Comparison of Models for Quantification of Tomato Brown Rugose Fruit Virus Based on a Bioassay Using a Local Lesion Host
Abstract
1. Introduction
2. Results
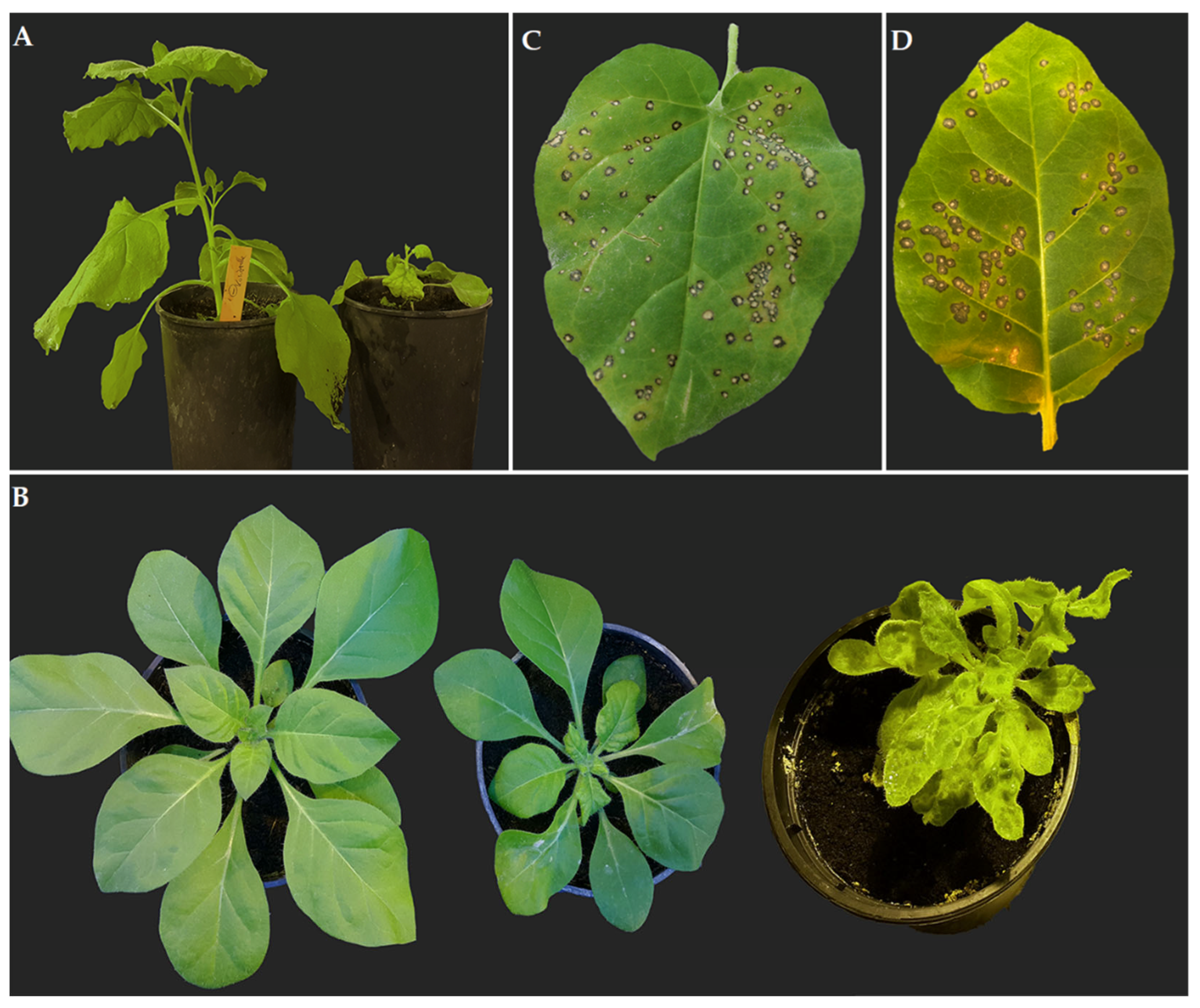
2.1. Propagation Hosts and Biotest Plants for ToBRFV Isolate DSMZ PV-1236
2.2. Fitting of the Models to Bioassay Data
2.3. Prediction of ToBRFV Concentration in a Spike Sample
2.4. Effects of Leaf Position and Local Lesion Host Species
3. Discussion
4. Materials and Methods
4.1. Virus Source, Systemic and Local Lesions Hosts
4.2. Mechanical Inoculation and Experimental Design of Local Lesion Bioassays
4.3. Purification of ToBRFV
4.4. Spectrophotometry
4.5. Correlation of Local Lesion with ToBRFV Concentration and Statistical Analyses
4.6. Detection of ToBRFV
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Luria, N.; Smith, E.; Reingold, V.; Bekelman, I.; Lapidot, M.; Levin, I.; Elad, N.; Tam, Y.; Sela, N.; Abu-Ras, A.; et al. A New Israeli Tobamovirus Isolate Infects Tomato Plants Harboring Tm-22 Resistance Genes. PLoS ONE 2017, 12, e0170429. [Google Scholar] [CrossRef] [PubMed]
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Alkowni, R.; Alabdallah, O.; Fadda, Z. Molecular identification of tomato brown rugose fruit virus in tomato in Palestine. J. Plant Pathol. 2019, 101, 719–723. [Google Scholar] [CrossRef]
- Camacho-Beltrán, E.; Pérez-Villarreal, A.; Leyva-López, N.E.; Rodríguez-Negrete, E.A.; Ceniceros-Ojeda, E.A.; Méndez-Lozano, J. Occurrence of Tomato brown rugose fruit virus Infecting Tomato Crops in Mexico. Plant Dis. 2019, 103, 1440. [Google Scholar] [CrossRef]
- Fidan, H.; Sarikaya, P.; Yildiz, K.; Topkaya, B.; Erkis, G.; Calis, O. Robust molecular detection of the new Tomato brown rugose fruit virus in infected tomato and pepper plants from Turkey. J. Integr. Agric. 2021, 20, 2170–2179. [Google Scholar] [CrossRef]
- Ling, K.S.; Tian, T.; Gurung, S.; Salati, R.; Gilliard, A. First Report of Tomato Brown Rugose Fruit Virus Infecting Greenhouse Tomato in the United States. Plant Dis. 2019, 103, 1439. [Google Scholar] [CrossRef]
- Menzel, W.; Knierim, D.; Winter, S.; Hamacher, J.; Heupel, M. First report of Tomato brown rugose fruit virus infecting tomato in Germany. New Dis. Rep. 2019, 39, 1. [Google Scholar] [CrossRef]
- Panno, S.; Caruso, A.G.; Davino, S. First Report of Tomato Brown Rugose Fruit Virus on Tomato Crops in Italy. Plant Dis. 2019, 103, 1443. [Google Scholar] [CrossRef]
- Skelton, A.; Buxton-Kirk, A.; Ward, R.; Harju, V.; Frew, L.; Fowkes, A.; Long, M.; Negus, A.; Forde, S.; Adams, I.P.; et al. First report of Tomato brown rugose fruit virus in tomato in the United Kingdom. New Dis. Rep. 2019, 40, 12. [Google Scholar] [CrossRef]
- Skelton, A.; Gentit, P.; Porcher, L.; Visage, M.; Fowkes, A.; Adams, I.P.; Harju, V.; Webster, G.; Pufal, H.; McGreig, S.; et al. First report of Tomato brown rugose fruit virus in tomato in France. New Dis. Rep. 2022, 45, e12061. [Google Scholar] [CrossRef]
- Yan, Z.Y.; Ma, H.Y.; Han, S.L.; Geng, C.; Tian, Y.P.; Li, X.D. First Report of Tomato brown rugose fruit virus Infecting Tomato in China. Plant Dis. 2019, 103, 2973. [Google Scholar] [CrossRef]
- Alfaro-Fernandez, A.; Castillo, P.; Sanahuja, E.; Rodriguez-Salido, M.D.C.; Font, M.I. First report of Tomato brown rugose fruit virus in tomato in Spain. Plant Dis. 2020, 105, 515. [Google Scholar] [CrossRef] [PubMed]
- Hasan, Z.M.; Salem, N.M.; Ismail, I.D.; Akel, E.H.; Ahmad, A.Y. First Report of Tomato Brown Rugose Fruit Virus on Greenhouse Tomato in Syria. Plant Dis. 2022, 106, 772. [Google Scholar] [CrossRef] [PubMed]
- Sabra, A.; Al-Saleh, M.A.; Al-Shahwan, I.M.; Amer, M.A. First Report of Tomato Brown Rugose Fruit Virus Infecting the Tomato Crop in Saudi Arabia. Plant Dis. 2022, 106, 1310. [Google Scholar] [CrossRef] [PubMed]
- Hamborg, Z.; Blystad, D.R. First Report of Tomato Brown Rugose Fruit Virus in Tomato in Norway. Plant Dis. 2022, 106, 2004. [Google Scholar] [CrossRef]
- Orfanidou, C.G.; Cara, M.; Merkuri, J.; Papadimitriou, K.; Katis, N.I.; Maliogka, V.I. First report of tomato brown rugose fruit virus in tomato in Albania. J. Plant Pathol. 2022, 104, 855. [Google Scholar] [CrossRef]
- Mahillon, M.; Kellenberger, I.; Dubuis, N.; Brodard, J.; Bunter, M.; Weibel, J.; Sandrini, F.; Schumpp, O. First report of Tomato brown rugose fruit virus in tomato in Switzerland. New Dis. Rep. 2022, 45, e12065. [Google Scholar] [CrossRef]
- Vučurović, A.; Brodarič, J.; Jakomin, T.; Pecman, A.; Benko Beloglavec, A.; Mehle, N. First report of Tomato brown rugose fruit virus in tomato in Slovenia. New Dis. Rep. 2022, 45, e12079. [Google Scholar] [CrossRef]
- Amer, M.A.; Mahmoud, S.Y. First report of Tomato brown rugose fruit virus on tomato in Egypt. New Dis. Rep. 2020, 41, 24. [Google Scholar] [CrossRef]
- Panno, S.; Caruso, A.G.; Blanco, G.; Davino, S. First report of Tomato brown rugose fruit virus infecting sweet pepper in Italy. New Dis. Rep. 2020, 41, 20. [Google Scholar] [CrossRef]
- Salem, N.M.; Cao, M.J.; Odeh, S.; Turina, M.; Tahzima, R. First Report of Tobacco Mild Green Mosaic Virus and Tomato Brown Rugose Fruit Virus Infecting Capsicum annuum in Jordan. Plant Dis. 2020, 104, 601. [Google Scholar] [CrossRef]
- Eldan, O.; Ofir, A.; Luria, N.; Klap, C.; Lachman, O.; Bakelman, E.; Belausov, E.; Smith, E.; Dombrovsky, A. Pepper Plants Harboring L Resistance Alleles Showed Tolerance toward Manifestations of Tomato Brown Rugose Fruit Virus Disease. Plants 2022, 11, 2378. [Google Scholar] [CrossRef] [PubMed]
- Esmaeilzadeh, F.; Koolivand, D. First report of tomato brown rugose fruit virus infecting bell pepper in Iran. J. Plant Pathol. 2022, 104, 893. [Google Scholar] [CrossRef]
- Salem, N.M.; Abumuslem, M.; Turina, M.; Samarah, N.; Sulaiman, A.; Abu-Irmaileh, B.; Ata, Y. New Weed Hosts for Tomato Brown Rugose Fruit Virus in Wild Mediterranean Vegetation. Plants 2022, 11, 2287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Griffiths, J.S.; Marchand, G.; Bernards, M.A.; Wang, A. Tomato brown rugose fruit virus: An emerging and rapidly spreading plant RNA virus that threatens tomato production worldwide. Mol. Plant Pathol. 2022, 23, 1262–1277. [Google Scholar] [CrossRef] [PubMed]
- Avni, B.; Gelbart, D.; Sufrin-Ringwald, T.; Zinger, A.; Chen, L.; MacHbash, Z.; Bekelman, I.; Segoli, M.; Dombrovsky, A.; Kamenetsky, R.; et al. Tomato genetic resistance to tobamoviruses is compromised. Acta Hortic. 2021, 1316, 89–98. [Google Scholar] [CrossRef]
- Jones, R.A.C. Global Plant Virus Disease Pandemics and Epidemics. Plants 2021, 10, 233. [Google Scholar] [CrossRef]
- Oladokun, J.O.; Halabi, M.H.; Barua, P.; Nath, P.D. Tomato brown rugose fruit disease: Current distribution, knowledge and future prospects. Plant Pathol. 2019, 68, 1579–1586. [Google Scholar] [CrossRef]
- EPPO. PM 7/146 (1) Tomato brown rugose fruit virus. EPPO Bull. 2021, 51, 178–197. [Google Scholar] [CrossRef]
- FAO. Agricultural Production Statistics 2000–2020; FAO: Rome, Italy, 2022. [Google Scholar]
- Quinet, M.; Angosto, T.; Yuste-Lisbona, F.J.; Blanchard-Gros, R.; Bigot, S.; Martinez, J.-P.; Lutts, S. Tomato fruit development and metabolism. Front. Plant Sci. 2019, 10, 1554. [Google Scholar] [CrossRef]
- Caruso, A.G.; Bertacca, S.; Parrella, G.; Rizzo, R.; Davino, S.; Panno, S. Tomato brown rugose fruit virus: A pathogen that is changing the tomato production worldwide. Ann. Appl. Biol. 2022, 181, 258–274. [Google Scholar] [CrossRef]
- EPPO. Tomato Brown Rugose Fruit Virus; EPPO datasheets on pests recommended for regulation; 9 September 2022; EPPO: Luxembourg, 2022. [Google Scholar]
- Balique, F.; Colson, P.; Raoult, D. Tobacco mosaic virus in cigarettes and saliva of smokers. J. Clin. Virol. 2012, 55, 374–376. [Google Scholar] [CrossRef] [PubMed]
- Castello, J.D.; Rogers, S.O.; Starmer, W.T.; Catranis, C.M.; Ma, L.; Bachand, G.D.; Zhao, Y.; Smith, J.E. Detection of tomato mosaic tobamovirus RNA in ancient glacial ice. Polar Biol. 1999, 22, 207–212. [Google Scholar] [CrossRef]
- Ilyas, R.; Rohde, M.J.; Richert-Poggeler, K.R.; Ziebell, H. To Be Seen or Not to Be Seen: Latent Infection by Tobamoviruses. Plants 2022, 11, 2166. [Google Scholar] [CrossRef] [PubMed]
- Creager, A.N.; Scholthof, K.-B.G.; Citovsky, V.; Scholthof, H.B. Tobacco mosaic virus: Pioneering research for a century. Plant Cell 1999, 11, 301–308. [Google Scholar] [CrossRef]
- Mrkvová, M.; Hančinský, R.; Grešíková, S.; Kaňuková, Š.; Barilla, J.; Glasa, M.; Hauptvogel, P.; Kraic, J.; Mihálik, D. Evaluation of new polyclonal antibody developed for serological diagnostics of tomato mosaic virus. Viruses 2022, 14, 1331. [Google Scholar] [CrossRef] [PubMed]
- Panno, S.; Caruso, A.G.; Barone, S.; Lo Bosco, G.; Rangel, E.A.; Davino, S. Spread of Tomato Brown Rugose Fruit Virus in Sicily and Evaluation of the Spatiotemporal Dispersion in Experimental Conditions. Agronomy 2020, 10, 834. [Google Scholar] [CrossRef]
- Ehlers, J.; Nourinejhad Zarghani, S.; Kroschewski, B.; Büttner, C.; Bandte, M. Cleaning of Tomato brown rugose fruit virus (ToBRFV) from Contaminated Clothing of Greenhouse Employees. Horticulturae 2022, 8, 751. [Google Scholar] [CrossRef]
- Panno, S.; Ruiz-Ruiz, S.; Caruso, A.G.; Alfaro-Fernandez, A.; Font San Ambrosio, M.I.; Davino, S. Real-time reverse transcription polymerase chain reaction development for rapid detection of Tomato brown rugose fruit virus and comparison with other techniques. PeerJ 2019, 7, e7928. [Google Scholar] [CrossRef]
- EPPO. EPPO A2 List of Pests Recommended for Regulation as Quarantine Pests; 2022-09; EPPO: Luxembourg, 2022. [Google Scholar]
- Chanda, B.; Gilliard, A.; Jaiswal, N.; Ling, K.S. Comparative Analysis of Host Range, Ability to Infect Tomato Cultivars with Tm-22 Gene, and Real-Time Reverse Transcription PCR Detection of Tomato Brown Rugose Fruit Virus. Plant Dis. 2021, 105, 3643–3652. [Google Scholar] [CrossRef]
- Dombrovsky, A.; Mor, N.; Gantz, S.; Lachman, O.; Smith, E. Disinfection Efficacy of Tobamovirus-Contaminated Soil in Greenhouse-Grown Crops. Horticulturae 2022, 8, 563. [Google Scholar] [CrossRef]
- Davino, S.; Caruso, A.G.; Bertacca, S.; Barone, S.; Panno, S. Tomato brown rugose fruit virus: Seed transmission rate and efficacy of different seed disinfection treatments. Plants 2020, 9, 1615. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, S.; Matsushita, Y.; Usugi, T.; Tsuda, S. Disinfection of Tomato chlorotic dwarf viroid by chemical and biological agents. Crop Prot. 2010, 29, 1157–1161. [Google Scholar] [CrossRef]
- Macias, W. Methods of disinfecting cucumber seeds that originate from plants infected by cucumber green mottle mosaic tobamovirus (CGMMV). Veg. Crops Res. Bull. 2000, 53, 75–82. [Google Scholar]
- Marcussen, K.; Meyer-Kahsnitz, S. Effectiveness of disinfectants against plant viruses. Nachrichtenblatt des Deutschen Pflanzenschutzdienstes 1991, 43, 165–169. [Google Scholar]
- Holmes, F.O. Local lesions in tobacco mosaic. Bot. Gaz. 1929, 87, 39–55. [Google Scholar] [CrossRef]
- Bald, J.G. TThe use of numbers of infections for comparing the concentration of plant virus suspensions: Dilution experiments with purified suspensions. Ann. Appl. Biol. 1937, 24, 33–55. [Google Scholar] [CrossRef]
- Kleczkowski, A. Interpreting relationships between the concentrations of plant viruses and numbers of local lesions. Microbiology 1950, 4, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Furumoto, W.A.; Mickey, R. A mathematical model for the infectivity-dilution curve of tobacco mosaic virus: Theoretical considerations. Virology 1967, 32, 216–223. [Google Scholar] [CrossRef]
- Furumoto, W.A.; Mickey, R. A mathematical model for the infectivity-dilution curve of tobacco mosaic virus: Experimental tests. Virology 1967, 32, 224–233. [Google Scholar] [CrossRef]
- Gokhale, D.; Bald, J. Relationship between plant virus concentration and infectivity: A ‘growth curve’model. J. Virol. Methods 1987, 18, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Bald, J.; Iltis, R.; Schneider, S.; Gokhale, D.; Desjardins, P. Association of logistic and Poisson models of infection with some physical characteristics of a single component plant virus. J. Virol. Methods 1990, 27, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, A. The statistical analysis of plant virus assays: A transformation to include lesion numbers with small means. Microbiology 1955, 13, 91–98. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dawson, W.O.; Hilf, M.E. Host-range determinants of plant viruses. Annu. Rev. Plant Biol. 1992, 43, 527–555. [Google Scholar] [CrossRef]
- McLeish, M.J.; Fraile, A.; García-Arenal, F. Evolution of plant–virus interactions: Host range and virus emergence. Curr. Opin. Virol. 2019, 34, 50–55. [Google Scholar] [CrossRef]
- Fermin, G. Chapter 5—Host Range, Host–Virus Interactions, and Virus Transmission. In Viruses; Tennant, P., Fermin, G., Foster, J.E., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 101–134. [Google Scholar]
- Wetter, C.; Conti, M.; Altschuh, D.; Tabillion, R.; Van Regenmortel, M. Pepper mild mottle virus, a tobamovirus infecting pepper cultivars in Sicily. Phytopathology 1984, 74, 405–410. [Google Scholar] [CrossRef]
- Horvath, J. Host plants in diagnosis. In Diagnosis of Plant Virus Diseases; CRC Press: Boca Raton, FL, USA, 2019; pp. 15–48. [Google Scholar]
- Schneider, W.L.; Roossinck, M.J. Genetic diversity in RNA virus quasispecies is controlled by host-virus interactions. J. Virol. 2001, 75, 6566–6571. [Google Scholar] [CrossRef]
- Nourinejhad Zarghani, S.; Dupuis-Maguiraga, L.; Bassler, A.; Wetzel, T. Mapping of the exchangeable and dispensable domains of the RNA 2-encoded 2AHP protein of arabis mosaic nepovirus. Virology 2014, 458–459, 106–113. [Google Scholar] [CrossRef]
- Dommes, A.B.; Gross, T.; Herbert, D.B.; Kivivirta, K.I.; Becker, A. Virus-induced gene silencing: Empowering genetics in non-model organisms. J. Exp. Bot. 2019, 70, 757–770. [Google Scholar] [CrossRef]
- Roth, B.M.; Pruss, G.J.; Vance, V.B. Plant viral suppressors of RNA silencing. Virus Res. 2004, 102, 97–108. [Google Scholar] [CrossRef]
- Torre, C.; Agüero, J.; Gómez-Aix, C.; Aranda, M.A. Comparison of DAS-ELISA and qRT-PCR for the detection of cucurbit viruses in seeds. Ann. Appl. Biol. 2020, 176, 158–169. [Google Scholar] [CrossRef]
- Vargas-Hernández, B.Y.; Ramírez-Pool, J.A.; Núñez-Muñoz, L.A.; Calderón-Pérez, B.; De La Torre-Almaráz, R.; Hinojosa-Moya, J.; Xoconostle-Cázares, B.; Ruiz-Medrano, R. Development of a droplet digital polymerase chain reaction (ddPCR) assay for the detection of Tomato brown rugose fruit virus (ToBRFV) in tomato and pepper seeds. J. Virol. Methods 2022, 302, 114466. [Google Scholar] [CrossRef] [PubMed]
- Leggett, H.C.; Cornwallis, C.K.; West, S.A. Mechanisms of pathogenesis, infective dose and virulence in human parasites. PLoS Pathog. 2012, 8, e1002512. [Google Scholar] [CrossRef] [PubMed]
- Wastie, R. Mechanism of action of an infective dose of Botrytis spores on bean leaves. Trans. Br. Mycol. Soc. 1962, 45, 465–473. [Google Scholar] [CrossRef]
- Bawden, F. Quantitative methods of assaying for viruses. In Plant Viruses and Plant Diseases, 3rd ed.; Chronica Botanica Company: Waltham MA, USA, 1956; pp. 150–166. [Google Scholar]
- Roberts, D. Local-lesion assay of plant viruses. In Plant Virology; Corbett, M.K., Sisler, H.D., Eds.; University of Florida Press: Gainesville, FL, USA, 1964; pp. 194–210. [Google Scholar]
- Samuel, G.; Bald, J.G. On the use of the primary lesions in quantitative work with two plant viruses. Ann. Appl. Biol. 1933, 20, 70–99. [Google Scholar] [CrossRef]
- Spencer, E.L.; Price, W. Accuracy of the local-lesion method for measuring virus activity. I. Tobacco-mosaic virus. Am. J. Bot. 1943, 30, 280–290. [Google Scholar] [CrossRef]
- Beale, H.P. The serum reactions as an aid in the study of filterable viruses of plants. Contrib. Boyce Thompson Inst. 1934, 6, 407–435. [Google Scholar]
- Fraenkel-Conrat, H. The chemical basis of the infectivity of tobacco mosaic virus and other plant viruses. In General Virology; Elsevier: Amsterdam, The Netherlands, 1959; pp. 429–457. [Google Scholar]
- Dijkstra, J.; de Jager, C. Practical Plant Virology: Protocols and Exercises; Springer: Berlin/Heidelberg, Germany, 1998. [Google Scholar]

| Name of the Model | Hypothesis/Assumption | Equation | Model Parameters |
|---|---|---|---|
| Bald 1937 model [50] * | (i) one-particle-one-region (ii) same susceptibility of regions | ) | a |
| Kleczkowski model [51] ** | (i) minimal dose of virus is needed (ii) variable susceptibility of regions | N, ξ, λ | |
| Furumoto and Mickey model I [52,53] *** | (i) one-particle-one-region (ii) each cell is a susceptible region with max. of 400,000 (iii) susceptibility of the regions follows beta distribution | Nα and c/β | |
| Furumoto and Mickey model II [52,53] *** | (i) one-particle-one-region (ii) each cell is a susceptible region (iii) follows beta distribution | Y = N (1) (model II) | N and c |
| Gokhale and Bald model or Growth curve model [54] **** | (i) no explicit physical assumption (ii) growth curve relation between expected proportion of infected regions and the log of virus concentration | N, | |
| Bald et al., 1990 model or Modified Poisson model [55] ***** | modified Poisson model | A = 10a | A, m, b |
| Virus Concentration (mg/mL) | Mean No. of Observed LL * | ** | Computed Based on | ||||
|---|---|---|---|---|---|---|---|
| Kleczkowski Model | Furumoto and Mickey Model I | Furumoto and Mickey Model II | Growth Curve Model | Modified Poisson Model | |||
| 2 | 89.58 | 45.24 | 44.07 | 46.94 | 44.84 | 44.65 | 41.333 |
| 0.4 | 30.45 | 13.69 | 15.08 | 14.06 | 15.25 | 14.49 | 16.169 |
| 0.08 | 10.45 | 4.43 | 4.61 | 3.18 | 3.44 | 4.53 | 5.190 |
| 0.016 | 4.66 | 2.07 | 1.26 | 0.65 | 0.71 | 1.40 | 1.563 |
| 0.0032 | 0.87 | 0.37 | 0.30 | 0.13 | 0.14 | 0.43 | 0.462 |
| 0.00064 | 0.5 | 0.11 | 0.07 | 0.03 | 0.03 | 0.13 | 0.136 |
| 0.000128 | 0.79 | 0.06 | 0.01 | 0.01 | 0.01 | 0.04 | 0.040 |
| Model parameters *** | N: 9266.07 | N: 45.24 | N: 56.65 | N: 581.86 | N: 69.44 | ||
| λ: 2.00 | C: 0.91 | c/β: 0.78 | β: 20 | c: 0.78 | |||
| ξ: 5.49 | γ: 1.69 | A: 0.64 | |||||
| b: 0.76 | |||||||
| Error X2 | 0.53 | 4.88 | 4.20 | 0.4 | 1.06 | ||
| Virus Concentration (mg/mL) | Mean of Observed No. LL | * | Computed Based on | ||||
|---|---|---|---|---|---|---|---|
| Kleczkowski Model | Furumoto and Mickey Model I | Furumoto and Mickey Model II | Growth Curve Model | Modified Poisson Model | |||
| 2 | 101.59 | 87.13 | 84.09 | 91.12 | 85.95 | 85.79 | 83.78 |
| 0.4 | 34.86 | 25.91 | 30.35 | 27.37 | 30.32 | 28.84 | 36.58 |
| 0.08 | 16.21 | 12.48 | 9.79 | 6.21 | 6.92 | 9.36 | 11.39 |
| 0.016 | 4.91 | 2.62 | 2.82 | 1.28 | 1.42 | 3.00 | 3.19 |
| 0.0032 | 2.13 | 1.13 | 0.72 | 0.26 | 0.29 | 0.96 | 0.86 |
| 0.00064 | 1.09 | 0.49 | 0.16 | 0.05 | 0.06 | 0.31 | 0.23 |
| 0.000128 | 0.17 | 0.03 | 0.03 | 0.01 | 0.01 | 0.10 | 0.06 |
| Model parameters ** | N: 11,064.25 | N: 87.13 | N: 105.14 | N: 1134.53 | N: 105.14 | ||
| λ: 2.00 | C: 0.92 | c/β: 0.85 | β: 20 | A: 1.03 | |||
| ξ: 5.16 | γ: 1.64 | b: 0.82 | |||||
| c: 0.82 | |||||||
| Error X2 | 2.4 | 14.78 | 11.97 | 1.60 | 3.84 | ||
| Host | Kleczkowski Model | Furumoto and Mickey Model I | Furumoto and Mickey Model II | Growth Curve Model | Modified Poisson Model | |
|---|---|---|---|---|---|---|
| N. tabacum cv. Xanthi nc | 28.302 | 1.008 mg/mL | 0.954 mg/mL | 0.883 mg/mL | 1.030 mg/mL | 0.977 mg/mL |
| N. glutinosa | 34.867 | 0.49 mg/mL | 0.533 mg/mL | 0.473 mg/mL | 0.5266/mg/mL | 0.372 mg/mL |
| Source | df * | Sum of Squares | F Value | Pr > F |
|---|---|---|---|---|
| Treatment | 47 | 40.72 | 11.01 | <0.0001 |
| Host species (a) | 1 | 0.93 | 11.83 | 0.0007 |
| Position of the leaf (b) | 2 | 0.24 | 1.55 | 0.1890 |
| Virus concentration (c) | 7 | 36.38 | 66.03 | <0.0001 |
| a × b | 2 | 0.03 | 0.19 | 0.8252 |
| a × c | 7 | 0.69 | 1.25 | 0.27 |
| b × c | 14 | 1.44 | 1.31 | 0.20 |
| a × b × c | 14 | 1.00 | 0.91 | 0.54 |
| Error | 336 | 26.45 | ||
| Corrected Total | 383 | 67.17 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nourinejhad Zarghani, S.; Monavari, M.; Ehlers, J.; Hamacher, J.; Büttner, C.; Bandte, M. Comparison of Models for Quantification of Tomato Brown Rugose Fruit Virus Based on a Bioassay Using a Local Lesion Host. Plants 2022, 11, 3443. https://doi.org/10.3390/plants11243443
Nourinejhad Zarghani S, Monavari M, Ehlers J, Hamacher J, Büttner C, Bandte M. Comparison of Models for Quantification of Tomato Brown Rugose Fruit Virus Based on a Bioassay Using a Local Lesion Host. Plants. 2022; 11(24):3443. https://doi.org/10.3390/plants11243443
Chicago/Turabian StyleNourinejhad Zarghani, Shaheen, Mehran Monavari, Jens Ehlers, Joachim Hamacher, Carmen Büttner, and Martina Bandte. 2022. "Comparison of Models for Quantification of Tomato Brown Rugose Fruit Virus Based on a Bioassay Using a Local Lesion Host" Plants 11, no. 24: 3443. https://doi.org/10.3390/plants11243443
APA StyleNourinejhad Zarghani, S., Monavari, M., Ehlers, J., Hamacher, J., Büttner, C., & Bandte, M. (2022). Comparison of Models for Quantification of Tomato Brown Rugose Fruit Virus Based on a Bioassay Using a Local Lesion Host. Plants, 11(24), 3443. https://doi.org/10.3390/plants11243443








