Characterization of Rhizosphere and Endophytic Microbial Communities Associated with Stipa purpurea and Their Correlation with Soil Environmental Factors
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites and Sample Collection
2.2. Determination of Soil Physicochemical Properties
2.3. Determination of Soil Enzymatic Activities
2.4. Genomic DNA Extraction and PCR Amplification
2.5. Illumina MiSeq Sequencing and Processing
2.6. Bioinformatics and Statistical Analysis
3. Results
3.1. Soil Physicochemical Properties Subsection
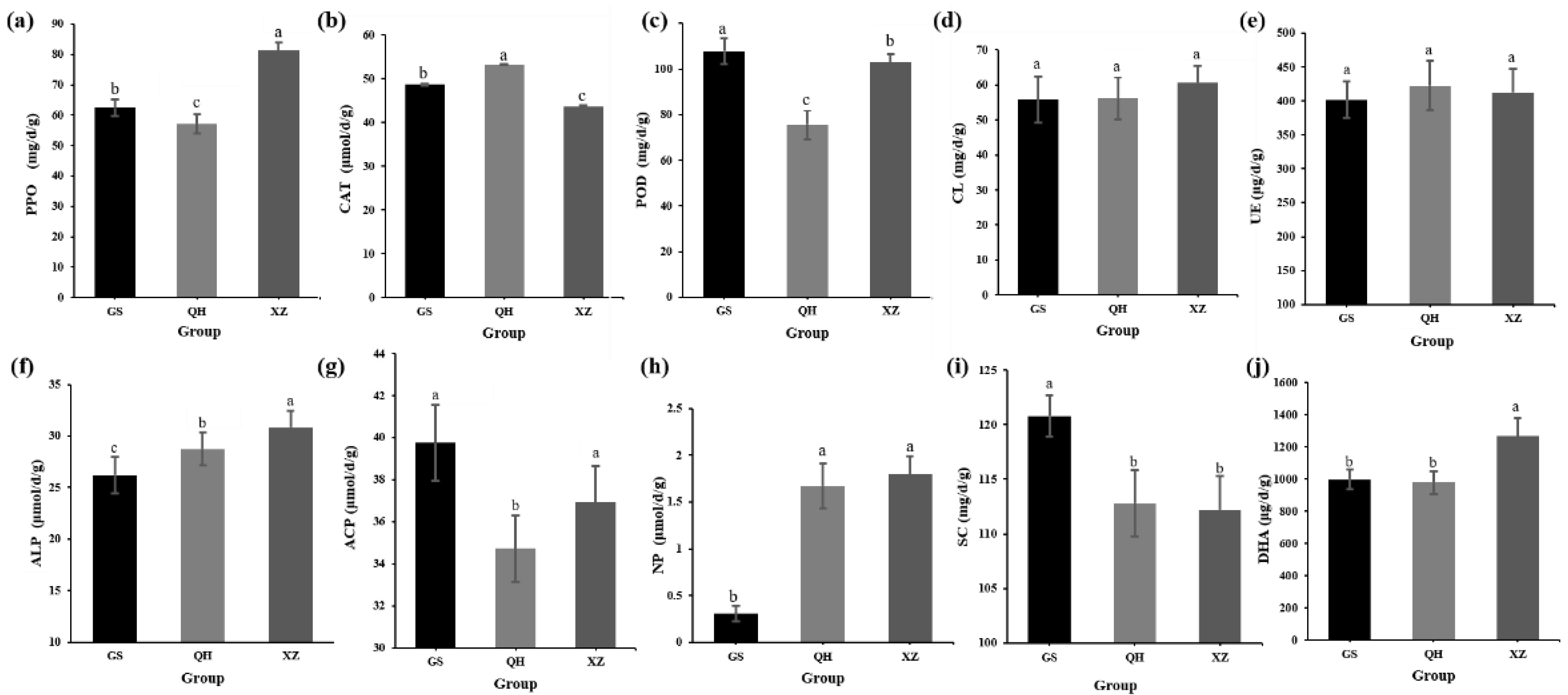
3.2. Soil Enzymatic Activity
3.3. Sequencing and Microbial Community Alpha Diversity
3.4. The Degree of Microbial Species (OTU) Overlap and Dissimilarity in Microbial Community Composition within and between Habitats
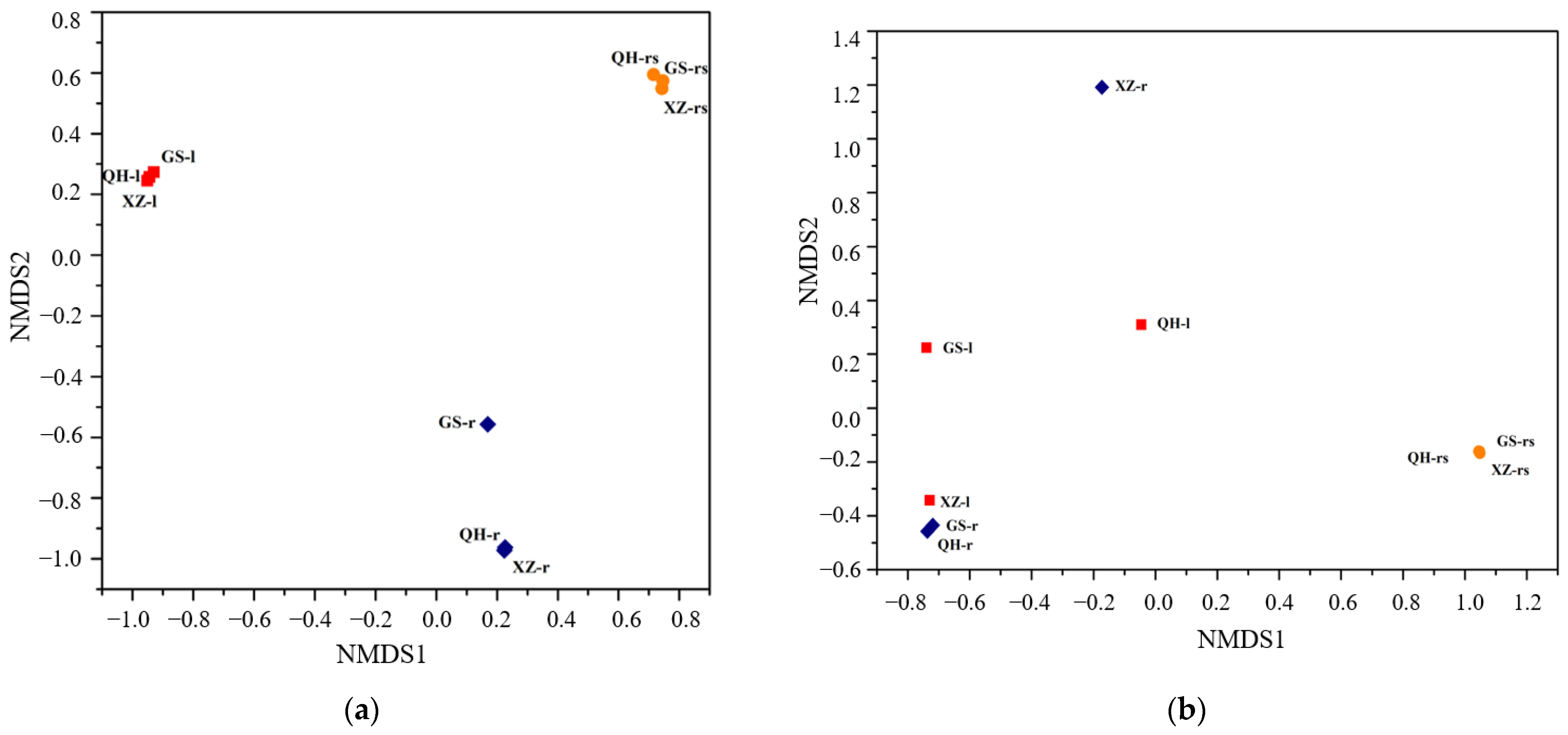
3.5. Microbial Community Similarities in Different Samples
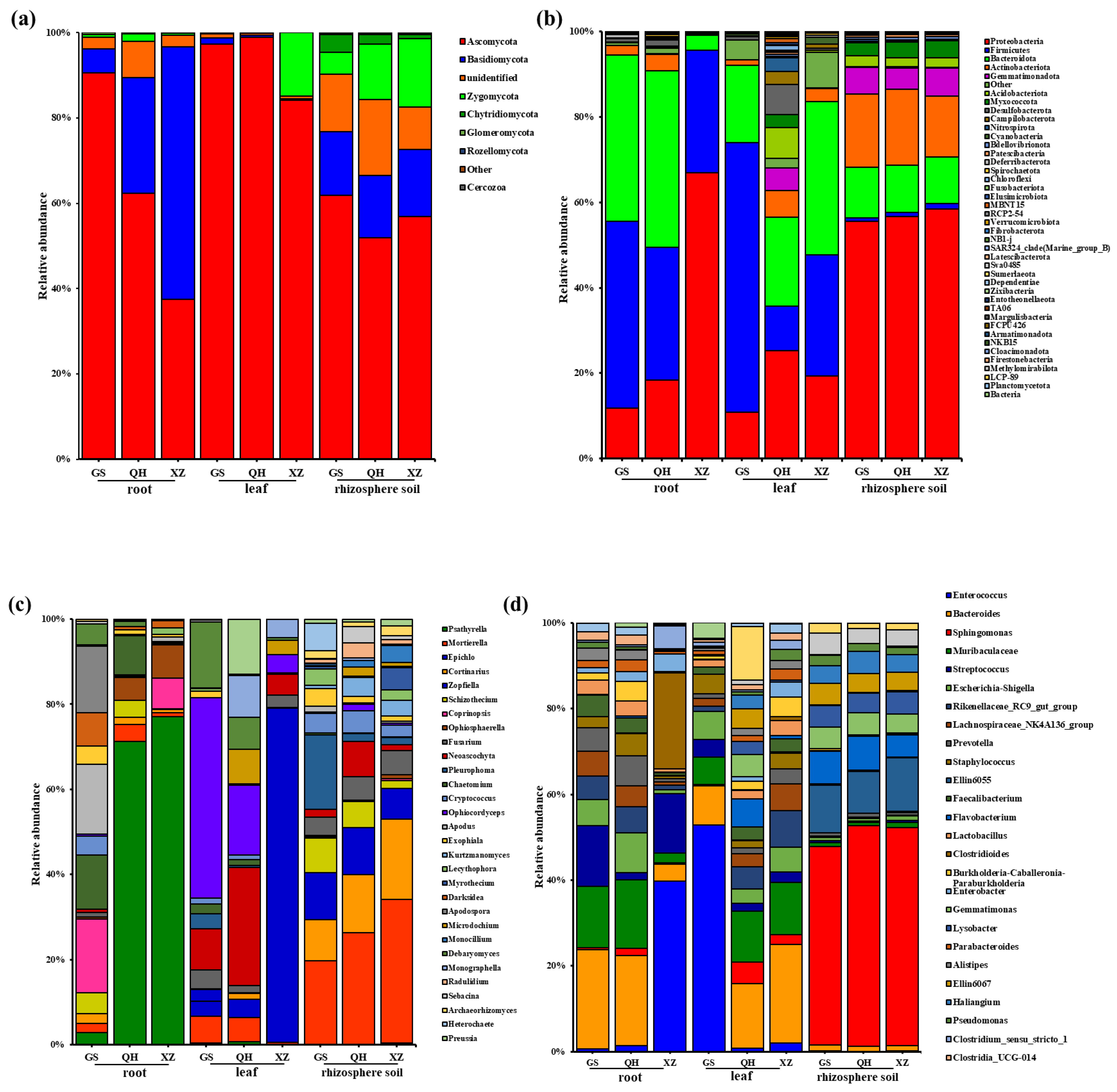
3.6. Microbial Community Composition Analysis
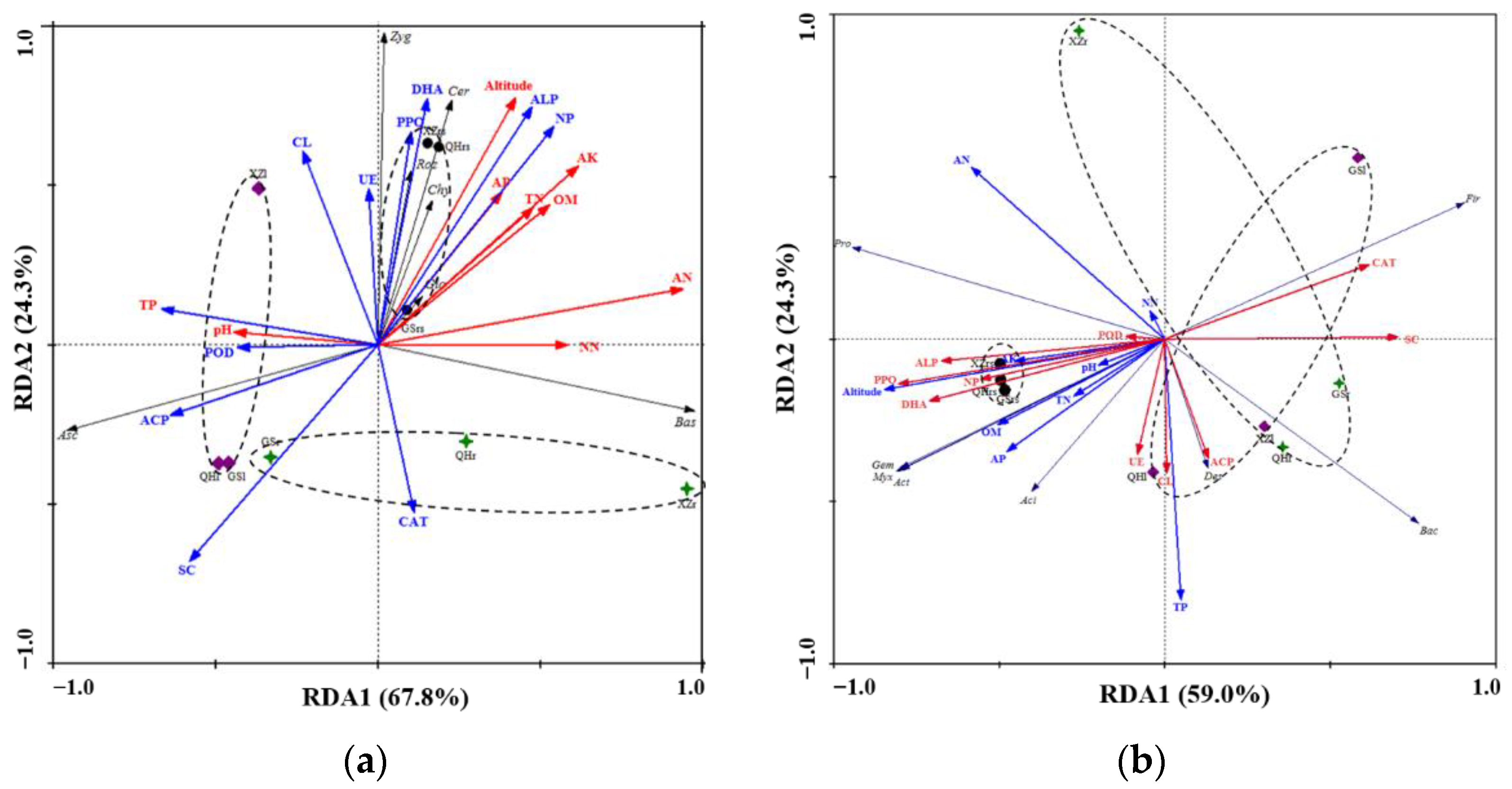
3.7. Relationship between Microbial Community and Environmental Variables
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| GS | QH | XZ | ||||||||||||||||||||||
| Ascomycota | Basidiomycota | Zygomycota | Shannon | Ascomycota | Basidiomycota | Zygomycota | Shannon | Ascomycota | Basidiomycota | Zygomycota | Shannon | |||||||||||||
| CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | |
| Altitude | −0.378 | 0.753 | 0.117 | 0.925 | 0.449 | 0.703 | −0.774 | 0.437 | −0.917 | 0.261 | 0.398 | 0.739 | 0.845 | 0.359 | −0.998 | 0.041 | −0.796 | 0.414 | 0.855 | 0.348 | 0.097 | 0.938 | −0.117 | 0.925 |
| pH | −0.133 | 0.915 | −0.136 | 0.913 | 0.660 | 0.542 | −0.590 | 0.599 | −0.954 | 0.193 | 0.901 | 0.285 | 0.289 | 0.814 | −0.796 | 0.414 | −0.178 | 0.886 | −0.851 | 0.353 | 0.843 | 0.362 | −0.832 | 0.375 |
| AN | 0.948 | 0.206 | −0.999 | 0.034 | 0.863 | 0.337 | 0.682 | 0.522 | −0.962 | 0.175 | 0.518 | 0.653 | 0.765 | 0.445 | −0.997 | 0.045 | −1.000 | 0.009 | 0.379 | 0.753 | 0.669 | 0.533 | −0.684 | 0.520 |
| TN | 0.948 | 0.206 | −0.999 | 0.034 | 0.863 | 0.337 | 0.682 | 0.522 | 0.973 | 0.149 | −0.870 | 0.329 | −0.354 | 0.770 | 0.836 | 0.370 | −0.056 | 0.965 | 0.950 | 0.203 | −0.695 | 0.511 | 0.680 | 0.524 |
| TP | −0.991 | 0.085 | 0.991 | 0.087 | −0.752 | 0.458 | −0.808 | 0.401 | 0.994 | 0.072 | −0.803 | 0.406 | −0.464 | 0.693 | 0.896 | 0.293 | −0.627 | 0.568 | 0.954 | 0.193 | −0.145 | 0.907 | 0.125 | 0.920 |
| AK | 0.662 | 0.539 | −0.838 | 0.367 | 1.000 | 0.004 | 0.225 | 0.856 | 0.997 | 0.050 | −0.782 | 0.428 | −0.495 | 0.670 | 0.911 | 0.270 | −0.283 | 0.817 | −0.789 | 0.421 | 0.896 | 0.293 | −0.887 | 0.306 |
| NN | −0.991 | 0.085 | 0.991 | 0.087 | −0.752 | 0.458 | −0.808 | 0.401 | −0.499 | 0.668 | −0.227 | 0.854 | 0.997 | 0.047 | −0.763 | 0.447 | 0.907 | 0.277 | 0.061 | 0.961 | −0.926 | 0.247 | 0.933 | 0.234 |
| OM | −0.998 | 0.036 | 0.977 | 0.136 | −0.699 | 0.507 | −0.851 | 0.352 | 0.937 | 0.227 | −0.923 | 0.252 | −0.238 | 0.847 | 0.763 | 0.447 | 0.550 | 0.629 | 0.576 | 0.609 | −0.986 | 0.105 | 0.983 | 0.118 |
| AP | 0.844 | 0.360 | −0.956 | 0.189 | 0.959 | 0.183 | 0.486 | 0.677 | 0.607 | 0.584 | −0.986 | 0.106 | 0.316 | 0.795 | 0.302 | 0.805 | 0.656 | 0.544 | 0.462 | 0.694 | −1.000 | 0.020 | 0.999 | 0.033 |
| PPO | −0.542 | 0.635 | 0.299 | 0.807 | 0.277 | 0.822 | −0.877 | 0.319 | −0.558 | 0.623 | 0.974 | 0.145 | −0.373 | 0.757 | −0.243 | 0.843 | 0.523 | 0.650 | −0.985 | 0.112 | 0.270 | 0.826 | −0.250 | 0.839 |
| CAT | 0.343 | 0.777 | −0.581 | 0.606 | 0.933 | 0.234 | −0.147 | 0.906 | −0.065 | 0.958 | −0.633 | 0.563 | 0.862 | 0.338 | −0.400 | 0.738 | 0.753 | 0.458 | −0.888 | 0.304 | −0.028 | 0.982 | 0.049 | 0.969 |
| POD | 0.995 | 0.064 | −0.986 | 0.108 | 0.730 | 0.479 | 0.827 | 0.381 | 0.997 | 0.052 | 0.784 | 0.427 | 0.493 | 0.672 | −0.910 | 0.272 | −0.754 | 0.456 | 0.887 | 0.306 | 0.031 | 0.980 | −0.051 | 0.967 |
| CL | 0.662 | 0.539 | −0.439 | 0.711 | −0.129 | 0.918 | 0.939 | 0.223 | 1.000 | 0.012 | −0.744 | 0.466 | −0.546 | 0.633 | 0.934 | 0.233 | 0.556 | 0.625 | 0.977 | 0.137 | −0.232 | 0.851 | 0.212 | 0.864 |
| UE | −0.921 | 0.255 | 0.784 | 0.427 | −0.311 | 0.798 | −0.995 | 0.061 | 0.819 | 0.389 | −0.207 | 0.868 | −0.935 | 0.231 | 0.965 | 0.169 | 0.451 | 0.702 | −0.996 | 0.059 | 0.349 | 0.773 | −0.329 | 0.786 |
| sDHA | 0.386 | 0.748 | −0.126 | 0.920 | −0.442 | 0.709 | 0.779 | 0.431 | 0.548 | 0.631 | 0.170 | 0.891 | −1.000 | 0.010 | 0.800 | 0.410 | −0.458 | 0.697 | −0.660 | 0.541 | 0.963 | 0.173 | −0.958 | 0.186 |
| ACP | −0.852 | 0.351 | 0.681 | 0.523 | −0.165 | 0.895 | −0.999 | 0.035 | 0.673 | 0.530 | −0.997 | 0.052 | 0.234 | 0.849 | 0.381 | 0.751 | −0.021 | 0.987 | −0.923 | 0.252 | 0.748 | 0.462 | −0.734 | 0.475 |
| NP | 0.536 | 0.640 | −0.292 | 0.811 | −0.284 | 0.817 | 0.874 | 0.323 | 0.895 | 0.295 | −0.349 | 0.773 | −0.872 | 0.326 | 0.993 | 0.074 | 0.277 | 0.821 | 0.793 | 0.417 | −0.893 | 0.297 | 0.884 | 0.310 |
| ALP | 0.807 | 0.402 | −0.935 | 0.230 | 0.975 | 0.141 | 0.428 | 0.719 | 0.707 | 0.500 | −0.999 | 0.022 | 0.188 | 0.880 | 0.425 | 0.721 | −0.909 | 0.274 | −0.055 | 0.965 | 0.923 | 0.251 | −0.931 | 0.238 |
| SC | 0.737 | 0.472 | −0.530 | 0.644 | −0.025 | 0.984 | 0.970 | 0.156 | 0.229 | 0.853 | −0.832 | 0.375 | 0.677 | 0.527 | −0.115 | 0.927 | −0.162 | 0.897 | 0.978 | 0.135 | −0.614 | 0.579 | 0.598 | 0.592 |
Appendix B
| GS | QH | XZ | ||||||||||||||||||||||
| Proteobacteria | Firmicutes | Bacteroidota | Shannon | Proteobacteria | Firmicutes | Bacteroidota | Shannon | Proteobacteria | Firmicutes | Bacteroidota | Shannon | |||||||||||||
| CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | CC | P | |
| Altitude | 0.198 | 0.873 | −0.060 | 0.962 | 0.160 | 0.897 | −0.774 | 0.437 | −0.933 | 0.233 | 0.733 | 0.477 | 0.233 | 0.850 | −0.147 | 0.906 | 0.750 | 0.460 | −0.367 | 0.761 | 0.183 | 0.883 | −0.117 | 0.925 |
| pH | 0.439 | 0.711 | −0.309 | 0.800 | 0.404 | 0.736 | −0.590 | 0.599 | −0.940 | 0.221 | 0.999 | 0.022 | −0.461 | 0.695 | 0.827 | 0.381 | 0.248 | 0.840 | −0.662 | 0.539 | −0.959 | 0.183 | −0.832 | 0.375 |
| AN | 0.965 | 0.168 | −0.992 | 0.079 | 0.975 | 0.143 | 0.682 | 0.522 | −0.973 | 0.148 | 0.818 | 0.391 | 0.100 | 0.936 | 0.939 | 0.223 | 0.996 | 0.055 | −0.848 | 0.355 | −0.437 | 0.712 | −0.684 | 0.520 |
| TN | 0.965 | 0.168 | −0.992 | 0.079 | 0.975 | 0.143 | 0.682 | 0.522 | 0.962 | 0.177 | −0.995 | 0.066 | 0.399 | 0.739 | −0.995 | 0.061 | −0.016 | 0.990 | 0.470 | 0.689 | 0.867 | 0.332 | 0.680 | 0.524 |
| TP | −0.899 | 0.289 | 0.951 | 0.200 | −0.915 | 0.264 | −0.808 | 0.401 | 0.988 | 0.100 | −0.975 | 0.143 | 0.285 | 0.816 | 0.779 | 0.431 | 0.570 | 0.614 | −0.133 | 0.915 | 0.414 | 0.728 | 0.125 | 0.920 |
| AK | 0.966 | 0.165 | −0.921 | 0.254 | 0.956 | 0.190 | 0.225 | 0.856 | 0.993 | 0.077 | −0.966 | 0.166 | 0.251 | 0.839 | −0.999 | 0.035 | 0.351 | 0.771 | −0.739 | 0.471 | −0.984 | 0.114 | −0.887 | 0.306 |
| NN | −0.899 | 0.289 | 0.951 | 0.200 | −0.915 | 0.264 | −0.808 | 0.401 | −0.536 | 0.640 | 0.183 | 0.883 | 0.767 | 0.444 | 0.874 | 0.323 | −0.935 | 0.231 | 0.994 | 0.070 | 0.784 | 0.426 | 0.933 | 0.234 |
| OM | −0.862 | 0.338 | 0.924 | 0.249 | −0.881 | 0.313 | −0.851 | 0.352 | 0.921 | 0.254 | −1.000 | 0.011 | 0.507 | 0.662 | 0.428 | 0.719 | −0.609 | 0.583 | 0.903 | 0.283 | 0.993 | 0.074 | 0.983 | 0.118 |
| AP | 1.000 | 0.014 | −0.993 | 0.075 | 1.000 | 0.011 | 0.486 | 0.677 | 0.572 | 0.612 | −0.837 | 0.369 | 0.888 | 0.304 | 0.970 | 0.156 | −0.709 | 0.499 | 0.952 | 0.198 | 0.969 | 0.159 | 0.999 | 0.033 |
| PPO | 0.014 | 0.991 | 0.125 | −0.716 | −0.025 | 0.984 | −0.877 | 0.319 | −0.522 | 0.651 | 0.802 | 0.407 | −0.914 | 0.266 | −0.877 | 0.319 | −0.460 | 0.695 | 0.006 | 0.996 | −0.526 | 0.647 | −0.250 | 0.839 |
| CAT | 0.806 | 0.404 | −0.716 | 0.492 | 0.782 | 0.428 | −0.147 | 0.906 | −0.109 | 0.931 | −0.270 | 0.826 | 0.971 | 0.153 | −0.147 | 0.906 | −0.703 | 0.504 | 0.303 | 0.804 | −0.250 | 0.839 | 0.049 | 0.969 |
| POD | 0.884 | 0.310 | −0.940 | 0.221 | 0.901 | 0.285 | 0.827 | 0.381 | −0.992 | 0.079 | 0.967 | 0.164 | −0.253 | 0.837 | 0.827 | 0.381 | 0.705 | 0.502 | −0.305 | 0.802 | 0.247 | 0.841 | −0.051 | 0.967 |
| CL | 0.136 | 0.913 | −0.273 | 0.824 | 0.174 | 0.888 | 0.939 | 0.223 | 0.998 | 0.040 | −0.949 | 0.203 | 0.193 | 0.876 | 0.939 | 0.223 | 0.494 | 0.671 | −0.045 | 0.972 | 0.493 | 0.672 | 0.212 | 0.864 |
| UE | −0.550 | 0.629 | 0.661 | 0.540 | −0.582 | 0.604 | −0.995 | 0.061 | 0.843 | 0.362 | −0.582 | 0.605 | −0.423 | 0.722 | −0.995 | 0.061 | −0.385 | 0.748 | −0.077 | 0.951 | −0.595 | 0.594 | −0.329 | 0.786 |
| sDHA | −0.190 | 0.878 | 0.052 | 0.967 | −0.152 | 0.903 | 0.779 | 0.431 | 0.584 | 0.603 | −0.240 | 0.846 | −0.728 | 0.481 | 0.779 | 0.431 | 0.521 | 0.651 | −0.852 | 0.350 | −1.000 | 0.006 | −0.958 | 0.186 |
| ACP | −0.419 | 0.725 | 0.541 | 0.636 | −0.453 | 0.700 | −0.999 | 0.035 | 0.640 | 0.558 | −0.880 | 0.315 | 0.846 | 0.358 | −0.999 | 0.035 | 0.093 | 0.941 | −0.536 | 0.640 | −0.903 | 0.283 | −0.734 | 0.475 |
| NP | −0.021 | 0.986 | −0.118 | 0.925 | 0.017 | 0.989 | 0.874 | 0.323 | 0.913 | 0.267 | −0.696 | 0.510 | −0.283 | 0.817 | 0.874 | 0.323 | −0.345 | 0.775 | 0.735 | 0.475 | 0.983 | 0.118 | 0.884 | 0.310 |
| ALP | 0.999 | 0.028 | −0.983 | 0.117 | 0.997 | 0.053 | 0.428 | 0.719 | 0.676 | 0.528 | −0.902 | 0.284 | 0.820 | 0.388 | 0.428 | 0.719 | 0.937 | 0.228 | −0.993 | 0.073 | −0.781 | 0.430 | −0.931 | 0.238 |
| SC | 0.239 | 0.846 | −0.372 | 0.757 | 0.276 | 0.822 | 0.970 | 0.156 | 0.187 | 0.881 | −0.539 | 0.637 | 0.998 | 0.036 | 0.970 | 0.156 | 0.090 | 0.942 | 0.373 | 0.757 | 0.809 | 0.400 | 0.598 | 0.592 |
References
- Gunatilaka, A.A.L. Natural products from plant-associated microorganisms: Distribution, structural diversity, bioactivity, and implications of their occurrence. J. Nat. Prod. 2006, 69, 509–526. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, C.E.; Gundel, P.E.; Helander, M.; Saikkonen, K. Endophytic mediation of reactive oxygen species and antioxidant activity in plants: A review. Fungal Divers. 2012, 54, 1–10. [Google Scholar] [CrossRef]
- Yuan, Z.L.; Zhang, C.L.; Lin, F.C. Recent advances on physiological and molecular basis of fungal endophyte-plant interactions. Acta Ecol. Sinica 2008, 28, 4430–4439. [Google Scholar]
- Badri, D.V.; Weir, T.L.; Lelie, D.; Vivanco, J.M. Rhizosphere chemical dialogues: Plant–microbe interactions. Curr. Opin. Biotechnol. 2009, 20, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Bakker, P.A.H.M.; Doornbos, R.F.; Christos, Z.; Berendsen, R.L.; Pieterse, C.M.J. Induced systemic resistance and the rhizosphere microbiome. Plant Pathol. J. 2013, 29, 136–143. [Google Scholar] [CrossRef]
- Qu, T.B.; Zhang, J.F.; Du, W.C.; Pang, S.; Xia, Y. Research progress on effects of grazing on diversity of microorganisms in grassland soil. Contemp. Eco-Agric. 2012, 14–20. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?dbcode=CJFD&dbname=CJFD2012&filename=DDSN2012Z2004&uniplatform=NZKPT&v=9XB6KdzfkJXUzWiU3GhGWOF8aVGUhkT15Ay5b-ZGK4R7WEdgjzI7dke0gC-wBB6G (accessed on 29 December 2021). (In Chinese).
- Singh, D.; Shi, L.; Adams, J.M. Bacterial diversity in the mountains of South-west China: Climate dominates over soil parameters. J. Microbiol. 2013, 51, 439–447. [Google Scholar] [CrossRef]
- Xu, M.; Li, X.L.; Cai, X.B.; Gai, J.P.; Li, X.L.; Christie, P.; Zhang, J.L. Soil microbial community structure and activity along a montane elevational gradient on the Tibetan Plateau. Eur. J. Soil Biol. 2014, 64, 6–14. [Google Scholar] [CrossRef]
- Bowles, T.M.; Acostamartinez, V.; Calderon, F.J.; Jackson, L.E. Soil enzyme activities, microbial communities, and carbon and nitrogen availability in organic agroecosystems across an intensively-managed agricultural landscape. Soil Biol. Biochem. 2014, 68, 252–262. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Yang, X.Y.; Liu, H.Y.; Zhao, Y.H.; Zheng, W.L.; Cheng, J.N.; Min, D.; Xu, X.X.; Qin, B. The structure of rhizosphere soil and endophytic fungal communities associated with Stipa purpurea and their correlation with soil environmental factors. Acta Microbiol. Sinica 2021, 1–34. [Google Scholar] [CrossRef]
- Liu, W.S.; Dong, M.; Song, Z.P.; Wei, W. Genetic diversity pattern of Stipa purpurea populations in the hinterland of Qinghai-Tibet Plateau. Annals Appl. Biol. 2010, 154, 57–65. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Li, X.; Kong, X.X.; Ma, L.; Hu, X.Y.; Yang, Y.P. Transcriptome analysis reveals diversified adaptation of Stipa purpurea along a drought gradient on the Tibetan Plateau. Funct. Integr. Genom. 2015, 15, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.T.; Wu, J.B.; Wang, X.D. Effects of grazing and fencing on Stipa purpurea community biomass allocation and carbon, nitrogen and phosphorus pools on the northern Tibet Plateau alpine. Pratacultural Sci. 2015, 32, 1878–1886. [Google Scholar] [CrossRef]
- Li, X.; Yang, S.H.; Yang, Y.Q.; Yin, X.; Sun, X.D.; Yang, Y.P. Comparative physiological and molecular analyses of intraspecific differences of Stipa purpurea (Poaceae) Response to Drought. Plant Divers. Resour. 2015, 37, 439–452. [Google Scholar] [CrossRef]
- Lu, D.X.; Jin, H.; Yang, X.Y.; Zhang, D.H.; Yan, Z.Q.; Li, X.Z.; Zhao, Y.H.; Han, R.B.; Qin, B. Characterization of rhizosphere and endophytic fungal communities from roots of Stipa purpurea in alpine steppe around Qinghai Lake. Canad. J. Microbiol. 2016, 62, 643–656. [Google Scholar] [CrossRef]
- Bao, G.S.; Li, C.J. Isolation and identification of endophytes infecting Stipa purpurea, a dominant grass in meadows of the Qinghai-Tibet Plateau. Acta Prataculturae Sin. 2016, 25, 32–42. [Google Scholar] [CrossRef]
- Wang, J.; Wang, G.Q.; Li, F.; Peng, Y.F.; Yang, G.B.; Yu, J.C.; Zhou, G.Y.; Yang, Y.H. Effects of short-term experimental warming on soil microbes in a typical alpine steppe. Chin. J. Plant Ecol. 2018, 42, 116–125. [Google Scholar] [CrossRef][Green Version]
- Li, H.Y.; Yao, T.; Gao, Y.M.; Zhang, J.G.; Ma, Y.C.; Lu, X.W.; Yang, X.L.; Zhang, H.R.; Xia, D.H. Relationship between soil fungal community and soil environmental faxtors in degraded alpine grassland. Acta Microbiol. Sin. 2019, 59, 678–688. [Google Scholar] [CrossRef]
- Koranda, M.; Schnecker, J.; Kaiser, C.; Fuchslueger, L.; Richter, A. Microbial processes and community composition in the rhizosphere of European beech-the influence of plant C. exudates. Soil Biol. Biochem. 2011, 43, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Cízkova, B.; Wos, B.; Pietrzykowski, M.; Frouz, J. Development of soil chemical and microbial properties in reclaimed and unreclaimed grasslands in heaps after opencast lignite mining. Ecol. Eng. 2018, 123, 103–111. [Google Scholar] [CrossRef]
- Lu, R.K. Soil and Agro-Chemistry Analytical Methods; Agriculture Science and Technology Press of China: Beijing, China, 1999; Volume 15, pp. 223–227. [Google Scholar]
- Wu, W.; Tang, X.P.; Yang, C.; Liu, H.B.; Guo, N.J. Investigation of ecological factors controlling quality of flue-cured tobacco (Nicotiana tabacum L.) using classification methods. Ecol. Inform. 2013, 16, 53–61. [Google Scholar] [CrossRef]
- Zibilsk, L.M.; Weaver, R.W.; Angle, S.; Bottomley, P.; Bezdicek, D.; Smith, S. Methods of Soil Analysis Part2-Microbiological and Biochemical Properties; Soil Science Society of America: Madison, WI, USA, 1994; pp. 835–859. [Google Scholar]
- Sinha, A.K. Colorimetric assay of catalase. Anal. Biochem. 1972, 47, 389–394. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; An, S.; Zeng, Q. Effects of stem and leaf decomposition in typical herbs on soil enzyme activity and microbial diversity in the south Ningxia loess hilly region of Northwest China. Ying Yong Sheng Tai Xue Bao 2016, 27, 3182–3188. [Google Scholar] [CrossRef]
- van Wyk, D.A.B.; Adeleke, R.; Rhode, O.H.J.; Bezuidenhout, C.C.; Mienie, C. Ecological guild and enzyme activities of rhizosphere soil microbial communities associated with Bt-maize cultivation under field conditions in North West Province of South Africa. J. Basic Microbiol. 2017, 57, 781–792. [Google Scholar] [CrossRef]
- Friedel, J.; Mölter, K.; Fischer, W. Comparison and improvement of methods for determining soil dehydrogenase activity by using triphenyltetrazolium chloride and iodonitrotetrazolium chloride. Biol. Fertil. Soils. 1994, 18, 291–296. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Chandra, J.; Retuerto, M.; Sikaroodi, M.; Jurevic, R.; Salata, R.A.; Lederman, M.M.; Gillevet, P.M.; Ghannoum, M.A. Oral mycobiome analysis of HIV-infected patients: Identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog. 2014, 10, e1003996. [Google Scholar] [CrossRef]
- Nossa, C.W.; Oberdorf, W.E.; Yang, L.; Aas, J.A.; Paster, B.J.; DeSantis, T.Z.; Brodie, E.L.; Malamud, D.; Poles, M.A.; Pei, Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010, 16, 4135–4144. [Google Scholar] [CrossRef]
- Bolger, A.M.; Marc, L.; Bjoern, U. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Kelley, S.T.; Huttley, G.A.; Knight, R.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. Vsearch: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Altschul, S.F. Basic local alignment search tool (blast). J. Mol. Biol. 2012, 215, 403–410. [Google Scholar] [CrossRef]
- Wu, X.F.; Yang, J.J.; Ruan, H.; Wang, S.N.; Yang, Y.R.; Naeem, I.; Wang, L.; Liu, L.; Wang, D. The diversity and co-occurrence network of soil bacterial and fungal communities and their implications for a new indicator of grassland degradation. Ecol. Indic. 2021, 129, 107989. [Google Scholar] [CrossRef]
- Galperin, M.Y. Genome Diversity of Spore-Forming Firmicutes. Microbiol. Spectrum. 2013, 1, TBS–0015–2012. [Google Scholar] [CrossRef]
- Xia, F.; Liu, Y.; Guo, M.Y.; Shen, G.R.; Lin, J.; Zhou, X.W. Pyrosequencing analysis revealed complex endogenetic microorganism community from natural Dong Chong Xia Cao and its microhabitat. BMC Microbiol. 2016, 16, 196. [Google Scholar] [CrossRef]
- Jin, H.; Yang, X.Y.; Liu, R.T.; Yan, Z.Q.; Li, X.D.; Li, X.Z.; Su, A.X.; Zhao, Y.H.; Qin, B. Bacterial community structure associated with the rhizosphere soils and roots of stellera chamaejasme l. along a tibetan elevation gradient. Annals Microbiol. 2018, 68. [Google Scholar] [CrossRef]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B.H. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef]
- Bardgett, R.D. The Biology of Soil: A Community and Ecosystem Approach; Oxford University Press: New York, NY, USA, 2005; pp. 57–85. [Google Scholar]
- Li, C.C.; Zhu, B.J.; Xu, L.; Li, X.Z.; Yao, M.J. Differentiations of prokaryotic communities in leaf and root endosphere of dominant plants and bulk soils in alpine meadows. Acta Ecol. Sinica 2020, 40, 4942–4953. [Google Scholar] [CrossRef]
- Cregger, M.A.; Veach, A.M.; Yang, Z.K.; Crouch, M.; Vilgalys, R.; Tuskan, G.A.; Schadt, C.W. The Populus holobiont: Dissecting the effects of plant niches and genotype on the microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Garrido-Oter, R.; González, A.; Spaepen, S.; Ackermann, G.; Lebeis, S.; McHardy, A.C.; Dangl, J.L.; Knight, R.; Ley, R.; et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Bonito, G.; Reynolds, H.; Robeson, M.S.; Nelson, J.; Hodkinson, B.P.; Tuskan, G.; Schadt, C.W.; Vilgalys, R. Plant host and soil origin influence fungal and bacterial assemblages in the roots of woody plants. Mol. Ecol. 2014, 23, 3356–3370. [Google Scholar] [CrossRef]
- Zhang, G.; Hu, A.; Yuan, Y.; Han, C. Microbial community in alpine forest soils along an altitudinal gradient on the Tibetan Plateau. In Proceedings of the American Geophysical Union, Fall Meeting, San Francisco, CA, USA, 14–18 December 2015. [Google Scholar]
- Wang, J.T.; Zheng, Y.M.; Hu, H.W.; Zhang, L.M.; Li, J.; He, J.Z. Soil pH determines the alpha diversity but not beta diversity of soil fungal community along altitude in a typical Tibetan forest ecosystem. J. Soils Sediments 2015, 15, 1224–1232. [Google Scholar] [CrossRef]
- Wang, J.T.; Cao, P.; Hu, H.W.; Li, J.; Han, L.L.; Zhang, L.M.; Zheng, Y.M.; He, J.Z. Altitudinal distribution patterns of soil bacterial and archaeal communities along Mt.Shegyla on the Tibetan Plateau. Microb. Ecol. 2015, 69, 135–145. [Google Scholar] [CrossRef]
- Deforest, J. Atmospheric nitrate deposition and the microbial degradation of cellobiose and vanillin in a northern hardwood forest. Soil Biol. Biochem. 2004, 36, 965–971. [Google Scholar] [CrossRef]
- Peng, Y.L.; Cai, X.B. Changes of arbuscular mycorrhizal fungal community in an alpine grassland altitudinal gradient. Acta Ecol. Sin. 2015, 35, 7475–7484. [Google Scholar] [CrossRef]
- Acostamartinez, V.; Zobeck, T.M.; Gill, T.E.; Kennedy, A.C. Enzyme activities and microbial community structure in semiarid agricultural soils. Biol. Fertil. Soils 2003, 38, 216–227. [Google Scholar] [CrossRef]
- Sundqvist, M.K.; Liu, Z.F.; Giesler, R.; Wardle, D.A. Plant and microbial responses to nitrogen and phosphorus addition across an elevational gradient in subarctic tundra. Ecology 2014, 95, 1819–1835. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.C.R.S.; Peixoto, R.S.; Cury, J.C.; Sul, W.J.; Pellizari, V.H.; Tiedje, J.; Rosado, A.S. Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J. 2010, 4, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Li, Z.; Li, Z.W.; Jiang, Y.H.; Weng, B.Q.; Lin, W.X. Variations of rhizosphere bacterial communities in tea (Camellia sinensis L.) continuous cropping soil by high-throughput pyrosequencing approach. J. Appl. Microbiol. 2016, 121, 787–799. [Google Scholar] [CrossRef] [PubMed]


| Samples | Vaild Tags | OUT | Chao1 | Shannon Index | Simpson Index | Coverage (%) | |
|---|---|---|---|---|---|---|---|
| fungi | GSrs | 64113 | 1037 | 1129.63 ± 9.75 aA | 7.21 ± 0.19 aA | 0.98 ± 0.01 aA | 99.76 |
| GSr | 64583 | 227 | 254.50 ± 10.99 bB | 2.81 ± 0.01 bB | 0.55 ± 0.00 bB | 99.95 | |
| GSl | 66038 | 172 | 225.22 ± 6.90 cB | 1.06 ± 0.03 cC | 0.20 ± 0.01 cC | 99.94 | |
| QHrs | 66395 | 824 | 904.02 ± 27.80 aA | 6.78 ± 0.13 aA | 0.98 ± 0.00 aA | 99.80 | |
| QHr | 65696 | 208 | 246.75 ± 17.51 bB | 4.22 ± 0.01 bA | 0.89 ± 0.00 bB | 99.93 | |
| QHl | 66323 | 133 | 180.19 ± 27.34 cAB | 0.68 ± 0.05 cB | 0.13 ± 0.01 cC | 99.94 | |
| XZrs | 63755 | 907 | 1005.35 ± 16.59 aA | 6.93 ± 0.08 aA | 0.98 ± 0.00 aA | 99.77 | |
| XZr | 66549 | 232 | 256.44 ± 22.38 bB | 3.62 ± 0.04 bB | 0.78 ± 0.00 bB | 99.96 | |
| XZl | 66234 | 149 | 208.64 ± 20.68 cC | 2.18 ± 0.01 cC | 0.63 ± 0.00 cC | 99.93 | |
| bacteria | GSrs | 60309 | 3131 | 3651.95 ± 64.65 aA | 9.50 ± 0.05 aA | 0.99 ± 0.00 aA | 98.62 |
| GSr | 60100 | 1109 | 1411.07 ± 42.62 bB | 8.06 ± 0.00 bB | 0.99 ± 0.00 bB | 99.49 | |
| GSl | 62605 | 1056 | 1370.27 ± 15.11 cB | 5.49 ± 0.06 cC | 0.83 ± 0.01 cC | 99.56 | |
| QHrs | 62815 | 2926 | 3491.50 ± 14.90 aA | 9.33 ± 0.03 bB | 0.99 ± 0.00 aA | 98.68 | |
| QHr | 58611 | 1074 | 1398.16 ± 20.88 cB | 8.30 ± 0.02 cC | 0.99 ± 0.00 bB | 99.51 | |
| QHl | 53525 | 2858 | 3048.95 ± 49.73 bAB | 10.15 ± 0.02 aA | 1.00 ± 0.00 cC | 99.43 | |
| XZrs | 59571 | 3201 | 3731.81 ± 79.12 aA | 9.34 ± 0.06 bA | 0.99 ± 0.00 aA | 98.59 | |
| XZr | 64531 | 699 | 946.17 ± 174.39 cB | 2.58 ± 0.09 cC | 0.58 ± 0.01 bB | 99.58 | |
| XZl | 60692 | 1096 | 1439.46 ± 64.45 bB | 8.07 ± 0.02 bB | 0.99 ± 0.00 cC | 99.45 |
| Samples a | Pairwise Comparison b | Kruskal–Wallis One-Way ANOVA on Ranks | Tukey Test | Spearman Rank Order Correlation | |||
|---|---|---|---|---|---|---|---|
| H | p | q | p | CC c | p | ||
| Rs | GS vs. QH | 0.00197 | 0.965 | 0.624 | 0.000232 | ||
| GS vs. XZ | 0.813 | 0.813 | 0.7 | ≤0.0001 | |||
| QH vs. XZ | 0.0289 | 0.865 | 0.761 | ≤0.0001 | |||
| L | GS vs. QH | 0.139 | 0.709 | 0.467 | 0.00961 | ||
| GS vs. XZ | 1.779 | 0.182 | 0.532 | 0.00265 | |||
| QH vs. XZ | 3.221 | 0.073 | 0.732 | ≤0.0001 | |||
| R | GS vs. QH | 1.977 | 0.16 | 0.508 | 0.00436 | ||
| GS vs. XZ | 3.221 | 0.073 | 0.462 | 0.0105 | |||
| QH vs. XZ | 0.395 | 0.53 | 0.604 | ≤0.0001 | |||
| GS | Rs vs. L | 3.162 | 0.075 | 0.0809 | 0.669 | ||
| Rs vs. R | 0.527 | 0.468 | −0.137 | 0.466 | |||
| L vs. R | 0.578 | 0.447 | 0.205 | 0.274 | |||
| QH | Rs vs. L | 1.385 | 0.239 | 0.116 | 0.539 | ||
| Rs vs. R | 5.857 | 0.016 | 3.419 | <0.05 | 0.0692 | 0.714 | |
| L vs. R | 1.039 | 0.308 | 0.463 | 0.0103 | |||
| XZ | Rs vs. L | 10.687 | 0.001 | 4.6 | <0.05 | −0.00771 | 0.966 |
| Rs vs. R | 10.207 | 0.001 | 4.495 | <0.05 | −0.0712 | 0.705 | |
| L vs. R | 0.0819 | 0.775 | 0.168 | 0.373 | |||
| Samples a | Pairwise Comparison b | Kruskal–Wallis One-Way ANOVA on Ranks | Tukey Test | Spearman Rank Order Correlation | |||
|---|---|---|---|---|---|---|---|
| H | p | q | p | CC c | p | ||
| Rs | GS vs. QH | 0.0315 | 0.859 | 0.972 | ≤0.0001 | ||
| GS vs. XZ | 0.0219 | 0.882 | 0.947 | ≤0.0001 | |||
| QH vs. XZ | 0.184 | 0.668 | 0.951 | ≤0.0001 | |||
| L | GS vs. QH | 6.694 | 0.01 | 3.659 | <0.05 | 0.063 | 0.739 |
| GS vs. XZ | 3.925 | 0.048 | 2.802 | <0.05 | 0.746 | ≤0.0001 | |
| QH vs. XZ | 0.0789 | 0.779 | 0.187 | 0.319 | |||
| R | GS vs. QH | 0.0919 | 0.762 | 0.904 | ≤0.0001 | ||
| GS vs. XZ | 1.181 | 0.277 | 0.575 | 0.000958 | |||
| QH vs. XZ | 2.078 | 0.149 | 0.583 | 0.000779 | |||
| GS | Rs vs. L | 0.0177 | 0.894 | −0.443 | 0.0145 | ||
| Rs vs. R | 0.184 | 0.668 | −0.27 | 0.147 | |||
| L vs. R | 0.735 | 0.391 | 0.802 | ≤0.0001 | |||
| QH | Rs vs. L | 7.4 | 0.007 | 3.847 | <0.05 | 0.495 | 0.00568 |
| Rs vs. R | 1.399 | 0.237 | −0.345 | 0.0619 | |||
| L vs. R | 0.614 | 0.433 | 0.196 | 0.296 | |||
| XZ | Rs vs. L | 2.941 | 0.086 | −0.355 | 0.0538 | ||
| Rs vs. R | 0.184 | 0.668 | −0.523 | 0.00315 | |||
| L vs. R | 4.918 | 0.027 | 3.136 | <0.05 | 0.695 | ≤0.0001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Cheng, J.; Jin, H.; Xu, Z.; Yang, X.; Min, D.; Xu, X.; Shao, X.; Lu, D.; Qin, B. Characterization of Rhizosphere and Endophytic Microbial Communities Associated with Stipa purpurea and Their Correlation with Soil Environmental Factors. Plants 2022, 11, 363. https://doi.org/10.3390/plants11030363
Liu H, Cheng J, Jin H, Xu Z, Yang X, Min D, Xu X, Shao X, Lu D, Qin B. Characterization of Rhizosphere and Endophytic Microbial Communities Associated with Stipa purpurea and Their Correlation with Soil Environmental Factors. Plants. 2022; 11(3):363. https://doi.org/10.3390/plants11030363
Chicago/Turabian StyleLiu, Haoyue, Jinan Cheng, Hui Jin, Zhongxiang Xu, Xiaoyan Yang, Deng Min, Xinxin Xu, Xiangfeng Shao, Dengxue Lu, and Bo Qin. 2022. "Characterization of Rhizosphere and Endophytic Microbial Communities Associated with Stipa purpurea and Their Correlation with Soil Environmental Factors" Plants 11, no. 3: 363. https://doi.org/10.3390/plants11030363
APA StyleLiu, H., Cheng, J., Jin, H., Xu, Z., Yang, X., Min, D., Xu, X., Shao, X., Lu, D., & Qin, B. (2022). Characterization of Rhizosphere and Endophytic Microbial Communities Associated with Stipa purpurea and Their Correlation with Soil Environmental Factors. Plants, 11(3), 363. https://doi.org/10.3390/plants11030363






