Abstract
In recent years, leaf rust (LR) and stem rust (SR) have become a serious threat to bread wheat production in Kazakhstan. Most local cultivars are susceptible to these rusts, which has affected their yield and quality. The development of new cultivars with high productivity and LR and SR disease resistance, including using marker-assisted selection, is becoming an important priority in local breeding projects. Therefore, the search for key genetic factors controlling resistance in all plant stages, including the seedling stage, is of great significance. In this work, we applied a genome-wide association study (GWAS) approach using 212 local bread wheat accessions that were phenotyped for resistance to specific races of Puccinia triticina Eriks. (Pt) and Puccinia graminis f. sp. tritici (Pgt) at the seedling stages. The collection was genotyped using a 20 K Illumina iSelect SNP assay, and 11,150 polymorphic SNP markers were selected for the association mapping. Using a mixed linear model, we identified 11 quantitative trait loci (QTLs) for five out of six specific races of Pt and Pgt. The comparison of the results from this GWAS with those from previously published work showed that nine out of eleven QTLs for LR and SR resistance had been previously reported in a GWAS study at the adult plant stages of wheat growth. Therefore, it was assumed that these nine common identified QTLs were effective for all-stage resistance to LR and SR, and the two other QTLs appear to be novel QTLs. In addition, five out of these nine QTLs that had been identified earlier were found to be associated with yield components, suggesting that they may directly influence the field performance of bread wheat. The identified QTLs, including novel QTLs found in this study, may play an essential role in the breeding process for improving wheat resistance to LR and SR.
1. Introduction
Bread wheat, or common wheat (Triticum aestivum L.), is one of the main cereal crops cultivated around the world and is, thus, important for food security. Kazakhstan is among the ten largest exporters of wheat, with a volume of 11.4 thousand tons produced in 2019 [1]. According to the Bureau of National Statistics of Kazakhstan, the sown area under wheat was 12.2 million hectares in 2021, which represents about 76.7% of the total area used for cereal crops in the country [2]. One of the largest problems in wheat production all over the world is foliar diseases. Among fungal pathogens of wheat worldwide, Puccinia graminis f. sp. tritici (Pgt), causing stem rust (SR), and Puccinia triticina Eriks. (Pt), causing leaf rust (LR), are the most common [3,4,5]. LR and SR may cause more than 50% grain yield losses in susceptible wheat cultivars [6], while some aggressive strains, such as Pgt race Ug99, cause severe wheat yield losses of up to 90% [5,7].
Pt is an obligate biotrophic fungus that mainly infects the leaves at various growth stages, but it can also infect the leaf sheath and glumes [8]. Pt is a significant hindrance for wheat production, generally causing yield losses from 1% to 20% over a large area. However, if severe disease occurs prior to heading time, up to 90% of the wheat crop may be destroyed [9]. The occurrence of LR in Kazakhstan has been observed ever since wheat started to be cultivated on a wider scale in the early 1900s. The cultivation of susceptible cultivars resulted in epidemics of LR in 1 year out of 4 on average, affecting up to 5 million ha with yield losses of up to 25–30% [10,11]. Pgt causing SR is another important rust disease that is often considered as the most devastating of the wheat rust diseases, because it may cause complete crop loss over a large area within a short period of time [7]. In 2015 and 2016, a major SR epidemic occurred in the northern regions of Kazakhstan, as well as in the adjacent Omsk region of Russia, affecting approximately two million ha of wheat [11,12]. An SR epidemic occurred again in 2017–2018 in the northern and eastern regions of Kazakhstan, and it resulted not only in severe yield reduction, but also in lower grain quality [11,12,13,14]. During 2015–2018, disease severity and incidence in the main wheat-growing regions of Kazakhstan were up to 90% and 70%, respectively, and were substantially higher than in previous years [11,15].
One of the most effective ways to prevent wheat rust epidemics is the development of cultivars with durable resistance to pathogens. LR and SR resistance is controlled by a diverse group of genes, designated as Lr and Sr, respectively [16]. Nowadays, approximately 80 leaf rust resistance genes (Lr) and about 60 stem rust resistance genes (Sr) have been identified and described in bread wheat, durum wheat, and diploid wheat species [16]. Wheat rust resistance genes belong to one of two classes: seedling resistance genes (R) or the adult plant resistance (APR) genes, which are active only at the adult plant stage [16,17]. APR genes are considered potentially more durable, while R genes have a lack of durability [16,18]. R genes often encode nucleotide-binding site proteins and recognize specific pathogen effectors [16]. Due to their selective specificity to effectors, R genes are usually called race-specific genes. This specificity to particular races results in the greater effectiveness of R genes. R genes mostly have stronger effects but, on the other hand, they lose their strength after several years in the field [16]. There is also frequent emergence of new virulent pathogen races that overcome even the strongest resistance genes. For example, Pgt race Ug99 (also known as TTKSK) is insensitive to the Sr31 resistance gene, which is highly effective against almost all Pgt races [7]. This means that the emergence of new virulent pathogen races restricts the durability and effectiveness of R and APR genes. It has been shown that the pathogen population structure on a particular territory is not constant and may change from year to year [15,19]. Therefore, there is a constant need for new sources of resistance in the wheat genome, including both R and APR genes.
Pathogen race composition is distinct in each region of the world and changes depending on many factors, including climatic conditions and human factors. Numerous studies on the race composition of Pt and Pgt in different countries [13,14,15,20,21,22] have been conducted to search for resistant germplasms [23,24,25] and QTLs [26,27,28].
In Kazakhstan, 38 Pgt races were identified and described in major wheat-growing areas in the country in 2015–2018 [15]. Among them, the most virulent races RFRTF and TKRTF were observed in the Kostanay and Akmola regions and, at the same time, were registered in the neighboring Omsk region in the Russian Federation [14]. The stem rust resistance genes Sr11, Sr13, Sr22, Sr26, Sr31, Sr33, and Sr35 were confirmed to be effective against all Pgt races found in Kazakhstan. Although Ug99 has not been observed in Kazakhstan yet [15], recent reports have indicated the spreading of this race in the Middle East and a potential scenario of migration to Central and South Asia [5,29]. In Kazakhstan, 25 races of Pt were identified based on the assessments in all major wheat-growing regions of the country in 2018. The TQTMQ, TQKHT, and TRTHT races were the most common and were found in all studied populations [30].
Previously, QTLs for wheat seedling resistance to three common Pgt (TKRTF, PKCTC, and RKRTF) and three common Pt races (TQKHT, TRTHT, and TQTMQ) in Kazakhstan were identified using bi-parental mapping population Pamyati Aziaeva × Paragon [31]. Some of those QTLs were associated with known Lr and Sr genes, but several QTLs were novel, with high breeding potential. However, the linkage mapping (LM) used in that study has certain limitations that were attributed to a restricted level of genetic diversity defined by a pedigree of parental lines [32]. In comparison with linkage mapping, a genome-wide association study (GWAS) approach uses large germplasm collections with high genetic diversity. The wheat collection in this study was previously used for GWAS analysis of APR to leaf and stem rusts in southern Kazakhstan [33]. Therefore, an interesting question is - whether GWAS would facilitate the identification of unique race-specific QTLs not identified by LM? Additionally, the other aim of the study is to search for commonality/differences in APR and R genes using GWAS for the same studied spring wheat collection. The identification of race-specific QTLs would be beneficial for future pyramiding of rust resistance genes in order to extend the effectiveness of wheat production and prevent rust epidemics in the territory of Kazakhstan.
2. Results
2.1. Screening of Infection Type of Pt and Pgt Races
In general, the genotypes of the studied collection were moderately susceptible to three Pt races (Figure 1): 55.7% for the race TQTMQ, and 56.6% and 59.9% for races TRTHT and TQKHT, respectively. Susceptible infection type was observed in 8.9%, 10.9%, and 12.3% of the collection. The cultivars Saratovskaya 29 (Russia) and Lutescens 1082 (Kazakhstan) were susceptible to all three races of Pt that were tested. Resistant infection type was observed for 15 (7.1%) accessions to the race TQKHT and for 24 (11.3%) accessions to the races TQTMQ and TRTHT. Cultivar Lutescens 1193 (Russia) demonstrated complete resistance to all three races of Pt.
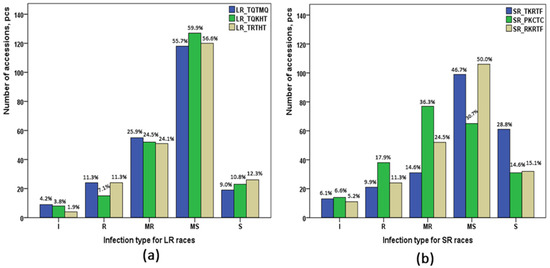
Figure 1.
Summary of the reactions of 212 wheat cultivars and breeding lines to races of Puccinia triticina (a) and Puccinia graminis f. sp. tritici (b). I: immune; R: resistant; MR: moderately resistant; MS: moderately susceptible; S: susceptible.
Evaluation of resistance to three Pgt races resulted in responses similar to those observed for Pt races. The majority of the collection was moderately susceptible for races TKRTF (46.7%) and RKRTF (50%) (Figure 1). As for the race PKCTC, most of the genotypes were of a moderately resistant infection type (36.3%, 77 accessions). Six cultivars revealed immunity to all three races of Pgt. These are accessions IR-38, IR-53, and E-795 of local breeds; Agent and Gatcher from America; and Seri 82 from Australia. Screening of seedling resistance identified three local cultivars (Karabalykskaya 25, Karabalykskaya 92, and Oskemen), which had a susceptible infection type to all three races of Pgt.
Pearson correlation analysis revealed a strong positive correlation (p < 0.001) of infection type among all Pt and Pgt races (Figure 2).
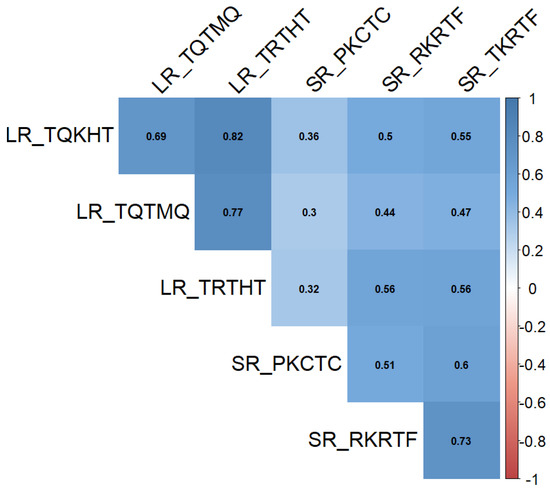
Figure 2.
Pairwise correlation analysis of leaf rust (LR) and stem rust (SR) infection type.
Two-way ANOVA revealed a strong significant effect (p < 0.001) of two factors (genotype and race), both separately and combined, on the resistance to LR and SR (Table 1). The broad-sense heritability (H2) of resistance to LR was higher than for resistance to SR (Table 1).

Table 1.
ANOVA analyses of infection type of LR and SR.
The correlations of LR and SR resistances between seedling and adult stages [33] were highly significant (Table 2). The average correlation index value for Pt races was higher for APR_LR (0.642) than for APR_SR (0.458). Similarly, the average correlation index value for Pgt races was higher for APR_SR (0.634) than for APR_LR (0.468).

Table 2.
Correlation among race-specific seedling resistance and adult plant resistance (APR) to leaf rust (LR) and stem rust (SR) in the studied collection.
2.2. Genotyping Results and Analysis of the Population Structure
Genotypic data of 212 common wheat accessions were compiled for 11,510 polymorphic SNP markers that were selected for the GWAS. The distribution of SNP markers among genomes was 2186 for A, 2955 for B, and 414 for D. The remaining 5955 markers in the 20 K array had unknown genomic positions. Chromosome 2B had the largest number of markers (640 SNP), and chromosome 7A was the longest chromosome (216.0 cM). The average SNP density for the three genomes was 1.6 markers/cM. The highest density was observed for genome B, with an average distance of 0.3 cM between neighboring markers. Generally, the density of the D genome was about seven times less than those of genomes A and B [33]. In average, linkage disequilibrium (LD) decayed at 14.9 cM for the whole genome at R2 of 0.1. For the subgenomes, the LD decay at 7.1 and 5.3 cM in the A and B genomes, respectively; for the D genome, the LD extends to 19.2 cM [33]. Based on the results of STRUCTURE and STRUCTURE Harvester analyses, the Q matrix was developed for three subpopulations, as suggested in Genievskaya et al. [33]. The generated Q matrix was used as a covariate matrix for MLM + Q + K in TASSEL.
2.3. Association Mapping
In total, 11 marker-trait associations with significant p-values were identified for the race-specific seedling resistance to LR and SR. The identified marker-trait associations were located on eight chromosomes (1A, 1B, 1D, 2A, 4B, 5B, 6A, and 7A). Manhattan and QQ plots for all races are provided in Table S1. Physical positions, effects, and R2 values for identified SNPs associated with race-specific seedling resistances to LR and SR were given in Table 3. All marker-trait associations were designated as QTLs and positioned on the genetic map along with approximate positions of potential candidate genes for LR and SR resistance (Table 3 and Table S2; Figure S1). Among the identified QTLs, three had associations with resistance to only LR. The other eight identified QTLs were associated with resistance for both diseases (LR and SR).

Table 3.
List of quantitative trait loci (QTLs) for race-specific seedling resistance to leaf rust (LR) and stem rust (SR).
All eleven QTLs responsible for the resistance to LR were associated with the race TRTHT. One of the QTLs was associated with the combination of races TQKHT and TRTHT, and six QTLs with a combination of all three races (Table 3). Eight out of eleven LR QTLs were associated with Pgt race TKRTF, and two with the combination of races TKRTF and RKRTF (Table 3). None of the QTLs were associated with PKCTC (Table 3). Notably, there were no genetic factors common to this work and that of a report that was based on the study of the same Pt and Pgt races using bi-parental mapping population Pamyati Azieva × Paragon [31].
The QTLs identified in this study were analyzed and compared with the QTLs previously reported for APR resistance to LR and SR that were identified using GWAS data from RIBSP 2018–2019 [33] and the QTLs previously reported to be associated with yield-related traits identified using GWAS data from northern Kazakhstan 2018–2020 [34] (Table 4). In addition, the location of each identified QTL was compared to the genetic positions of known Lr and Sr genes (Table 4). In total, three candidate Lr genes and 9 QTLs were found for 11 QTLs associated with LR resistance in this study. In the analysis of QTLs for SR resistance, there were no similarities among the genetic locations of known Sr genes and one similarity with previously identified QTLs (Table 4).

Table 4.
Comparison of quantitative trait loci (QTLs) of seedling resistance to leaf rust (LR) and stem rust (SR) identified in this study with previously described QTLs and candidate Lr and Sr genes.
3. Discussion
The results indicate a lack of QTLs identified both at the seedling stage in this GWAS study and a previous LM study using Pamyati Azieva × Paragon RILs [31], suggesting different responses between two types of genetic materials with the same Pt and Pgt races. Notably, both the APR for LR and SR in LM and GWAS populations were tested in the same environment and for the same years (RIBSP, 2018–2019). It can be hypothesized that the difference was probably determined by the specificity of the LM population’s reaction due to the pedigree restricted by two parental pools. Additionally, unlike in the LM study [31] where no correlation was registered between race-specific seedling resistance and APR, the correlation of resistance between two growth stages was found to be highly significant (p < 0.0001) in this study.
By contrast, it was found that 9 out of 11 QTLs identified in this study had been previously identified in an APR GWAS using data from RIBSP in 2018–2019 [33] (Table 4), suggesting that these are QTLs for all-stage resistance. Notably, all nine QTLs in this comparative assessment were characterized by similar directions of QTL effects, i.e., toward either resistance or susceptibility. In addition, five out of these nine QTLs were earlier found to be associated with yield components [34] (Table 4), suggesting that these QTLs might directly influence the field performance of bread wheat. The comparative evaluation of all eleven identified QTLs suggests that two QTLs (QLr.ipbb-1A.3 and QLr.ipbb-1B.5) were presumably novel, as they were not reported in previously published LR and SR studies (Table 4).
The majority of the studied collection had shown moderately susceptible IT at the seedling stage to all races of Pt and Pgt, except for the Pgt race PKCTC, where most of the accessions were moderately resistant (Figure 1). The ANOVA showed a more significant influence of wheat genotype as compared to race type on the resistance to both diseases (Table 1), suggesting the significant involvement of genetic factors in the resistance to all six studied races.
3.1. Patterns of Identified QTLs for Leaf and Stem Rust Resistances
Multiple occurrences of most QTLs were associated with LR resistance in different growth stages and environments. In particular, six identified QTLs appeared to be efficient in LR resistance to all three specific races, which is an indication of the broad stability of these loci. Three of those six QTLs (QLr.ipbb-2A.2, QLr.ipbb-4B.2, and QLr.ipbb-6A.2) showed effects toward LR resistance, while the effects in the remaining three QTLs (QLr.ipbb-1B.2, QLr.ipbb-6A.6, and QLr.ipbb-7A.2) were toward susceptibility (Table 3). The latter three QTLs can be selected for the rapid elimination of susceptible cultivars via breeding efforts. Interestingly, the QLr.ipbb-4B.2 was positioned in the vicinity of genes Lr12 (70.9 cM) [35], Lr31 (70.9 cM) [36], and Lr49 (81.5 cM) [37] on chromosome 4B (Table 5). This result confirms the previously reported effects of Lr12 [35] in northern and southeastern Kazakhstan [11]. QLr.ipbb-6A.2 was closely mapped to the SNP marker S16_50275005 of LR identified by Juliana et al. [38], which is adjacent to the Traes_6AS_EB7270F83 gene with a predicted LRR (leucine-rich repeat) receptor-like STPK (serine/threonine-protein kinase) function. The QTLs QLr.ipbb-1B.5, QLr.ipbb-2A.2, and QLr.ipbb-4B.2 are located at the same position as QTLs 1B_1, 2A_2, and 4B_3 identified in the GWAS for LR by Gao et al. [39]. Additionally, QLr.ipbb-7A.2 was in the vicinity of QTL 7A_3 (~2 cM) [39]. The QTL QLr.ipbb-2A.2 was positioned approximately 1.4 cM away from SNP IWA574, which was previously found to be associated with seedling resistance to Pt race TBDJ [40]. The QLr.ipbb-5B.1 was in a similar genetic position to QLr.fcu-5BL associated with LR field resistance [26].

Table 5.
Virulence/avirulence pattern of pathogen races used in the study based on the nomenclature by Long and Kolmer [41] for LR and by Roelfs and Martens [42] for SR.
None of the eight SR QTLs identified in this study (Table 4) were located in the vicinity of known Sr genes. Additionally, all eight SR QTLs at the seedling stage conferred seedling resistance to LR and adult resistance to SR, confirming that the identified QTLs are expressed in both the seedling and adult stages. The comparative evaluation of the identified SR QTLs in this study with known SR resistant factors also identified several examples of pleiotropic effects. For instance, the location of QSr.ipbb-1B.2 (race TKRTF) was adjacent to the genetic position of IWB42604 that was associated with seedling resistance to the TRTTF race [43]. Two QTLs responsible for the resistance to SR were previously identified by Genievskaya et al. [33] at the adult plant stage (Table 4). A survey of the associated literature comprising many studies on bread and durum wheat and their resistance to LR and SR suggests that pleiotropy is a common scenario [44,45]. Hence, the finding in this study may positively impact the development of high-yielding wheat germplasm with the resistance to Pt and Pgt races via the application of marker-assisted selection.
3.2. Comparison of the Physical Positions of SNPs in Quantitative Trait Loci and Protein-Coding Genes
Among eleven marker-trait associations identified in this work (Table 4), three protein-coding genes are potentially directly involved in determining the resistance to rust pathogens. The position of one of those genes, TraesCS4B02G328500, coding for the major facilitator superfamily (MFS) domain-containing protein, overlapped with the positions of QLr.ipbb-4B.2 and QSr.ipbb-4B.1. The MFS transporter has previously been reported to participate in the secretion of fungi toxin, which affects host species [46]. The position of the TraesCS7A02G250500 gene, coding for L-ascorbate peroxidase 6, overlaps with those of QLr.ipbb-7A.2 and QSr.ipbb-7A.2. Gou and co-authors (2015) proposed that the phosphorylation of ascorbate peroxidase by the Wheat Kinase START 1 (WKS1.1) gene reduces the ability of the cells to detoxify reactive oxygen species, thus contributing to promoting cell death [47]. This response takes several days longer than typical hypersensitive cell death responses, thus allowing the limited pathogen growth and restricted sporulation that is characteristic of the WKS1 partial resistance response to Puccinia striiformis [47]. The position of the other gene, TraesCS7A02G389100, coding for the Rab-GAP TBC domain-containing protein, physically overlapped with the genetic positions of QLr.ipbb-7A.2 and QSr.ipbb-7A.3. This protein has positive or negative effects on the immune response of wheat to infection by rust pathogens depending on its levels [48]. Although the functions of the protein-coding genes associated with SNPs, as mentioned above, do not directly explain the genetic mechanism of resistance to the studied rusts, they still indicate their potential involvement in the complex processes of plant resistance.
4. Materials and Methods
4.1. Genetic Material
A total of 212 (Table S3) wheat cultivars and breeding lines were selected and evaluated for their response upon exposure to Pt and Pgt races at the seedling stage. The collection included 88 commercial and prospective breeding cultivars from Kazakhstan and Russia, including 64 cultivars approved by the State Seed Trials Commission for use in the territory of Kazakhstan; 38 cultivars from Europe provided by the John Innes Centre, United Kingdom; and 86 cultivars and lines from Kazakhstan, Russia, USA, Canada, Mexico, Germany, and Australia provided by the Research Institute of Biological Safety Problems (RIBSP, Gvardeisky, South Kazakhstan) [33]. Most of the cultivars and lines from Kazakhstan and Russia originated from locally made crosses, though a few originated from the Kazakhstan–Siberia Network for Spring Wheat Improvement shuttle breeding program.
4.2. Seedling LR and SR Evaluation
The evaluation of race-specific resistance was conducted at the seedling stage under greenhouse conditions at the RIBSP in 2019. For the resistance assessment, seedlings of the studied collection were separately inoculated with three races of P. triticina (TQTMQ, TQKHT, and TRTHT) and three races of P. graminis (TKRTF, PKCTC, and RKRTF) with different levels of virulence to Lr and Sr genes, respectively [15,30]. Seeds of each accession were sown in plastic pots (6 seeds per pot) in two replicates for each rust race. Before the inoculation, urediniospores of pathogen races (stored in a refrigerator at −80 °C) were heated at 40 °C for 10 min, followed by watering in a humid chamber at 20 °C for 2 h, containing a 23.5% KOH solution (80% relative humidity) [49]. Urediniospores were then suspended in light mineral oil (Soltrol 170), and each pot with wheat seedlings was individually inoculated by spraying with races of Pt and Pgt onto the fully expanded primary leaves of 7–9-day-old seedlings. Seedlings were incubated in a humid chamber in the dark at 18 ± 2 °C and 100% humidity for 14 h and then exposed to fluorescent light for 3–4 h. The inoculated plants were placed in greenhouse boxes, with favorable conditions (22 ± 2 °C for stem rust, 18 ± 2 °C for leaf rust) and illumination (10–15 thousand lux, light period 16 h) [26,50,51]. The resistance of the studied collection was assessed two weeks after inoculation according to the Stakman et al. infection type scale [52]. The infection type values for each combination of wheat accession and pathogen race was determined as an average for 6 plants in the pot. In order to use the Stakman et al. scale in the GWAS, the 0–4 scale was converted into a 0–9 linear scale as proposed by Zhang et al. [53]. The average resistant values for two replications were further used in GWAS.
The statistical analysis included correlation analysis and analysis of variance (two-way ANOVA) using the SPSS 22.0 (https://www.ibm.com/support/pages/spss-statistics-220-available-download, accessed on 13 July 2021) and STATISTICA 10.0 (http://statsoft.ru/resources/support/download.php, accessed 21 July 2021) software packages. Variance components (%) were determined by the division of phenotypic variance due to each component on the total phenotypic variance. The broad-sense heritability (H2), describing the proportion of phenotypic variation due to genetic factors, was calculated by the following formula:
where is phenotypic variance explained by the genotype and is the total phenotypic variance (sum of genotype variance, race variance, genotype × race variance, and residuals variance) [41].
4.3. DNA Extracting and Genotyping
Total DNA was isolated from the seedlings of wheat accessions according to Dellaporta et al. [42]. The DNA concentration for each sample was adjusted to 50 ng/mL. The panels of the studied collection were genotyped using 20K Illumina iSelect SNP assay at the TraitGenetics GmbH (Gatersleben, Germany). SNP genotyping was performed using the Illumina Genome Studio software version V2011.1 (Illumina Inc., San Diego, CA, USA, 2018). A total of 11,510 SNP markers [33] were selected after removing all monomorphic markers and markers with a minor allele frequency (MAF) <0.05. Accessions with more than 10% missing data were also removed.
4.4. Association Mapping
The analysis of the population structure was carried out using STRUCTURE (v2.3.4.) software with a Bayesian Markov chain Monte Carlo (MCMC) approach based on the admixture and correlated frequency models [54]. The numbers of hypothetical groups ranging from K = 1 to K = 10 were assessed using 50,000 burn-in iterations, followed by 100,000 recorded iterations. The output from STRUCTURE was analyzed for the delta K value (ΔK) in STRUCTURE HARVESTER [55].
Using K = 5 values, the Q-matrix for the five identified clusters was developed. GWAS was conducted using TASSEL 5.0 (v20191212) [56] based on the mixed linear model (MLM) with the kinship (K) and Q matrices (MLM + K + Q) [57]. For confirmation of the correction due to K and Q matrices, the distribution lines in each quantile–quantile plot were analyzed. Significant marker-trait associations were selected after the application of a threshold at p < 0.0001. The positions and sequences of SNP markers were obtained from the 90K Array Consensus map of the common wheat genome [58]. For markers with unknown positions in the 90K Array Consensus map, the CSS POPSEQ 2014 map [59], available at the Triticeae Toolbox (2020), was used. For several significant marker-trait associations linked to each other, the SNP with the lowest p-value was chosen. For the search of protein-coding genes that overlap with identified significant marker–trait associations, the sequence of each marker was inserted into the BLAST tool [60] of Ensembl Plants [61] and compared with the reference genome of T. aestivum. The genetic map was constructed using MapChart v.2.3 software [62].
5. Conclusions
A GWAS of 212 bread wheat accessions inoculated with three races of P. triticina (TQTMQ, TQKHT, and TRTHT) and three races of P. graminis (TKRTF, PKCTC, and RKRTF) at the seedling stage of growth, resulted in the identification of eleven marker-trait associations for LR and SR resistance. Nine out of the eleven identified QTLs were previously reported in a GWAS using the same collection with assessment at the adult plant stage in a natural infection field of southern Kazakhstan in 2018–2019. Correspondingly, it was concluded that these nine identified QTLs were effective for all-stage resistance to LR and SR, and the two other QTLs appear to be novel and were effective at the seedling growth stage for the LR resistance. Five out of these nine QTLs were earlier found to be associated with yield components, suggesting that these QTLs might directly influence the field performance of bread wheat. In addition, the alignment of SNPs in QTLs to the sequencing data of a hexaploid wheat physical map using the Ensemble platform suggests the direct involvement of at least three protein-coding genes in determining the resistance to rust pathogens. The obtained results can be further validated and potentially used in marker-assisted selection for the construction of new highly productive cultivars resistant to LR and SR.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants11010074/s1; Figure S1: Genetic map of the location of the identified QTLs associated with seedling resistance to Pt and Pgt races. Table S1: Graphical representation of GWAS results using TASSEL software based on MLM+Q+K method, Table S2: Complete data of GWAS results for race-specific seedling resistance to leaf rust and stem rust, Table S3: List of common wheat cultivars and breeding lines used in the current study.
Author Contributions
Conceptualization, Y.T. and S.A.; methodology, A.R. and Y.T.; formal analysis, A.Z., G.Y. and A.M.; investigation, A.R., T.S. and S.A.; resources, Y.T. and A.R.; data curation, A.Z. and Y.G.; writing—original draft preparation, Y.T.; writing—review and editing, A.Z., Y.G., T.S., S.A., A.R. and Y.T.; supervision, Y.T. and S.A.; project administration, S.A. and Y.T.; funding acquisition, T.S., A.R. and Y.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Agriculture of the Republic of Kazakhstan, grants number BR10765056 and BR06249329, and by the Ministry of Education and Science of the Republic of Kazakhstan grant number AP08855387.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No data available.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Food and Agriculture Organization Corporate Statistical Database (FAOSTAT). Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 20 August 2021).
- Statistics Committee. Ministry of National Economy of the Republic of Kazakhstan. Available online: https://stat.gov.kz/ (accessed on 13 September 2021).
- Roelfs, A.P.; Singh, R.P.; Saari, E.E. Rust Diseases of Wheat: Concepts and Methods of Disease Management; CIMMYT: Mexico City, Mexico, 1992; p. 81. [Google Scholar]
- Kolmer, J. Leaf rust of wheat: Pathogen biology, variation and host resistance. Forests 2013, 4, 70–84. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.P.; Hodson, D.P.; Jin, Y.; Lagudah, E.S.; Ayliffe, M.A.; Bhavani, S.; Rouse, M.N.; Pretorius, Z.A.; Szabo, L.J.; Huerta-Espino, J.; et al. Emergence and spread of new races of wheat stem rust fungus: Continued threat to food security and prospects of genetic control. Phytopathology 2015, 10, 872–884. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Espino, J.; Singh, R.; German, S.; Mccallum, B.; Park, R.; Chen, W.Q.; Bhardwaj, S.C.; Goyeau, H. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 2011, 179, 143–160. [Google Scholar] [CrossRef]
- Singh, R.P.; Hodson, D.P.; Huerta-Espino, J.; Jin, Y.; Bhavani, S.; Njau, P.; Herrera-Foessel, S.; Singh, P.K.; Singh, S.; Velu, G. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 2011, 49, 465–481. [Google Scholar] [CrossRef] [Green Version]
- Figlan, S.; Ntushelo, K.; Mwadzingeni, L.; Terefe, T.; Tsilo, T.J.; Shimelis, H. Breeding Wheat for Durable Leaf Rust Resistance in Southern Africa: Variability; Distribution; Current Control Strategies; Challenges and Future Prospects. Front. Plant Sci. 2020, 11, 549. [Google Scholar] [CrossRef] [PubMed]
- Samborski, D.J. Wheat leaf rust. In The Cereal Rusts Vol. II: Diseases, Distribution, Epidemiology and Control; Roelfs, A.P., Bushnell, W.R., Eds.; Academic Press: Orlando, FL, USA, 1985; pp. 39–59. [Google Scholar]
- Morgounov, A.; Rosseeva, L.; Koyshibayev, M. Leaf rust of spring wheat in Northern Kazakhstan and Siberia: Incidence, virulence, and breeding for resistance. Aust. J. Agric. Res. 2007, 58, 847. [Google Scholar] [CrossRef]
- Koyshybaev, M. Wheat Diseases, 1st ed.; FAO: Ankara, Turkey, 2018; p. 365. [Google Scholar]
- Shamanin, V.; Salina, E.; Wanyera, R.; Zelenskiy, Y.; Olivera, P.; Morgounov, A.I. Genetic diversity of spring wheat from Kazakhstan and Russia for resistance to stem rust Ug99. Euphytica 2016, 212, 287–296. [Google Scholar] [CrossRef]
- Rsaliyev, A.S.; Rsaliyev, S.S. Principal approaches and achievements in studying race composition of wheat stem rust. Vavilov J. Genet. Breed. 2018, 22, 967–977. [Google Scholar] [CrossRef]
- Shamanin, V.P.; Pototskaya, I.V.; Shepelev, S.S.; Pozherukova, V.E.; Salina, E.A.; Skolotneva, E.S.; Morgounov, A.I. Stem rust in Western Siberia–race composition and effective resistance genes. Vavilov J. Genet. Breed. 2020, 24, 131–138. [Google Scholar] [CrossRef] [Green Version]
- Rsaliyev, A.; Yskakova, G.; Maulenbay, A.; Zakarya, K.; Rsaliyev, S. Virulence and race structure of Puccinia graminis f. sp. tritici in Kazakhstan. Plant Prot. Sci. 2020, 56, 275–284. [Google Scholar] [CrossRef]
- McIntosh, R.A.; Dubcovsky, J.; Rogers, W.J.; Morris, C.F.; Xia, X.C. Catalogue of Gene Symbols for Wheat: 2017 Supplement. Available online: https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2017.pdf (accessed on 23 September 2021).
- Ellis, J.G.; Lagudah, E.; Spielmeyer, W.; Dodds, P.N. The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 2014, 5, 641. [Google Scholar] [CrossRef] [Green Version]
- Bajgain, P.; Rouse, M.; Bulli, P.; Bhavani, S.; Gordon, T.; Wanyera, R.; Njau, P.N.; Legesse, W.; Andersen, J.A.; Pumphrey, M.O. Association mapping of North American spring wheat breeding germplasm reveals loci conferring resistance to Ug99 and other African stem rust races. BMC Plant Biol. 2015, 15, 249. [Google Scholar] [CrossRef]
- Bariana, H.S.; Hayden, M.J.; Ahmed, N.U.; Bell, J.A.; Sharp, P.J.; McIntosh, R.A. Mapping of durable adult plant and seedling resistances to stripe rust and stem rust diseases in wheat. Aust. J. Agric. Res. 2001, 52, 1247. [Google Scholar] [CrossRef]
- Olivera, P.D.; Sikharulidze, Z.; Dumbadze, R.; Szabo, L.J.; Newcomb, M.; Natsarishvili, K.; Rouse, M.N.; Luster, D.G.; Jin, Y. Presence of a sexual population of Puccinia graminis f.sp. tritici in Georgia provides a hotspot for genotypic and phenotypic diversity. Phytopathology 2019, 109, 2152–2160. [Google Scholar] [CrossRef] [Green Version]
- Li, T.Y.; Ma, Y.C.; Wu, X.X.; Chen, S.; Xu, X.F.; Wang, H.; Cao, Y.Y.; Xuan, Y.H. Race and virulence characterization of Puccinia graminis f. sp. tritici in China. PLoS ONE 2018, 13, e0197579. [Google Scholar] [CrossRef] [PubMed]
- Omara, R.I.; Nehela, Y.; Mabrouk, O.I.; Elsharkawy, M.M. The Emergence of New Aggressive Leaf Rust Races with the Potential to Supplant the Resistance of Wheat Cultivars. Biology 2021, 10, 925. [Google Scholar] [CrossRef]
- Admass, B.; Friedt, W.; Ordon, F. Stem rust seedling resistance genes in Ethiopian wheat cultivars and breeding lines. Afr. Crop Sci. J. 2012, 20, 149–162. [Google Scholar]
- Zhang, P.P.; Gebrewahid, T.W.; Yue, Z.; Li, Q.L.; Li, Z.F.; Liu, D.Q. Seedling and adult plant resistance to leaf rust in 46 Chinese bread wheat landraces and 39 wheat lines with known Lr genes. J. Integr. Agric. 2019, 18, 1014–1023. [Google Scholar] [CrossRef]
- Atia, M.A.; El-Khateeb, E.A.; El-Maksoud, A.; Reem, M.; Abou-Zeid, M.A.; Salah, A.; Abdel-Hamid, A.M. Mining of Leaf Rust Resistance Genes Content in Egyptian Bread Wheat Collection. Plants 2021, 10, 1378. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.G.; Friesen, T.L.; Xu, S.S.; Faris, J.D.; Kolmer, J.A. Identification of novel QTLs for seedling and adult plant leaf rust resistance in a wheat doubled haploid population. Theor. Appl. Genet. 2009, 119, 263–269. [Google Scholar] [CrossRef]
- Li, G.; Xu, X.; Bai, G.; Carver, B.F.; Hunger, R.; Bonman, J.M.; Kolmer, J.; Dong, H. Genome-wide association mapping reveals novel QTL for seedling leaf rust resistance in a worldwide collection of winter wheat. Plant Genome 2016, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Rollar, S.; Serfling, A.; Geyer, M.; Hartl, L.; Mohler, V.; Ordon, F. QTL mapping of adult plant and seedling resistance to leaf rust (Puccinia triticina Eriks.) in a multiparent advanced generation intercross (MAGIC) wheat population. Theor. Appl. Genet. 2021, 134, 37–51. [Google Scholar] [CrossRef]
- Afzal, A.; Ali, S.R.; Ijaz, M.; Saeed, M. Combating Ug99-Current Scenario. Int. J. Phytopathol. 2021, 10, 57–70. [Google Scholar] [CrossRef]
- Maulenbay, A.D.; Yskakova, G.S.; Rsaliyev, A. Virulence and racial composition of Puccina triticina in Kazakhstan in 2018. Her. Sci. S. Seifullin Kazakh Agro Tech. Univ. 2020, 3, 25–35. (In Russian) [Google Scholar]
- Genievskaya, Y.; Abugalieva, S.; Rsaliyev, A.; Yskakova, G.; Turuspekov, Y. QTL Mapping for Seedling and Adult Plant Resistance to Leaf and Stem Rusts in Pamyati Azieva à Paragon Mapping Population of Bread Wheat. Agronomy 2020, 10, 1285. [Google Scholar] [CrossRef]
- Semagn, K.; Bjørnstad, Å.; Ndjiondjop, M.N. Principles; requirements and prospects of genetic mapping in plants. Afr. J. Biotechnol. 2006, 5, 2569–2587. [Google Scholar]
- Genievskaya, Y.; Turuspekov, Y.; Rsaliyev, A.; Abugalieva, S. Genome-wide association mapping for resistance to leaf; stem; and yellow rusts of common wheat under field conditions of South Kazakhstan. PeerJ 2020, 8, e9820. [Google Scholar] [CrossRef]
- Amalova, A.; Abugalieva, S.; Babkenov, A.; Babkenova, S.; Turuspekov, Y. Genome-wide association study of yield components in spring wheat collection harvested under two water regimes in Northern Kazakhstan. PeerJ 2021, 9, e11857. [Google Scholar] [CrossRef]
- Dyck, P.L.; Samborski, D.J.; Anderson, R.G. Inheritance of adult-plant leaf rust resistance derived from the common wheat varieties Exchange and Frontana. Can. J. Genet. Cytol. 1966, 8, 665–671. [Google Scholar] [CrossRef]
- Singh, R.P.; McIntosh, R.A. Complementary genes for reaction to Puccinia recondita tritici in Triticum aestivum. II. Cytogenetic studies. Can. J. Genet. Cytol. 1984, 26, 736–742. [Google Scholar] [CrossRef]
- Bansal, U.K.; Hayden, M.J.; Venkata, B.P.; Khanna, R.; Saini, R.G.; Bariana, H.S. Genetic mapping of adult plant leaf rust resistance genes Lr48 and Lr49 in common wheat. Theor. Appl. Genet. 2008, 117, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Juliana, P.; Singh, R.P.; Singh, P.K.; Poland, J.A.; Bergstrom, G.C.; Huerta-Espino, J.; Bhavani, S.; Crossa, J.; Sorrells, M.E. Genome-wide association mapping for resistance to leaf rust; stripe rust and tan spot in wheat reveals potential candidate genes. Theor. Appl. Genet. 2018, 131, 1405–1422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Turner, M.K.; Chao, S.; Kolmer, J.; Anderson, J.A. Genome Wide Association Study of Seedling and Adult Plant Leaf Rust Resistance in Elite Spring Wheat Breeding Lines. PLoS ONE 2016, 11, e0148671. [Google Scholar] [CrossRef] [Green Version]
- Kertho, A.; Mamidi, S.; Bonman, J.M.; McClean, P.E.; Acevedo, M. Genome-Wide Association Mapping for Resistance to Leaf and Stripe Rust in Winter-Habit Hexaploid Wheat Landraces. PLoS ONE 2015, 10, e0129580. [Google Scholar] [CrossRef] [Green Version]
- Covarrubias-Pazaran, G.E. Optimizing Breeding Schemes. Manual. Heritability: Meaning and Computation, 1st ed.; CGIAR Excellence in Breeding Platform (EiB): Montpellier, France, 2019; p. 10. [Google Scholar]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation. Version II. Plant Mol. Biol. Report. 1983, 4, 19–21. [Google Scholar] [CrossRef]
- Saccomanno, A.; Matny, O.; Marone, D.; Laidò, G.; Petruzzino, G.; Mazzucotelli, E.; Desiderio, F.; Blanco, A.; Gadaleta, A.; Pecchioni, N.; et al. Genetic Mapping of Loci for Resistance to Stem Rust in a Tetraploid Wheat Collection. Int. J. Mol. Sci. 2018, 19, 3907. [Google Scholar] [CrossRef] [Green Version]
- Dakouri, A.; McCallum, B.D.; Radovanovic, N.; Cloutier, S. Molecular and phenotypic characterization of seedling and adult plant leaf rust resistance in a world wheat collection. Mol. Breed. 2013, 32, 663–677. [Google Scholar] [CrossRef] [Green Version]
- Aoun, M.; Kolmer, J.A.; Rouse, M.N.; Elias, E.M.; Breiland, M.; Bulbula, W.D.; Chao, S.; Acevedo, M. Mapping of Novel Leaf Rust and Stem Rust Resistance Genes in the Portuguese Durum Wheat Landrace PI 192051. Genes Genomes Genet. 2019, 9, 2535–2547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perlin, M.H.; Andrews, J.; Toh, S.S. Essential Letters in the Fungal Alphabet: ABC and MFS Transporters and Their Roles in Survival and Pathogenicity. Adv. Genet. 2014, 85, 201–253. [Google Scholar] [CrossRef]
- Gou, J.Y.; Li, K.; Wu, K.; Wang, X.; Lin, H.; Cantu, D.; Uauy, C.; Dobon-Alonso, A.; Midorikawa, T.; Inoue, K.; et al. Wheat Stripe Rust Resistance Protein WKS1 Reduces the Ability of the Thylakoid-Associated Ascorbate Peroxidase to Detoxify Reactive Oxygen Species. Plant Cell 2015, 27, 1755–1770. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wang, B.; Zhang, Q.; Fu, Y.; Huang, L.; Wang, X.; Kang, Z. Exploration of microRNAs and their targets engaging in the resistance interaction between wheat and stripe rust. Front. Plant Sci. 2015, 6, 469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowell, J.B. Controlled infection by Puccinia graminis f.sp. tritici under artificial conditions. In The Cereal Rusts, Vol 1: Origins, Specificity, Structure and Physiology; Bushnell, W.R., Roelfs, A.P., Eds.; Academic Press: Orlando, FL, USA, 1984; pp. 292–332. [Google Scholar]
- Spanic, V.; Rouse, M.N.; Kolmer, J.A.; Anderson, J.A. Leaf and stem seedling rust resistance in wheat cultivars grown in Croatia. Euphytica 2015, 203, 437–448. [Google Scholar] [CrossRef]
- Pretorius, Z.A.; Jin, Y.; Bender, C.M.; Herselman, L.; Prin, R. Seedling resistance to stem rust race Ug99 and marker analysis for Sr2, Sr24 and Sr31 in South African wheat cultivars and lines. Euphytica 2012, 186, 15–23. [Google Scholar] [CrossRef]
- Stakman, E.C.; Stewart, D.M.; Loegering, W.Q. Identification of physiologic races of Puccinia graminis var. tritici. US Agric. Res. Serv. 1962, 617, 1–53. [Google Scholar]
- Zhang, D.; Bowden, R.; Bai, G. A method to linearize Stakman infection type ratings for statistical analysis. In Proceedings of the Borlaug Global Rust Initiative 2011 Technical Workshop, Saint Paul, MN, USA, 13–16 June 2011. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.; Todhunter, R.; Tiwari, H.; Gore, M.; Bradbury, P.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Edae, E.A.; Bowden, R.L.; Poland, J. Application of population sequencing (POPSEQ) for ordering and imputing genotyping-by-sequencing markers in hexaploid wheat. G3 Genes Genomes Genet. 2015, 5, 2547–2553. [Google Scholar] [CrossRef] [Green Version]
- BLAST Tool. Triticum Aestivum. Available online: https://plants.ensembl.org/Triticum_aestivum/Tools/Blast (accessed on 4 October 2021).
- EnsemblPlant. Triticum Aestivum. Available online: https://plants.ensembl.org/Triticum_aestivum/Info/Index (accessed on 7 October 2021).
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).