Identification of QTLs for Grain Protein Content in Russian Spring Wheat Varieties
Abstract
1. Introduction
2. Results
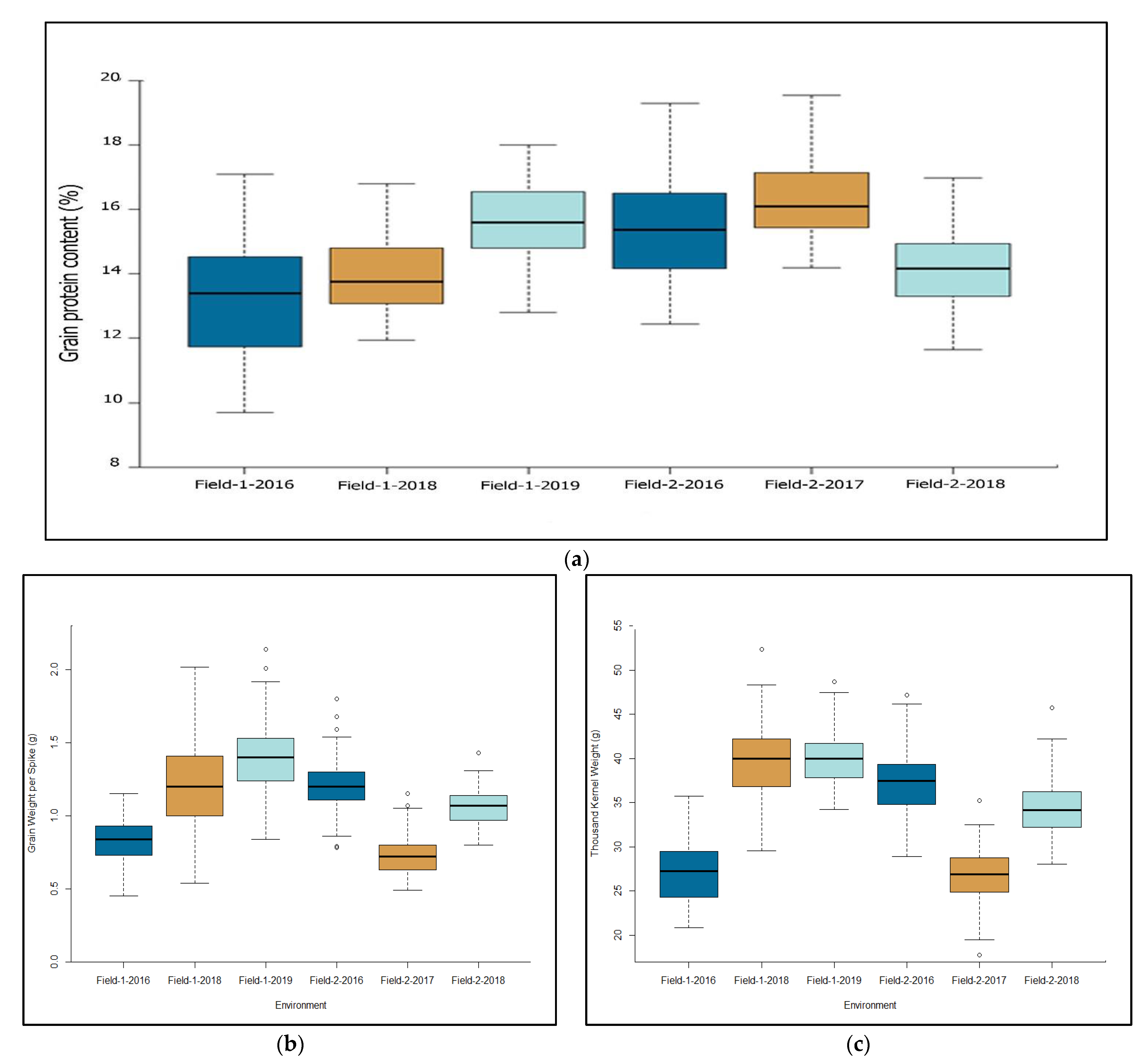
2.1. Phenotypic Variation
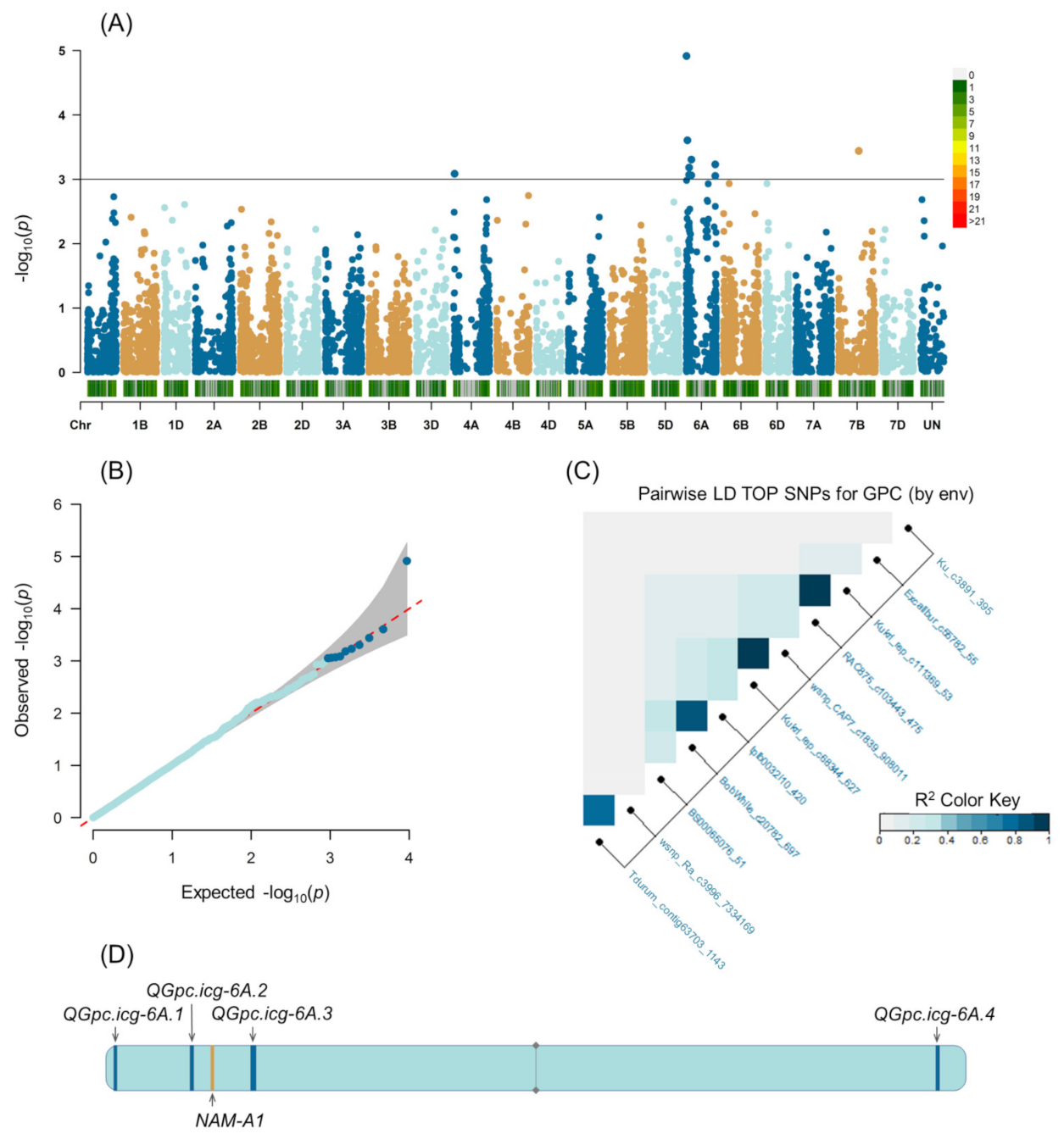
2.2. Population Structure and Genome-Wide Association Study
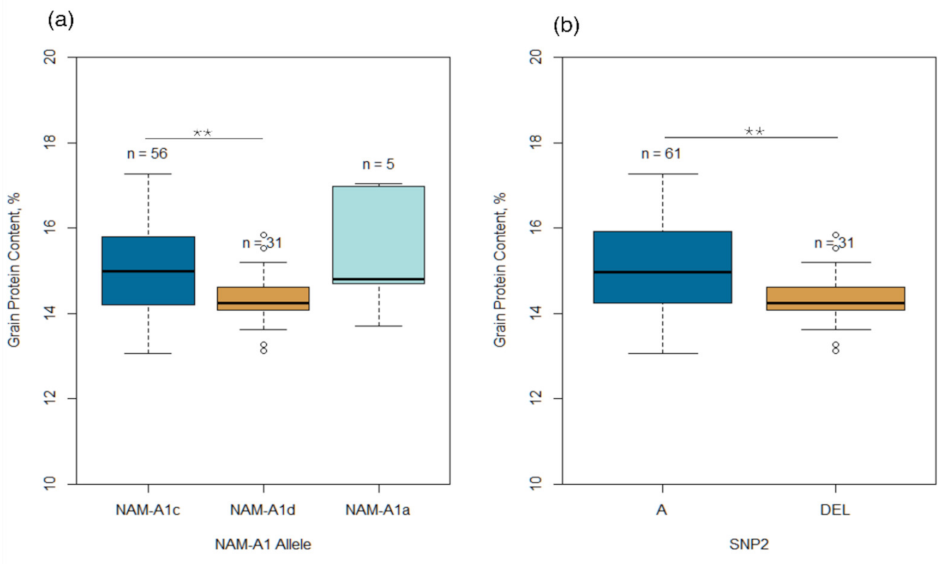
2.3. Allelic Polymorphism of NAM-A1 Gene
3. Discussion
3.1. Phenotypic Variation and MTAs
3.2. Effect of NAM-A1 Gene on GPC and Yield Components
4. Materials and Methods
4.1. Plant Materials and Phenotyping
4.2. Statistical Analysis of Phenotype Data
4.3. DNA Isolation, Genotyping, and Genome-Wide Association Study
4.4. Genotyping with NAM-A1 Specific SNPs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Cereal Supply and Demand Brief. Available online: https://www.fao.org/worldfoodsituation/csdb/en/ (accessed on 12 January 2021).
- Federal State Statistics Service. Available online: https://rosstat.gov.ru/compendium/document/13276 (accessed on 1 October 2021).
- Silaeva, L.P.; Barinova, E.V. Modern condition and conditions of rational placement of wheat production. Econ. J. 2019, 53, 33–42. [Google Scholar] [CrossRef]
- Pomeranz, Y.; Bolling, H.; Zwingelberg, H. Wheat hardness and baking properties of wheat flours. J. Cereal Sci. 1984, 2, 137–143. [Google Scholar] [CrossRef]
- MacRitchie, F. Evaluations of contributions from wheat protein fractions to dough mixing and bread making. J. Cereal Sci. 1987, 6, 259–268. [Google Scholar] [CrossRef]
- Huebner, F.R.; Bietz, J.A.; Nelson, T.; Bains, S.G.; Finney, P.L. Soft wheat quality as related to protein composition. Cereal Chem. 1999, 76, 650–655. [Google Scholar] [CrossRef]
- Kulwal, P.; Kumar, N.; Kumar, A.; Gupta, R.K.; Balyan, H.S.; Gupta, P.K. Gene networks in hexaploid wheat: Interacting quantitative trait loci for grain protein content. Funct. Integr. Genom. 2005, 5, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mantovani, E.E.; Simsek, S.; Jain, S.; Elias, E.M.; Mergoum, M. Genome wide genetic dissection of wheat quality and yield related traits and their relationship with grain shape and size traits in an elite × non-adapted bread wheat cross. PLoS ONE 2019, 14, e0221826. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.; Kumar, N.; Kulwal, P.L.; Röder, M.S.; Balyan, H.S.; Dhaliwal, H.S.; Gupta, P.K. QTL analysis for grain protein content using SSR markers and validation studies using NILs in bread wheat. Theor. Appl. Genet. 2003, 106, 659–667. [Google Scholar] [CrossRef]
- Balyan, H.S.; Gupta, P.K.; Kumar, S.; Dhariwal, R.; Jaiswal, V.; Tyagi, S.; Agarwal, P.; Gahlaut, V.; Kumari, S. Genetic improvement of grain protein content and other health-related constituents of wheat grain. Plant Breed. 2013, 132, 446–457. [Google Scholar] [CrossRef]
- Ahmed, H.G.M.; Sajjad, M.; Zeng, Y.; Iqbal, M.; Khan, S.H.; Ullah, A.; Akhtar, M.N. Genome-wide association mapping through 90K SNP array for quality and yield attributes in bread wheat against water-deficit conditions. Agriculture 2020, 10, 392. [Google Scholar] [CrossRef]
- Blanco, A.; de Giovanni, C.; Laddomada, B.; Sciancalepore, A.; Simeone, R.; Devos, K.M.; Gale, M.D. Quantitative trait loci influencing grain protein content in tetraploid wheats. Plant Breed. 1996, 115, 310–316. [Google Scholar] [CrossRef]
- Bogard, M.; Allard, V.; Martre, P.; Heumez, E.; Snape, J.W.; Orford, S.; Griffiths, S.; Gaju, O.; Foulkes, J.; Le Gouis, J. Identifying wheat genomic regions for improving grain protein concentration independently of grain yield using multiple inter-related populations. Mol. Breed. 2013, 31, 587–599. [Google Scholar] [CrossRef]
- Kumar, A.; Jain, S.; Elias, E.M.; Ibrahim, M.; Sharma, L.K. An overview of QTL identification and marker-assisted selection for grain protein content in wheat. In Eco-friendly Agro-biological Techniques for Enhancing Crop Productivity; Sengar, R., Singh, A., Eds.; Springer Nature: Singapore, 2018; pp. 245–273. [Google Scholar] [CrossRef]
- Zanetti, S.; Winzeler, M.; Feuillet, C.; Keller, B.; Messmer, M. Genetic analysis of bread-making quality in wheat and spelt. Plant Breed. 2001, 120, 13–19. [Google Scholar] [CrossRef]
- Bordes, J.; Ravel, C.; Jaubertie, J.P.; Duperrier, B.; Gardet, O.; Heumez, E.; Pissavy, A.L.; Charmet, G.; Le Gouis, J.; Balfourier, F. Genomic regions associated with the nitrogen limitation response revealed in a global wheat core collection. Theor. Appl. Genet. 2013, 126, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Joppa, L.R.; Du, C.; Hart, G.E.; Hareland, G.A. Mapping a QTL for grain protein in tetraploid wheat (Triticum turgidum L.) using a population of recombinant inbred chromosome lines. Crop Sci. 1997, 37, 1586–1589. [Google Scholar] [CrossRef]
- Hagenblad, J.; Asplund, L.; Balfourier, F.; Ravel, K.; Leino, M.W. Strong presence of the high grain protein content allele of NAM-B1 in Fennoscandian wheat. Theor. Appl. Genet. 2012, 125, 1677–1686. [Google Scholar] [CrossRef]
- Uauy, C.; Brevis, J.C.; Dubcovsky, J. The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J. Exp. Bot. 2006, 57, 2785–2794. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubkovsky, J.A. NAC gene regulating senescence improves grain protein, zink, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Saripalli, G.; Gahlaut, V.; Goel, N.; Meher, P.K.; Mishra, K.K.; Mishra, P.C.; Sehgal, D.; Vikram, P.; Sansaloni, C.; et al. Genetics of Fe, Zn, b-carotene, GPC and yield traits in bread wheat (Triticum aestivum L.) using multi-locus and multitraits GWAS. Euphytica 2018, 214, 219. [Google Scholar] [CrossRef]
- Carter, A.H.; Santra, D.K.; Kidwell, K.K. Assessment of the effects of the Gpc-B1 allele on senescence rate, grain protein concentration and mineral content in hard red spring wheat (Triticum aestivum L.) from the Pacific Northwest Region of the USA. Plant Breed. 2012, 131, 62–68. [Google Scholar] [CrossRef]
- Lundström, M.; Leino, M.W.; Hagenblad, J. Evolutionary history of the NAM-B1 gene in wild and domesticated tetraploid wheat. BMC Genet. 2017, 18, 118. [Google Scholar] [CrossRef]
- Chen, X.; Song, G.; Zhang, S.; Li, Y.; Gao, J.; Islam, S.; Ma, W.; Li, G.; Ji, W. The allelic distribution and variation analysis of the NAM-B1 gene in Chinese wheat cultivars. J. Integr. Agric. 2017, 16, 1294–1303. [Google Scholar] [CrossRef]
- Distelfeld, A.; Pearce, S.P.; Avni, R.; Scherer, B.; Uauy, C.; Piston, F.; Slade, A.; Zhao, R.; Dubcovsky, J. Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol. Biol. 2012, 78, 515–524. [Google Scholar] [CrossRef]
- Avni, R.; Zhao, R.; Pearce, S.; Jun, Y.; Uauy, C.; Tabbita, F.; Fahima, T.; Slade, A.; Dubcovsky, J.; Distelfeld, A. Functional characterization of GPC-1 genes in hexaploid wheat. Planta 2014, 239, 313–324. [Google Scholar] [CrossRef]
- Yang, R.; Juhasz, A.; Zhang, Y.; Chen, X.; Zhang, Y.; She, M.; Zhang, J.; Maddern, R.; Edwards, I.; Diepeveen, D.; et al. Molecular characterization of the NAM-1 genes in bread wheat in Australia. Crop Pasture Sci. 2018, 69, 1173. [Google Scholar] [CrossRef]
- Harrington, S.A.; Overend, L.E.; Cobo, N.; Borrill, P.; Uauy, C. Conserved residues in the wheat (Triticum aestivum) NAM-A1 NAC domain are required for protein binding and when mutated lead to delayed peduncle and flag leaf senescence. BMC Plant Biol. 2019, 19, 407. [Google Scholar] [CrossRef]
- Kiseleva, A.A.; Leonova, I.N.; Pshenichnikova, T.A.; Salina, E.A. Dissection of novel candidate genes for grain texture in Russian wheat varieties. Plant Mol. Biol. 2020, 104, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Blake, V.; Birkett, C.L.; Matthews, D.E.; Hane, D.; Bradbury, P.; Jannink, J. The Triticeae Toolbox: Combining phenotype and genotype data to advance small-grains breeding. Plant Genome 2016, 9, 1–10. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Feuillet, K.; Keller, B.; Rogers, J.; Stein, N. Shifting the limits in wheat research and breeding using a fully annotated reference genome International Wheat Genome Sequencing Consortium (IWGSC). Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Cormier, F.; Throude, M.; Ravel, C.; Gouis, J.L.; Leveugle, M.; Lafarge, S.; Exbrayat, F.; Duranton, N.; Praud, S. Detection of NAM-A1 natural variants in bread wheat reveals differences in haplotype distribution between a worldwide core collection and European elite germplasm. Agronomy 2015, 5, 143–151. [Google Scholar] [CrossRef]
- Blanco, A.; Pasqualone, A.; Troccoli, A.; Di Fonzo, N.; Simeone, R. Detection of grain protein content QTLs across environments in tetraploid wheats. Plant Mol. Biol. 2002, 48, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Kaya, Y.; Sahin, M. Non-parametric stability analysis of protein content in multi-environment trialsof wheat (T. aestivum L.). Genetica 2015, 47, 795–810. [Google Scholar] [CrossRef]
- Kondić-Špika, A.; Mladenov, N.; Grahovac, N.; Zorić, M.; Mikić, S.; Trkulja, D.; Marjanović-Jeromela, A.; Miladinović, D.; Hristov, N. Biometric analyses of yield, oil and protein contents of wheat (Triticum aestivum L.) genotypes in different environments. Agronomy 2019, 9, 270. [Google Scholar] [CrossRef]
- Muqaddasi, Q.H.; Brassac, J.; Ebmeyer, E.; Kollers, S.; Korzun, V.; Argillier, O.; Stiewe, G.; Plieske, J.; Ganal, M.W.; Röder, M.S. Prospects of GWAS and predictive breeding for European winter wheat’s grain protein content, grain starch content, and grain hardness. Sci. Rep. 2020, 10, 12541. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, L.; Wang, C.; Liu, Y.; Yan, Z.; Wang, Z.; Xiang, L.; Zhong, X.; Gong, F.; Zheng, Y.; et al. Genome-wide association study reveals novel genomic regions associated with high grain protein content in wheat lines derived from wild emmer wheat. Front. Plant Sci. 2019, 10, 464. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.H.; Fazal, M.; Bajgain, P.; Matthew, M.; Szinyei, T.; Conley, E.; Brown-Guedira, G.; Muehlbauer, G.J.; Anderson, J.A.; Steffenson, B.J. Genetic architecture of agronomic and quality traits in a nested association mapping population of spring wheat. Plant Genome 2020, 13, e20051. [Google Scholar] [CrossRef] [PubMed]
- Alemu, A.; Baouchi, A.E.; Hanafi, S.E.; Kehel, Z.; Eddakhir, K.; Tadesse, W. Genetic analysis of grain protein content and dough quality traits in elite spring bread wheat (Triticum aestivum) lines through association study. J. Cereal Sci. 2021, 100, 103214. [Google Scholar] [CrossRef]
- Lou, H.Y.; Zhang, R.Q.; Liu, Y.T.; Guo, D.D.; Zhai, S.S.; Chen, A.Y.; Zhang, Y.F.; Xie, C.J.; You, M.S.; Peng, H.R.; et al. Genome-wide association study of six quality-related traits in common wheat (Triticum aestivum L.) under two sowing conditions. Theor. Appl. Genet 2021, 134, 399–418. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Semagn, K.; Iqbal, M.; Moakhar, N.P.; Haile, T.; N’Diaye, A.; Yang, R.C.; Hucl, P.; Pozniak, C.; Spaner, D. Genome-wide association mapping of genomic regions associated with phenotypic traits in Canadian western spring wheat. Mol. Breed. 2017, 37, 141. [Google Scholar] [CrossRef]
- Zeng, B.; Yan, H.; Liu, X.; Zang, W.; Zhang, A.; Zhou, S.; Huang, L.; Liuet, J. Research open access genome-wide association study of rust traits in orchardgrass using SLAF-seq technology. Hereditas 2017, 154, 5. [Google Scholar] [CrossRef][Green Version]
- Mwadzingeni, L.; Shimelis, H.; Rees, D.J.G.; Tsilo, T.J. Genome-wide association analysis of agronomic traits in wheat under drought-stressed and non-stressed conditions. PLoS ONE 2017, 12, e0171692. [Google Scholar] [CrossRef]
- Leonova, I.N.; Skolotneva, E.S.; Salina, E.A. Genome-wide association study of leaf rust resistance in Russian spring wheat varieties. BMC Plant Biol. 2020, 20, 135. [Google Scholar] [CrossRef] [PubMed]
- Berezhnaya, A.; Kiseleva, A.; Leonova, I.; Salina, E. Allelic variation analysis at the vernalization response and photoperiod genes in Russian wheat varieties identified two novel alleles of Vrn-B3. Biomolecules 2021, 11, 1897. [Google Scholar] [CrossRef] [PubMed]
- Alhabbar, Z.; Yang, R.; Juhasz, A.; Xin, H.; She, M.; Anwar, M.; Sultana, N.; Diepeveen, D.; Ma, W.; Islam, S. NAM gene allelic composition and its relation to grain-filling duration and nitrogen utilization efficiency of Australian wheat. PLoS ONE 2018, 13, e0205448. [Google Scholar] [CrossRef]
- Alhabbar, Z.; Islam, S.; Yang, R.; Diepeveen, D.; Anwar, M.; Balotf, F.; Sultana, N.; Maddern, R.; She, M.; Zhang, J.; et al. Associations of NAM-A1 alleles with the onset of senescence and nitrogen use efficiency under Western Australian conditions. Euphytica 2018, 214, 180. [Google Scholar] [CrossRef]
- Shaukat, M.; Sun, M.; Ali, M.; Mahmood, T.; Naseer, S.; Maqbool, S.; Rehman, S.; Mahmood, Z.; Hao, Y.; Xia, X.; et al. Genetic gain for grain micronutrients and their association with phenology in historical wheat cultivars released between 1911 and 2016 in Pakistan. Agronomy 2021, 11, 1247. [Google Scholar] [CrossRef]
- Orlovsjaya, O.A.; Vakula, S.I.; Khotyleva, L.V.; Kilchevsky, A.V. Estimation of NAM-A1 haplotypes effects on the level of quantitative traits and grain protein content in wheat. Fakt. Eksperimentalnoi Evol. Org. 2020, 26, 114–119. [Google Scholar] [CrossRef]
- Genetic Resources Information System for Wheat and Triticale. Available online: http://wheatpedigree.net (accessed on 11 January 2022).
- Kiseleva, A.A.; Shcherban, A.B.; Leonova, I.N.; Frenkel, Z.; Salina, E.A. Identification of new heading date determinants in wheat 5B chromosome. BMC Plant Biol. 2016, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Gogarten, S.M.; Bhangale, T.; Conomos, M.P.; Laurie, C.A.; McHugh, C.P.; Painter, I.; Zheng, X.; Crosslin, D.R.; Levine, D.; Lumley, T.; et al. GWASTools: An R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics 2012, 28, 3329–3331. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Population Genetics. Available online: https://cran.r-project.org/web/packages/genetics/index.html (accessed on 15 October 2021).
- Shin, J.H.; Blay, S.; McNeney, B.; Graham, J. LDheatmap: An r function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Soft. 2006, 16, 1–10. [Google Scholar] [CrossRef]
| Genome | Chromosome | Unknown | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| A | 571 | 499 | 479 | 382 | 514 | 532 | 570 | - | 3547 |
| B | 567 | 747 | 535 | 215 | 681 | 600 | 463 | - | 3808 |
| D | 347 | 429 | 213 | 96 | 288 | 296 | 214 | - | 1883 |
| Unknown | - | - | - | - | - | - | - | 113 | 9351 |
| Marker | Chromosome Based on IWGSC Refseq v. 1.0 | Position | F * | p-Value | Favorable Allele | R2 ** | QTL | |
|---|---|---|---|---|---|---|---|---|
| IWGSC Refseq v. 1.0 (bp) | 90K Array Consensus Map, cM | |||||||
| Tdurum_contig63703_1143 | 6A | 6733494 | 16.58 | 9.75 | 0.00018 | T | 19.3 | QGpc.icg-6A.1 |
| wsnp_Ra_c3996_7334169 | 6A | 6733803 | 16.95 | 22.11 | 0.00001 | A | 21.8 | |
| BS00065076_51 | 6A | 23372012 | 43.09 | 9.16 | 0.00030 | A | 27.1 | |
| Kukri_rep_c68344_627 | 6A | 61037678 | 71.23 | 7.67 | 0.00095 | T | 26.8 | QGpc.icg-6A.2 |
| wsnp_CAP7_c1839_908011 | 6A | 61212495 | 71.23 | 12.79 | 0.00063 | A | 24.6 | |
| tplb0032i10_420 | 6A | 103760106 | 76.92 | 7.59 | 0.00100 | A | 23.6 | QGpc.icg-6A.3 |
| BobWhite_c20782_697 | 6A | 107113198 | 77.13 | 8.59 | 0.00044 | G | 22.8 | |
| RAC875_c103443_475 | 6A | 596903177 | 125.22 | 7.89 | 0.00078 | A | 27.7 | QGpc.icg-6A.4 |
| Kukri_rep_c111369_53 | 6A | 597031623 | 125.22 | 8.34 | 0.00054 | T | 27.7 | |
| Ku_c3891_395 | 4A | 27673099 | 37.05 | 12.19 | 0.00083 | C | 25.4 | |
| Excalibur_c55782_55 | Unknown | 5950 | 127.65 (7D) | 8.54 | 0.00047 | G | 18.7 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leonova, I.N.; Kiseleva, A.A.; Berezhnaya, A.A.; Stasyuk, A.I.; Likhenko, I.E.; Salina, E.A. Identification of QTLs for Grain Protein Content in Russian Spring Wheat Varieties. Plants 2022, 11, 437. https://doi.org/10.3390/plants11030437
Leonova IN, Kiseleva AA, Berezhnaya AA, Stasyuk AI, Likhenko IE, Salina EA. Identification of QTLs for Grain Protein Content in Russian Spring Wheat Varieties. Plants. 2022; 11(3):437. https://doi.org/10.3390/plants11030437
Chicago/Turabian StyleLeonova, Irina N., Antonina A. Kiseleva, Alina A. Berezhnaya, Anatoly I. Stasyuk, Ivan E. Likhenko, and Elena A. Salina. 2022. "Identification of QTLs for Grain Protein Content in Russian Spring Wheat Varieties" Plants 11, no. 3: 437. https://doi.org/10.3390/plants11030437
APA StyleLeonova, I. N., Kiseleva, A. A., Berezhnaya, A. A., Stasyuk, A. I., Likhenko, I. E., & Salina, E. A. (2022). Identification of QTLs for Grain Protein Content in Russian Spring Wheat Varieties. Plants, 11(3), 437. https://doi.org/10.3390/plants11030437







