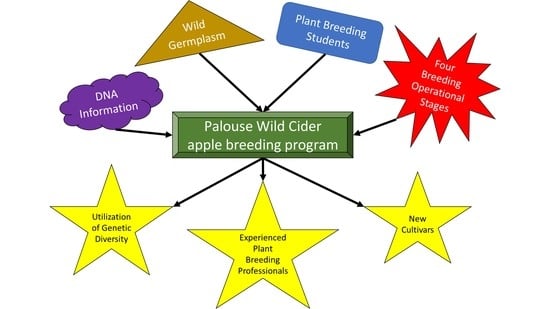
As It Stands: The Palouse Wild Cider Apple Breeding Program
Abstract
:1. Introduction
2. Results
2.1. Goal-Setting
2.2. New Genetic Variation
2.3. Selection
2.4. Commercialization
2.5. Student Outcomes
3. Discussion
4. Materials and Methods
4.1. Goal-Setting
4.2. New Genetic Variation
4.3. Selection
4.4. Commercialization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Duyvelshoff, C. Nova Scotia’s 2014 Fire Blight Epidemic Impacts & Outcome. CHC Apple Working Group. 2015. Available online: https://www.hortcouncil.ca/wp-content/uploads/2016/01/4-Nova-Scotias-2014-Fire-Blight-Epidemic.pdf (accessed on 20 June 2021).
- Baenziger, P.S.; Mumm, R.H.; Bernardo, R.; Brummer, E.C.; Langridge, P.; Simon, P.; Smith, S. Plant breeding and genetics: A paper in the series on The Need for Agricultural Innovation to Sustainably Feed the World by 2050. Issue Paper Council Agricult. Sci. Technol. 2017, 57, 24. [Google Scholar]
- Lee, E.A.; Dudley, J.W. Plant breeding education. In Plant Breeding: The Arnel R. Hallauer International Symposium; Lamkey, K.R., Hallauer, A.R., Eds.; Blackwell Publ.: Oxford, UK, 2006; pp. 120–126. [Google Scholar]
- Morris, M.; Edmeades, G.; Pehu, E. The global need for plant breeding capacity: What roles for the public and private sectors? HortScience 2006, 41, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Repinski, S.; Hayes, K.; Miller, J.; Trexler, C.; Bliss, F. Plant breeding graduate education: Opinions about critical knowledge, experience, and skill requirements from public and private stakeholders worldwide. Crop. Sci. 2011, 51, 2325–2336. [Google Scholar] [CrossRef] [Green Version]
- Bliss, F.A. Education and preparation of plant breeders for careers in global crop improvement. Crop. Sci. 2007, 47, S250–S261. [Google Scholar] [CrossRef]
- Waliczek, T.M.; Zajicek, J.M. School gardening: Improving environmental attitudes of children through hands-on learning. J. Environ. Hortic. 1999, 17, 180–184. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Feugey, L.; Poncet, C.; Lateur, M.; Houben, P.; Ordidge, M.; et al. Analysis of the genetic diversity and structure across a wide range of germplasm reveals prominent gene flow in apple at the European level. BMC Plant Biol. 2016, 16, 130. [Google Scholar] [CrossRef]
- Zhang, H.; Mittal, N.; Leamy, L.; Barazani, O.; Song, B. Back into the wild—Apply untapped genetic diversity of wild relatives for crop improvement. Evol. Appl. 2017, 10, 5–24. [Google Scholar] [CrossRef]
- Baumgartner, I.O.; Patocchi, A.; Lussi, L.; Kellerhals, M.; Peil, A. Accelerated introgression of fire blight resistance from Malus ×robusta 5 and other wild germplasm into elite apples. Acta Hortic. 2014, 1056, 281–287. [Google Scholar] [CrossRef]
- Luo, F.; Norelli, J.L.; Howard, N.P.; Wisniewski, M.; Flachowsky, H.; Hanke, M.-V.; Peace, C. Introgressing blue mold resistance into elite apple germplasm by rapid cycle breeding and foreground and background DNA-informed selection. Tree Genet. Genomes 2020, 16, 28. [Google Scholar] [CrossRef]
- Migicovsky, Z.; Myles, S. Exploiting wild relatives for genomics-assisted breeding of perennial crops. Front. Plant Sci. 2017, 8, 460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, N.P.; van de Weg, E.; Bedford, D.S.; Peace, C.P.; Vanderzande, S.; Clark, M.D.; Teh, S.L.; Cai, L.; Luby, J.J. Elucidation of the ‘Honeycrisp’ pedigree through haplotype analysis with a multi-family integrated SNP linkage map and a large apple (Malus × domestica) pedigree-connected SNP data set. Hortic. Res. 2017, 4, 17003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderzande, S.; Micheletti, D.; Troggio, M.; Davey, M.W.; Keulemans, J. Genetic diversity, population structure, and linkage disequilibrium of elite and local apple accessions from Belgium using the IRSC array. Tree Genet. Genomes 2017, 13, 125. [Google Scholar] [CrossRef]
- Cuevas, H.E.; Rosa-Valentin, G.; Hayes, C.M.; Rooney, W.L.; Hoffmann, L. Genomic characterization of a core set of the USDA-NPGS Ethiopian sorghum germplasm collection: Implications for germplasm conservation, evaluation, and utilization in crop improvement. BMC Genom. 2017, 18, 108. [Google Scholar] [CrossRef] [Green Version]
- Evans, K. Apple breeding in the pacific northwest. Acta Hortic. 2013, 976, 75–78. [Google Scholar] [CrossRef]
- Ru, S.; Hardner, C.; Evans, K.; Main, D.; Carter, P.A.; Harshman, J.; Sandefur, P.; Edge-Garza, D.; Peace, C. Empirical evaluation of multi-trait DNA testing in an apple seedling population. Tree Genet. Genomes 2021, 17, 13. [Google Scholar] [CrossRef]
- Peace, C. DNA-informed breeding of rosaceous crops: Promises, progress and prospects. Hortic. Res. 2017, 4, 17006. [Google Scholar] [CrossRef] [Green Version]
- Iezzoni, A.; McFerson, J.; Luby, J.; Gasic, K.; Whitaker, V.; Bassil, N.; Yue, C.; Gallardo, K.; McCracken, V.; Coe, M.; et al. RosBREED: Bridging the chasm between discovery and application to enable DNA-informed breeding in rosaceous crops. Hortic. Res. 2020, 7, 177. [Google Scholar] [CrossRef]
- Evans, K.; Peace, C. Advances in marker-assisted breeding of apples. In Achieving Sustainable Cultivation of Apples, 2nd ed.; Evans, K., Ed.; Burleigh Dodds Science Publishing: London, UK, 2017; pp. 165–194. [Google Scholar]
- National Research Council. Colleges of Agriculture at the Land Grant Universities: A Profile; National Academies Press: Washington, DC, USA, 1995. [Google Scholar]
- GRIN-Global. Available online: https://npgsweb.ars-grin.gov/gringlobal/search (accessed on 5 June 2021).
- Volk, G.; Richards, C.M.; Reilley, A.A.; Henk, A.D. Ex Situ conservation of vegetatively propagated species: Development of a seed-based core collection for Malus sieversii. J. Am. Soc. Hortic. Sci. 2005, 130, 203–210. [Google Scholar] [CrossRef] [Green Version]
- Fahey, J. Irrigation, apples, and the Spokane country. Pac. Northwest Q. 1993, 84, 7–18. [Google Scholar]
- Washington Grain Commission. Available online: https://wagrains.org/wheat/ (accessed on 6 June 2021).
- Miles, C.A.; Alexander, T.R.; Peck, G.; Galinato, S.P.; Gottschalk, C.; van Nocker, S. Growing apples for hard cider production in the United States—trends and research opportunities. HortTechnology 2020, 30, 148–155. [Google Scholar] [CrossRef]
- Degenhard, J. Alcoholic Drinks Report 2019—Cider, Perry & Rice Wine; Report 48819; Statista: Hamburg, Germany, 2019. [Google Scholar]
- Pashow, L.; Mahr, M.; Hard Cider Supply Chain Analysis. Cornell Cooperative Extension. 2018. Available online: https://harvestny.cce.cornell.edu/uploads/doc_48.pdf (accessed on 14 November 2021).
- Leforestier, D.; Ravon, E.; Muranty, H.; Cornille, A.; Lemaire, C.; Giraud, T.; Durel, C.-E.; Branca, A. Genomic basis of the differences between cider and dessert apple varieties. Evol. Appl. 2015, 8, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Liao, L.; Zheng, H.; Chen, J.; Wu, B.; Ogutu, C.; Li, S.; Korban, S.S.; Han, Y. Genes encoding aluminum-activated malate transporter II and their association with fruit acidity in apple. Plant Genome 2015, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradshaw, T.L.; Kingsley-Richards, S.L.; Foster, J. Apple cultivar evaluations for cider making in Vermont, USA. Acta Hortic. 2016, 1205, 453–460. [Google Scholar] [CrossRef]
- Gottschalk, C.; Rothwell, N.; van Nocker, S. Apple Cultivars for Production of Hard Cider in Michigan; Michigan State University Extension Bulletin E3364; Michigan State University: East Lansing, MI, USA, 2017. [Google Scholar]
- James, T.; Luo, F.; van Nocker, S.; Peace, C. A proposed natural genetic solution to the long juvenility problem in apple. J. Am. Pom. Soc. 2020, 74, 164–168. [Google Scholar]
- Zhu, Y.; Barritt, B.H. Md-ACS1 and Md-ACO1 genotyping of apple (Malus × domestica Borkh.) breeding parents and suitability for marker-assisted selection. Tree Genet. Genomes 2008, 4, 555–562. [Google Scholar] [CrossRef]
- Longhi, S.; Cappellin, L.; Guerra, W.; Costa, F. Validation of a functional molecular marker suitable for marker-assisted breeding for fruit texture in apple (Malus × domestica Borkh.). Mol. Breed. 2013, 32, 841–852. [Google Scholar] [CrossRef]
- Chagné, D.; Vanderzande, S.; Kirk, C.; Profitt, N.; Weskett, R.; Gardiner, S.E.; Peace, C.P.; Volz, R.K.; Bassil, N.V. Validation of SNP markers for fruit quality and disease resistance loci in apple (Malus × domestica Borkh.) using the OpenArray® platform. Hortic. Res. 2019, 6, 30. [Google Scholar] [CrossRef] [Green Version]
- Schaller, A. Method of Deducing Genotypes Using Curated and Uncurated SNP Data. Master’s Thesis, Washington State University, Pullman, WA, USA, 2019. [Google Scholar]
- Kwon, Y.S.; Kwon, S.; Kim, J.-H.; Park, M.Y.; Park, J.T.; Lee, J. ‘Arisoo’, a midseason apple. HortScience 2021, 56, 1139–1141. [Google Scholar] [CrossRef]
- Musacchi, S.; Hanrahan, I.; Lewis, K.; Evans, K.; DuPont, T. WA 38 Characteristics and Horticulture. 2017. Available online: https://s3-us-west-2.amazonaws.com/treefruit.wsu.edu/wp-content/uploads/2017/09/WA38_factsheet_2017.09.pdf (accessed on 20 December 2021).
- Edge-Garza, D.A.; Rowland, T.V.; Haendiges, S.; Peace, C.P. A high-throughput and cost-efficient DNA extraction protocol for the tree fruit crops of apple, sweet cherry, and peach relying on silica beads during tissue sampling. Mol. Breed. 2014, 34, 2225–2228. [Google Scholar] [CrossRef]
- Chagné, D.; Krieger, C.; Rassam, M.; Sullivan, M.; Fraser, J.; André, C.; Pindo, M.; Troggio, M.; Gardiner, S.E.; Henry, R.A.; et al. QTL and candidate gene mapping for polyphenolic composition in apple fruit. BMC Plant Biol. 2012, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Chibon, P.-Y.; de Vos, R.C.H.; Schipper, B.A.; Walraven, E.; Beekwilder, J.; van Dijk, T.; Finkers, R.; Visser, R.G.F.; van de Weg, E.W.; et al. Genetic analysis of metabolites in apple fruits indicates an mQTL hotspot for phenolic compounds on linkage group 16. J. Exp. Bot. 2012, 63, 2895–2908. [Google Scholar] [CrossRef] [PubMed]
- LGC Genomics. Available online: https://biosearch-cdn.azureedge.net/assetsv6/sbeadex-mini-plant.pdf (accessed on 14 December 2021).
- Bianco, L.; Cestaro, A.; Sargent, D.J.; Banchi, E.; Derdak, S.; di Guardo, M.; Salvi, S.; Jansen, J.; Viola, R.; Gut, I.; et al. Development and validation of a 20K single nucleotide polymorphism (SNP) whole genome genotyping array for apple (Malus × domestica Borkh). PLoS ONE 2014, 9, e110377. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Trait | 2014 Goal | 2019 Goal | Trait Level | Essential or Enhancing |
|---|---|---|---|---|
| Flesh color | X | X | Heterozygous for red flesh | Enhancing 1× |
| X | X | Homozygous for red flesh | Enhancing 2× | |
| Fruit size | X | >40 mm (i.e., at least golf ball size) | Essential | |
| X | >60 mm (i.e., at least tennis ball size) | Enhancing | ||
| Phenolic compound amount | X | X | 1.0–1.5 on a 0–2 sensory evaluation scale (bitter after blend) | Essential |
| X | X | >1.5 on a 0–2 sensory evaluation scale (extremely bitter but still blends well) | Enhancing | |
| Juiciness | X | 200 mL juice per lb fresh fruit | Essential | |
| X | >275 mL juice per lb fresh fruit | Enhancing | ||
| Seediness | X | Not seedy | Enhancing | |
| Nutritional components | X | Nutritional boost | Enhancing | |
| Juvenility duration | X | X | Fruit in fifth leaf | Essential |
| X | X | Fruit in fourth leaf | Enhancing 1× | |
| X | X | Fruit in third leaf | Enhancing 2× | |
| Fire blight resistance | X | X | Not susceptible | Essential |
| X | X | Highly tolerant | Enhancing 1× | |
| X | X | Fully resistant | Enhancing 2× | |
| Powdery mildew resistance | X | X | Not susceptible | Essential |
| Cedar apple rust resistance | X | Not susceptible | Essential | |
| X | Highly tolerant | Enhancing 1× | ||
| X | Fully resistant | Enhancing 2× | ||
| Fruit-bearing habit | X | X | Not tip bearing | Essential |
| X | Clearly spur-bearing type | Enhancing | ||
| Annual bearing tendency | X | X | Not biennial bearing | Essential |
| Leaves and bark | X | X | Novel shape, color, texture, etc. | Enhancing |
| Branch angle | X | X | >70° | Enhancing 1× |
| X | X | 90° | Enhancing 2× | |
| Yield | X | X | High yielding | Enhancing |
| Year of Crossing | No. Flowers Pollinated | No. M. sieversii Parents | No. Other Wild Parents Used | No. Hybrid Wild × Non-Wild Parents Used | No. Non-Wild Parents Used | No. of Families Attempted/ Successful | No. Attempted Families with Wild Background | Largest Family Created (no. Seeds) | No. Seeds Created * | No. Seeds Greenhouse Planted and Raised | No. Seedlings Planted in Field | Flowering and Fruiting Outcomes of Crosses |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2013 | OP | 7 | 5 | 4 | 5 | 30/30 | 24 | ‘Kerr Crab’ OP (337) | 1700 | 800 | 350 | Flowering and fruiting since 2016: “Big Blush”, “Yellow Tiny”, “Red-Yellow Tiny”, “Orange Tiny”, “Stripey” |
| 2014 | 574 | 2 | 6 | 1 | 19 | 69/18 | 66 | ‘Kerr Crab’ × PRI E14-32 (157) | 1028 | 865 | 530 | Flowering and fruiting since 2018: “Red 1” and “Red 2”; flowering and fruiting from 2019: “Red 3” to “Red 16” (except “Red 14”) |
| 2015 | 2867 | 0 | 6 | 3 | 14 | 79/21 | 75 | ‘Robert’s Crab’ OP (311) | 512 | n.d. | n.d. | One seedling flowering since 2021 |
| 2016 | 4146 | 0 | 8 | 0 | 2 | 25/15 | 25 | ‘Golden Delicious’ × M. × zumi (450) | 1612 | n.d. | n.d. | Two seedlings with M. × zumi mother flowering since 2021 |
| 2017 | 5879 | 4 | 5 | 2 | 3 | 29/20 | 29 | Golden ‘Delicious’ × “Big Blush” (356) | 1360 | 858 | 425 | ** |
| 2018 | 4319 | 0 | 6 | 2 | 1 | 24/19 | 24 | “Yellow Tiny” × “Big Blush” (615) | 3281 | 976 | 243 | ** |
| 2019 | >2500 | 1 | 0 | 11 | 19 | 23/20 | 20 | “Red 1” × ‘Wickson Crab’ (415) | 3727 | 1297 | 148 | ** |
| 2020 | 1916 | 1 | 0 | 13 | 6 | 27/18 | 22 | “Most Bitter” × “Red 11” (153) | 3054 | 757 | 130 | ** |
| 2021 | 1774 | 4 | 1 | 3 | 4 | 11/7 | 11 | ‘King-ston Black’ × “Pink Puma” (115) | 1606 | *** | *** | *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
James, T.; Johnson, A.; Schaller, A.; Vanderzande, S.; Luo, F.; Sandefur, P.; Ru, S.; Peace, C. As It Stands: The Palouse Wild Cider Apple Breeding Program. Plants 2022, 11, 517. https://doi.org/10.3390/plants11040517
James T, Johnson A, Schaller A, Vanderzande S, Luo F, Sandefur P, Ru S, Peace C. As It Stands: The Palouse Wild Cider Apple Breeding Program. Plants. 2022; 11(4):517. https://doi.org/10.3390/plants11040517
Chicago/Turabian StyleJames, Tymon, Alexandra Johnson, Alexander Schaller, Stijn Vanderzande, Feixiong Luo, Paul Sandefur, Sushan Ru, and Cameron Peace. 2022. "As It Stands: The Palouse Wild Cider Apple Breeding Program" Plants 11, no. 4: 517. https://doi.org/10.3390/plants11040517
APA StyleJames, T., Johnson, A., Schaller, A., Vanderzande, S., Luo, F., Sandefur, P., Ru, S., & Peace, C. (2022). As It Stands: The Palouse Wild Cider Apple Breeding Program. Plants, 11(4), 517. https://doi.org/10.3390/plants11040517