Metabolomics and Network Pharmacology in the Exploration of the Multi-Targeted Therapeutic Approach of Traditional Medicinal Plants
Abstract
1. Introduction
2. Methodology
2.1. Search Strategy
2.2. Inclusion Criteria
2.3. Exclusion Criteria
2.4. Study Selection
3. Review Findings
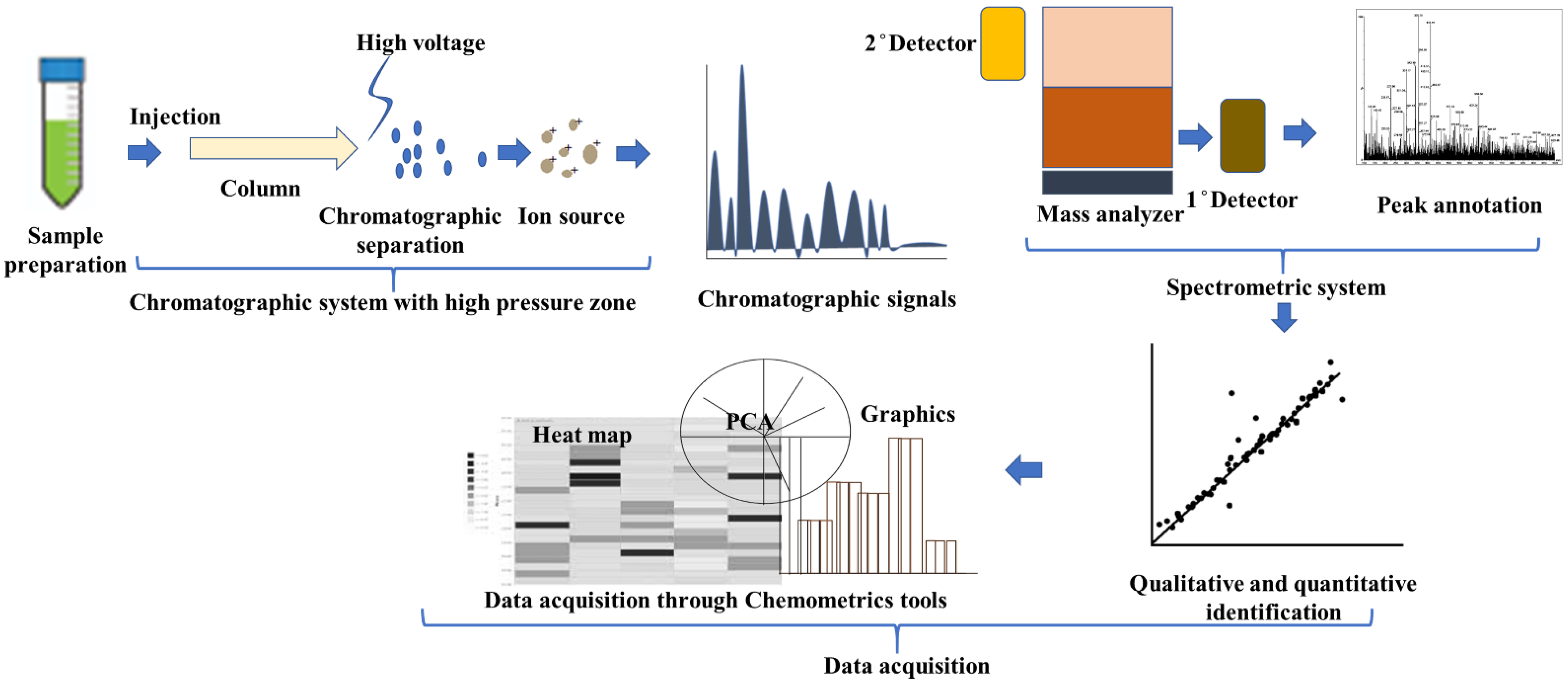
3.1. Need for Metabolomic Study for Medicinal Plants
Challenges in Metabolomics Analysis
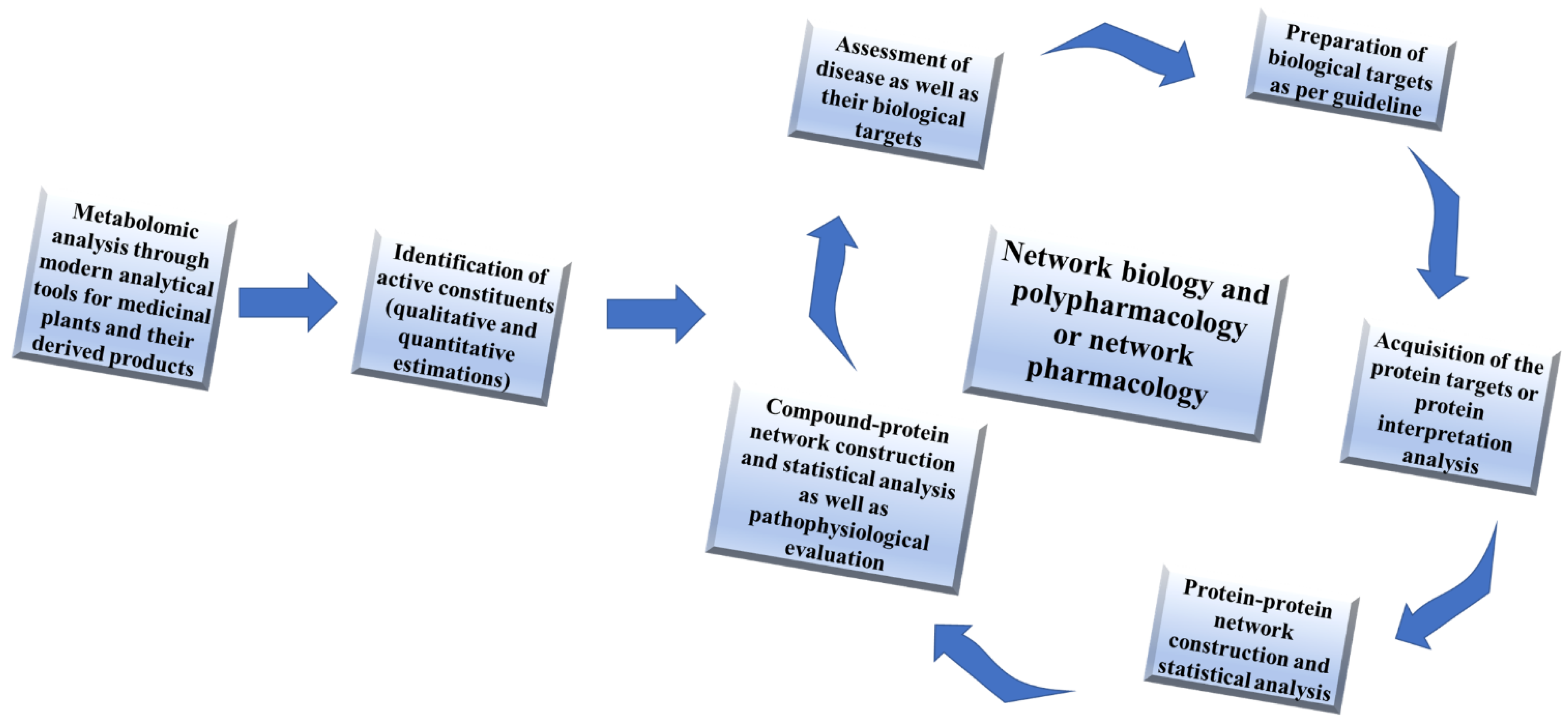
3.2. Needs of Network Pharmacology for Therapeutic Exploration of Medicinal Plants
3.2.1. Untargeted Network Pharmacology Analysis
3.2.2. Target Based Network Pharmacology Analysis
3.2.3. Gene Ontology or Gene-Disease Association Network Analysis
| Sr. No | Medicinal Plant and Part Used for Analysis | Technique | Method/Type of Analysis | Identified Metabolites (Major) | References |
|---|---|---|---|---|---|
| 1 | Achyranthes aspera (Leaf) | LITE-5MS GC-MS (Qualitative) | Column: 30 m × 0.25 mm I.D., 0.25 µm Medium: Helium as carrier gas, flow rate of 1.1 mL/min | Butanoic acid, Phytol, 1-Butanol, 6-Octen-1-ol, 3,7 Dimethyl-, Propanoate, Decanoic acid, 6-Methylfuro [2,3-C]pyrid-5-one, 3,7,11,15-Tetramethyl-2-hexadecen-1-ol, 2-Decen-1-ol, 3-Bromo-2-Methoxycyclohexanone, 2-Nonen-1-ol, Cyclohexane, 1-Hexadecyne (14.43%), 6-Octen-1-ol, Propanoate, 6-Octen-1-ol, 3,7 Dimethyl-,Propanoate. | [44] |
| 2 | Aerva javanica (Leaf) | GC-MS (Qualitative) | Column: HP-5MS (30 m × 0.025 mm) Medium: Helium as a carrier gas, flow rate of 13 mL/min | 3-allyl-6-methoxyphenol, dodecanal, (E)-6,10-dimethyl-5,9-undecadien-2-one, trans-β-ionone, β-panasinsene | [45,46] |
| 3 | Aerva lanata (Aerial parts and roots) | LC-ESI-MS/MS (Qualitative and quantitative) | Column: (30 m × 0.25 mm I.D., × 1 EMdf) Medium: Helium as carrier gas, flow rate of 1 mL/min. | Quercetin, kaempferol, and myricetin, Gallic acid, p-coumaric, Vanillic, Caffeic acid, Rutin, Ferulic acid, Astragalin | [47] |
| 4 | Allamanda cathartica (Leaf) | GC-MS (Qualitative) | Column: 30 mm in length, 0.25 mm I.D., and 1 µm Medium: Helium as a carrier gas, flow rate of 1 mL/min | Glycerin, n-hexadecanoic acid, Phytol, Thymine, Tetradecanoic acid, Dodecanoic acid, Octanoic acid, ethyl ester, | [48,49] |
| 5 | Artemisia absinthium (Aerial part) | GC/MS HS-SPME analysis (Qualitative and quantitative) | Colum: DB-5(30 × 0.2 mm, film thickness 0.32 µm) Medium: helium gas, flow rate of 1.7 mL/min | Camphor, p-cymene, Isoledene, Caryophyllene, Isopulegol Acetate, Hysterol, Isocaryophillene, Diisoamylene, β-farnesene, and Cyclohexane,2,4-diisopropyl-1,1-dimethyl | [50] |
| 6 | Aurantii fructus (Dry whole fruit part) | HP-5 MS (Qualitative and quantitative) | Column: (30.0 m × 250 μm × 0.25 μm Medium: Helium as a carrier gas, flow rate of 3 mL/min | p-Xylene, (−)-α-Pinene, α-Phellandrene, 3-Carene, d-Limonene, Ocimene, 4-Carene, Linalool, Terpineol, Limonene oxide | [51] |
| 7 | Bergenia ligulata (Rhizome) | GC-MS (Qualitative) | Colum: 100 m × 0.25 mm. I.D., 0.5 μm, Film thickness: 0.25 μm. Medium: Helium as a carrier gas, flow rate of 1 µL/min | Tadecanoic acid methyl ester, hexadecanoic acid, methyl ester, quinoline and phenol-2,4-bis (1,1- dimethylethyl), hexadecanoic acid, methyl ester, octadecanoic acid methyl ester, and 2-Propyl-5-oxohexanoic acid, aR Turmerone, 3,7,11,15-Tetramethyl-2-hexadecen-1-ol and Squalene | [52] |
| 8 | Boerhaavia diffusa (Root and whole plant) | GC-MS (Qualitative and quantitative) | Colum: 30 m × 0.25 mm I.D., × 0.25 μm Medium: Helium as a carrier gas, flow rate of 1 mL/min | Sucrose, l-Tyrosine, Malic acid, Uracil, Succinic acid, Fumaric acid, 4-Methylcatechol, D-Pinitol, | [53,54] |
| 9 | Boerhavia diffusai (Root and aerial parts) | Column diameter is 0.32 mm; column length is 30 m, column thickness 0.50 μm), Medium: Helium gas, flow rate of 1.73 mL/min | Boeravinone-B, eupalitin galactoside | [55] | |
| 10 | Carica papaya (Leaf) | GC-MS QP-2010 ULTRA | Column: 30.0 m × 0.25 mm × 0.25, Medium: Helium as carrier gas, flow rate of 1 mL/min. | Myricetin, caffeic acid, trans-ferulic acid, and kaempferol | [56] |
| 11 | Chlorophytum borivilianum (Tuber) | GC-MS (Qualitative and quantitative) | Column: 25 m × 0.25 mm I.D., × 0.25 um Medium: Helium as carrier gas, using 122.2 KPa (51.6 cm/s) | Chlorophytoside-I (3β, 5α, 22R, 25R)-26-(β-D-glucopyranosyloxy)-22-hydroxy-furostan-12-one-3 yl O-β-D-galactopyranosyl (1-4) glucopyranoside | [57] |
| 12 | Chrysanthemum morifolium (Flower) | LC-DAD-ESI/MS (Qualitative) | Column: 30 m × 0.32 mm × 0.5 µm Medium: Helium as carrier gas, flow rate of 1.5 mL/min | β-Humulene, Ledene oxide-(I), Caryophyllene, Eicosane, Heneicosane, Germacrene, Limonene, Borneol, α-Farnesol, Camphene etc. | [58] |
| 13 | Cichorium intybus (Root) | Rtx-5 MS GC-MS (Qualitative) | Column: 30.0 m × 0.20 mm I.D., 0.25 μm Medium: Helium as carrier gas, flow rate of 1 mL/min | 2-methoxy phenol, 2,3-butanedione, 2-furfurylthiol, 2-thenylthio, 1-octene-3-one, 2-ethyl-3,5-dimethylpyrazine, 3-methylbutanal, 2,3- butanedione | [59,60] |
| 14 | Dicentra spectabilis, | HP-5 MS GC-MS (Qualitative and quantitative) | Column: 30 m × 0.25 mm × 0.25 μm Medium: Helium as a carrier gas, flow rate of 1 mL/min | Hordenine, Trisphaeridine, Hamayne, Caranine, Galanthine, Homolycorine, Galwesine | [61] |
| 15 | Didymocarpus pedicellata (Leaf) | GC/FID and GC/MS (Qualitative) | Column: 30 mm in length, 0.25 mm i.d., and 1 µm Medium: Helium as carrier gas, flow rate of 1 µL/min | β-caryophyllene, α-humulene, β-selinene and α-selinene, caryophyllene oxide, veridiflorol, spathulenol, and humulene oxide | [62] |
| 16 | Dracocephalum moldavica, Ocimum americanum, Lophanthus anisatus, Monarda fistulosa, and Satureja hortensis (aerial parts) | HP-5MS GC/MS | Colum: HP-5 column (30 m × 0.25 mm i.d with 0.25 μm film thickness) Medium: Helium as carrier gas, flow rate of 1 mL/min. | Geraniol, geranyl acetate, thymol, citral, β-caryophyllene | [63] |
| 17 | Fragaria vesca (Berries) | GC-MS (Qualitative and quantitative) | Column: 0.25 mm ID 15 m length 1.0 µm Medium: Helium as carrier gas, flow rate of 1 mL/min | Eugenol, δ-Decalactone, Hexanoic acid, Terpinen-4-Ol, ϒ-Hexalactone, Ethyl pentanoate, Phenylethyl acetate | [64] |
| 18 | Genista tinctoria (NA) | HILIC (Qualitative) | Genistein, luteolin, naringenin, quercetin myricetin, apigenin, quercitrin | [65] | |
| 19 | Glycyrrhiza inflate and Glycyrrhiza echinata (Root) | GC-MS (Qualitative) | Column: (30 m × 0.32 mm × 0.25 μm) Medium: Helium as carrier gas, flow rate of 1 mL/min | Cadaverine and myo-inositol | [66] |
| 20 | Glycyrrhizae radix (Root) | GC-MS (Qualitative and quantitative) | Column: 30 m 0.25 mm Medium: Helium as carrier gas, flow rate of 1 mL/min | α-bisabolol, eudesmol, β-Pinene, Myrcenol, Germacreme, Limoneon monoxide, D-Limonene, β-Citronellal, Tricyclene | [67] |
| 21 | Gymnema sylvestre (Leaf) | GC-MS analysis was conducted using SHIMADZU, QP2010 PLUS (Qualitative) | Column: 29.3 m × 0.7 m, 320 μm. Medium: Helium as carrier gas, flow rate of 1.5 mL/min | Squalene, Tetratriacontane, Phytol, n-Hexadecanoic acid, Phthalic acid, di(2-propylpentyl) ester, Benzoic acid, 3,5-dicyclohexyl-4-hydroxy-, methyl ester | [68] |
| 22 | Hydrophilic and hydrophobic compounds mixture | HILIC-RPLC (Qualitative) | Pseudouridine, isonicotinic acid, palmatine, uracil, adenosine, nicotine, propranolol, Atenolol, 4-hydroxybenzoic Acid, hippuric acid, phenol, anisic acid, 4-nitrobenzoic acid, 4-chlorobenzoic acid, 2,6-dimethyl phenol, 2,3-dimethyl naphthalene, 4-chloro biphenyl, fluoranthene, pyrene. | [69,70] | |
| 23 | Lepidium sativum | TR5–MS GC-MS (Qualitative) | Column: (30 mm × 0.25 mm I.D., × 1 μMdf) Medium: Helium as carrier gas, flow rate of 1 mL/min | Eugenol, tigmastane-3,6-dione, n-Hexadecanoic acid, Dodecanoic acid, n-Hexadecanoic acid, Stigmasterol, etc. | [71] |
| 24 | Lolium perenne (Stem) | HP-5 MS GC–MS (Qualitative and quantitative) | Column: 30 m × 0.25 mm × 0.25 µm Medium: Helium as carrier gas, flow rate of 1 mL/min | Camphene, p-cymene, bornyl acetate, amorph-4-en-7-ol, cadinol, bisabolol, amorpha-4,7(11)-dien-8-one and 3-acetomorpha-4,7(11)-dien-8-one | [72] |
| 25 | Lupinus angustifolius (Aerial parts) | HP-5 GC-MS (Qualitative and quantitative | Column: 30 m 0.25 mm I.D., 0.25 μm) Medium: Helium as carrier gas, flow rate of 1 mL/min | 13-tigloyloxylupanine, lupanine 11,12-dehydrosparteine, and tetrahydrorhombifoline, 13-tigloyloxylupanine, tetrahydrorhombifoline, and lupanine | [73] |
| 26 | Mangifera indica (Leaf) | GC-MS (Qualitative and quantitative) | Column: PB-1 column (Supelco, USA, 30 × 0.32 mm I.D., DF = 1 μm) Medium: Helium as carrier gas, flow rate of 1 mL/min | α-thujene, camphene, R-terpinene, limonene, linalool, R-terpineol, eugenol | [74] |
| 27 | Mentha pulegium and Origanum majorana (Aerial parts) | GC-MS (Qualitative and quantitative) | Column: HP5LS (30 m × 0.025 mm) Medium: Helium as carrier gas, flow rate of 2 mL/min | α-pinene, β-pinene, 3-Octanol, Methyl cyclohexene, Menthone, Iso-menthol, Piperitenone, Dodecane, 2, 2-dimethyl propylidene | [75] |
| 28 | Morinda officinalis (Root) | HP-5 GC-MS (Qualitative and quantitative) | Colum: 30 m × 0.25 mm I.D., with 0.25 μm Medium: Helium as carrier gas, flow rate of 1 mL/min | Tecnazene, alpha-BHC, Hexachlorobenzene, beta-BHC, quintozene, gamma-BHC, delta-BHC, heptachlor, octachlorodipropyl ether, chlorothalonil, triadimefon, aldrin, dicofol, fenson, fipronil, chlorfenvinphos, heptachlor epoxide, | [76] |
| 29 | Moringa oleifera (Leaf) | GCMS (Qualitative and quantitative) | Colum: HP-5MS (30 m × 0.25 mm I.D., × 0.25 µm) Medium: Helium as carrier gas, flow rate of 1 mL/min | Propanamide, D-Mannoheptulose, N-Isopropyl-3-phenylpropanamid, 1,3-Propanediol, 2-ethyl-2- (hydroxymethyl), Propionic acid, 2-methyl-, octyl ester, Ethanamine, N-ethyl-N-nitroso, and 9,12,15-Octadecatrienoic acid, | [77] |
| 30 | Morus alba (Leaf) | HP-S GC-MS (Qualitative and quantitative) | Column: 30 m × 0.25 mm × 0.25 µm Medium: Helium as carrier gas, flow rate of 1 mL/min | 9,12,15-octadecatrienoic acidethyl ester, linolenic acid ethyl ester, gibberellic acid 4-methoxy phenol, ethyl isoallocholate, and octadecanoic acid | [78] |
| 31 | Musa paradisiaca (Fruit pulp oil) | HP-5 MS GCMS (Quantitative) | Column: (30 m × 0.25 mm × 0.25 μm. Medium: Helium as carrier gas, flow rate of 1 mL/min. | α-Thujene, γ-Terpinene, α-Pinene, 2-β-Pinene, Limonene, Butanoic acid, α-Terpinene | [79] |
| 32 | Myristica fragrans (Whole parts) | GC-MS (Qualitative and quantitative) | Colum: 30 m × 0.25 mm I.D., × 0.25 µL Medium: Helium as carrier gas, flow rate of 1.23 mL/min | Elemicin, isoelemicin, myristicin, surinamensin, malabaricone C, 2-(3′-allyl-2′,6′-dimethoxy-phenyloxy)-1-acetoxy-(3,4-dimethoxyphenyl)-propyl ester, methoxylicarin A, licarin A, malabaricone B, licarin C, 5′-methoxylicarin B, licarin B, and 2-(3′-allyl-2′,6′-dimethoxy-phenyloxy)-1-methyl-5-methoxy-1,2-dihydrobenzofuran | [80] |
| 33 | Nardostachys jatamansi (Root) | GC-MS-HT-TOF (Qualitative and quantitative) | Column: 29.3 m × 0.7 m, 320 μm. Medium: Helium as carrier gas, a flow rate of 1.5 mL/min | Dodecane, Linalool, l-calamenene, A-ionone, Oleic acid, Palmitic acid, Heptacosane, a-Muurolene, Tridecanoic acid, methyl ester. | [81] |
| 34 | Ornithogalum procerum (Aerial parts) | GC-MS (Qualitative) | Column: (60 m × 0.25 mm × 0.25 μm) Medium: Helium as carrier gas, flow rate of 1 mL/min | Linalool, Nonanal, γ-Terpinene, Octanal, Hexanal, Camphene, Furfural | [82] |
| 35 | Paederia foetida (Leaf) | GC-MS and NMR (Qualitative and quantitative) | Column: (30 m × 250 μm × 0.25 μm). Medium: Helium as carrier gas, a flow rate of 1 mL/min. | 1,3,5-benzenetriol, palmitic acid, cholesta-7,9(11)-diene-3-ol, 1-monopalmitin, β-tocopherol, α-tocopherol, 24-epicampesterol, stigmast-5-ene, 4-hydroxyphenylpyruvic acid, and glutamine | [83] |
| 36 | Potentilla anserine (Whole plant) | HP-5MS GC-MS (Qualitative | Column: 30 m × 0.25 mm × 0.25 µm Medium: Helium as carrier gas, flow rate of 1 mL/min | Glycolic acid, Alanine, Valine, Threonine, Malic acid, Aspartic acid, Shikimic acid, Pinitol, Quinic acid, etc. | [84] |
| 37 | Psidium guajava (Leaf) | GC-MS (Qualitative and quantitative) | Column: 30 m × 320 µm × 0.25 µm Medium: Helium as carrier gas, flow rate of 3.3245 mL/min | ascorbic acid, α-tocopherol, 3-hydroxyanthranilic acid, Cis-Zeatin-9-glucoside, Quercetin, 5-O-Galloylquinic acid, Ellagic acid Kaempferol, Methylsuccinic acid, Oxysterol (R)-3-Amino-4-phenylbutyric acid, etc. | [85] |
| 38 | Rostellularia diffusa (Whole plant) | GC-MS (Qualitative) | Column: (30 m × 0.32 mm × 0.25 μm) Medium: Helium as carrier gas, flow rate of 1 mL/min | 16-Hentriacontanone (22.59%), Hexadecanoic acid (11.23%), Stigmast-5-en-3-ol (6.78%), 9-Octadecenoic acid (n = 40) | [86] |
| 39 | Salix alba | GC-MS (Qualitative) | Column: 60 M TRX 5-MS (30 m × 0.25 mm × 0.25 µm), Medium: Helium as carrier gas, flow rate of 1.21 mL/min | Terpineol, 3-Thujanol, Eicosanoic acid, methyl ester, 1-Octadecyne, 1,2,3-Propanetriol, 2,2-Dimethylbutane, Acetic acid, butyl ester | [87] |
| 40 | Scutellaria barbata | 2-D HILIC × HILIC-MS (Qualitative) | Naringin, luteolin-7-O-glucoside, apigenin-7-O-glucoside, 5,7,8,2′-tetrahydroxyflavone-7-O-glucoside, scutellarin, carthamidin-7-O-glucuronide/isocarthamidin7-O-glucuronide, luteolin-7-O-glucuronide, | [88,89] | |
| 41 | Solanum nigrum (Aerial parts) | GC-MS (Qualitative and quantitative) | Colum: HP-5 column (30 m × 0.25 mm with 0.25 μm film thickness) Medium: Helium as carrier gas, flow rate of 1 mL/min. | 4-nitroguaiacol, 3-cyclohexen-1-ol,4-methyl-1-(methylethyl), Nonanoic acid,1-methyl ethyl ester, Flavone, 5Decenedioic acid,5,6dimethyl, dimethyl ester, Oleic acid, Z, E-2-methyl-3,13-octadecadien-1-ol, 2,4-ditertbutylphenol, 2,6,6,10-tetramethylundeca-8,10-diene-3,7-dione | [90] |
| GC-FID-MS (Qualitative) | Column: 30 m × 0.25 mm; 0.25 μm Medium: Helium as carrier gas, flow rate of 1 mL/min | Thymol, Carvacrol, Eugenol, Tetradecanal, iso-Longifolol | [91] | ||
| 42 | Sugarcane molasses (Plants) | Rtx-5MS GC-MS (Qualitative) | Column: 30 m × 0.25 mm × 0.25 µm Medium: Helium as carrier gas, flow rate of 1 mL/min | Ethylene glycol, Lactic acid, Acetic acid, Glyceric acid, Glyceric acid isomer, Ribitol, Glucitol, Malic acid, Citric acid, Threonic acid, Fructose, Inositol, etc. | [92] |
| 43 | Swertia chirata (Leaf) | Elite-5MS LC-MS (Qualitative and quantitative) | Column: 30 m × 0.25 mm I.D., 0.25 µm Medium: Helium as carrier gas, flow rate of 1.1 mL/min | Xanthone, Succinic acid, Viminalol, O, N-Permythylated N-Acetyllsine, Hexadecanoic acid, Cis-13-Octadecenoic acid, Tetracosanoic acid | [93] |
| 44 | Tagetes erecta (Flower) | GC-MS (Qualitative and quantitative) | Colum: HP-5MS (30 m × 0.25 mm ID × 0.25 µm) Medium: Helium as carrier gas, flow rate of 0.56 mL/min | Caryophyllene, Germacrene, Spathulenol, Palmitic acid, Heptadecanoic acid, Linolenic acid-metil ester, Bicyclogermacrene | [94] |
| 45 | Tarconanthus camphorantus (Leaf) | DB-1 GC-MS (Qualitative and quantitative) | Column: (0.25 µm film × 0.25 mm I.d. × 30 m Medium: Helium as carrier gas, flow rate of 1 mL/min | carvacrol, tetracontane, squalene, tetrapentacontane, and Phytol | [95] |
| 46 | Theobroma cacao (Seed) | GC-MS (Qualitative) | Column: 30 m × 0.25 mm × 0.25 µm Medium: Helium as carrier gas, flow rate of 1.5 mL/min | Dimethyl sulfone, 2-Cyclohexane-1-one, (1-methyl), hexadecanoic acid, methyl ester, Hexadecanoic acid, Octadecanoic acid, | [96] |
| 47 | Tinospora cordifolia (Root) | GC-MS (Qualitative and quantitative) | Column: 30 m × 0.25 mm I.D., × 0.25 mm Medium: Helium as carrier gas, flow rate of 1 mL/min | Isopinocarveol, α-ylangene, 1H-3a,7-Methanoazulene, octahydro-tetramethylCaryophyllene, trans-Z-α-Bisabolene epoxide, Benzene, 1-(1,5-dimethyl-4-hexenyl)-4-methyl- trans-α-Bergamotene, β-Bisabolene, β-Cubebene cubedol Sativen Methyl-hexadecatetraenoate, Alloaromadendrene oxide-(1) αacorenol, epi-cis sesquisabinene hydrate Octadecadiynoic acid, methyl ester, Phenol, 2-methyl trimethylcyclopentyl)-(S)-Isopropyl-2,8-dimethyl-9-Oxatricyclo decan-7-one, Hexadecanoic acid, ethyl ester, Octadecynoic acid | [97] |
| 48 | Trigonella foenum-graecum (Hairy roots) | GC-MS (Qualitative and quantitative) | Column: (20 m × 0.20 mm i.d., 0.5 μm film thickness Medium: Helium as carrier gas, flow rate of 0.6 mL/min. | Hexanal, γ-butyrolactone, cyclopenta-1,2-dieno, valeric acid, 3- hydroxy-4,5-dimethyl-2(5H)- furanone (sotolone), 2-(4-methylthiazo-5-yl)-ethanol, 6-methyl-2,3-dihydroxy-5,6-dihydropyran-4-one, 3-amino-4,5-dimethyl-2(5H)-furanone, 5-(hydroxymethyl) furan-2-carboxaldehyde | |
| Trigonella foenum-graecum (Seed) | HP5MS GC-MS (Qualitative) | Column: 30 mm in length, 0.25 mm i.d. and 0.25 µm Medium: Helium as carrier gas, flow rate of 1 mL/min | 3,5-Octadiene, p-Xylene, δ-3-Carene, Limonene, Decanal, β-Thujone, Hexadecanoic acid, Cis-calamenene | [98] | |
| 49 | Vaccinium angustifolium (Fruit) | P&T-GC-MS (Qualitative) | Column: (0.25 mm × 30 m × 0.25 m) Medium: Helium as carrier gas, flow rate of 1 mL/min | Ethyl caprylate, Linalool, 2-Nonanone, Ethyl acetate, 2-Methylbutyraldehyde | [99] |
| 50 | Withania somnifera (Root) | GC-MS (Qualitative) | Column: (30 m × 0.32 mm × 0.25 μm) Medium: Helium as carrier gas, flow rate of 1 mL/min | Oleic acid, phytol, n-hexadecanoic acid, 9-octadecenoicacid(z)-, methyl ester, hexadecanoic acid, methy ester, 2-methoxy-4-vinylphenol, azetidin-2-one3,3-dimethyl-4-(1-aminoethyl), 17-octadecynoicacid, o-bromoatropine, and sucrose | [100] |
| 51 | Xenostegiatridentata | LC-ESI-MS/MS | (A) water with 0.1% formic acid and solvent. (B) was acetonitrile with 0.1% formic acid. | 3,5-dicaffeoylquinic acid, luteolin-7-O-glucoside, quercetin-3-O-rhamnoside, kaempferol-3-O-rhamnoside. | [101] |
| 52 | Jasminum grandiflorum L. | HPLC-PDA-MS/MS-ESI. | Column: Gemini C18 110 Å (150 × 2 mm, 5 μm) Medium: A gradient of water and acetonitrile (ACN) (0.1% formic acid each) was applied from 5% to 30% ACN | Quercetin, kaempferol, myricitrin, laricitrin 3-o-glucoside, myricetin 3-xyloside, reynoutrin, kaempferitrin, oleuropein, multifloroside, isoquercitrin, oleuropein glucoside, verbascoside | [102] |
| 53 | Yucca gigantea (Leaves) | LC-MS/MS | Column: X select HSS T3 (2.5 µm, 2.1 mm × 150 mm) Medium: Buffer A (5 mM ammonium formate buffer pH 3 containing 1% methanol), Buffer B (5 mM ammonium formate buffer pH 8 containing 1% methanol), and buffer C (100% acetonitrile). Composition: 90 (A or B): 10 (C) | Okanin-4′-o-glucoside (marein), citraconic acid, muconic acid, spirostan-3-ol-glucoside-galactoside, hecogenin, kaempferol-7-o-neohesperidoside, kaempferol-3-o-α-l-rhamnoside, spirostan-3-ol-diglucoside, luteolin-7-o-β-d-glucoside, vitexin-2″-o-rhamnoside, hesperetin-7-o-neohesperidoside, isorhamnetin, quercetin-4′-o-glucoside, isorhamnetin-3-o-rutinoside, kaempferol-3-o-(6-p-coumaroyl)-glucopyranoside, poncirin, 4′,5,7-trihydroxyflavonol, hesperetin, naringenin, apigenin-7-o-β-d-glucoside, luteolin, caffeic acid, acacetin, 3,3′,4′,5-tetrahydroxy-7-methoxyflavone | [103] |
| 54 | Cycas thouarsii (Leaves) | LC-MS/MS | Column: X select HSS T3 (2.5 µm, 2.1 mm × 150 mm) Medium: Buffer A (5 mM ammonium formate buffer pH 3 containing 1% methanol), Buffer B (5 mM ammonium formate buffer pH 8 containing 1% methanol), and buffer C (100% acetonitrile). Composition: 90 (A or B): 10 (C) | Quinic acid, chlorogenic acid, trigonelline, piperidine, pantothenate, cinnamaldehyde, resveratrol, ferulic acid, quercetin 3-o-glucuronide, vitexin-2′′-o-rhamnoside, vitexin, hyperoside, luteolin-7-o-glucoside. |
| Sr. No. | Resources | Brief Summary | Application | Website/URL (Access Date: 7 October 2022) |
|---|---|---|---|---|
| 1 | BioCarta | Online maps of metabolic and signaling pathways | Database of gene interaction models | https://maayanlab.cloud/Harmonizome/dataset/Biocarta+Pathways |
| 2 | BioGRID | Biological General Repository for Interaction Datasets | Retrieval of protein–protein interaction network | http://thebiogrid.org/ |
| 3 | C2Maps | Computational Connectivity Maps | Annotation of drug–protein pairs | http://bio.informatics.iupui.edu/ |
| 4 | ChEMBL | Database of bioactive compounds | Retrieval of functional as well as binding information of active compounds | https://www.ebi.ac.uk/chembl/ |
| 5 | ChemProt | Chemical–protein–disease annotation database | Analysis of interaction between chemical and protein | http://www.cbs.dtu.dk/services/ChemProt-2.0/ |
| 6 | ChemSpider | Database of chemical structures | Retrieval of chemical structures | http://www.chemspider.com/ |
| 7 | COGs | Clusters of Orthologous Gene | Classification of proteins on phylogenetic basis | https://www.ncbi.nlm.nih.gov/COG/ |
| 8 | CPDB | Consensus Path DataBase | Molecular functional interaction database | http://cpdb.molgen.mpg.de/ |
| 9 | Cytoscape | Database for network construction and visualization | Network analysis | https://cytoscape.org/ |
| 10 | DAVID | Database for Annotation, Visualization & Integrated Discovery | Functional annotation | https://david.ncifcrf.gov/ |
| 11 | DIP | Database of Interacting proteins | Analysis of protein–protein interaction network | http://dip.doe-mbi.ucla.edu |
| 12 | DisGeNET | Database of Interacting proteins | Pathway analysis | (https://www.disgenet.org/) |
| 13 | GeneCards | Database of human genes | For identification of disease-related genes | https://www.genecards.org/ |
| 14 | Guess | Computer program for the analysis and visualization of networks | Network analysis | http://www.levmuchnik.net/Content/Networks/ComplexNetworksPackage.html |
| 15 | HAPPI | Human Annotated & Predicted Protein | Retrieval of protein–protein interaction network | http://bio.informatics.iupui.edu/HAPPI/ |
| 16 | HPRD | Human Protein Reference Database | Retrieval of protein–protein interaction network | http://www.hprd.org/ |
| 17 | InterPro | Integrative database of protein families | Collection of protein families | http://www.ebi.ac.uk/interpro/ |
| 18 | KEGG | Kyoto Encyclopedia of Genes and Genomes | Pathway analysis | http://www.genome.jp/kegg/ |
| 19 | MetaCoreTM | MetaCore (TM) | Pathway analysis | http://www.genego.com |
| 20 | Metascape | Computer program for the analysis and visualization of networks | Network and Pathway analysis | (https://metascape.org) |
| 21 | NetMiner | Computer program for the analysis and visualization of networks | Network analysis | http://graphexploration.cond.org/ |
| 22 | NetPath | Network pathway analysis | Pathway analysis | http://www.netpath.org/ |
| 23 | NetworkX | Computer program for the analysis and visualization of networks | Network analysis | http://www.analytictech.com/ucinet/ |
| 24 | Network analyst | Computer program for the analysis and visualization of networks | Network analysis | (https://www.networkanalyst.ca/), |
| 25 | OPHID | Online predicted human interaction database | Retrieval of protein–protein interaction network | http://ophid.utoronto.ca |
| 26 | Pajek | Computer program for the analysis and visualization of network | Network analysis | http://pajek.imfm.si/doku.php |
| 27 | PDB | Protein Data bank | Retrieval of protein-related information | http://www.rcsb.org/pdb/ |
| 28 | PharmGBK | Pharmacogenomics knowledge base | Analyze the genes’ response to drugs | http://www.pharmgkb.org/ |
| 29 | Reactome | Database of pathways, reactions, and biological processes | Pathway analysis | http://www.reactome.org |
| 30 | SignaLink | Signaling pathway analysis resource | Pathway analysis | http://signalink.org/ |
| 31 | STITCH | Search Tool for Interactions of Chemicals | Analysis of target–drug relationship and biological pathways | http://stitch.embl.de/ |
| 32 | STRING | Search Tool for the Retrieval of Interacting Genes/Proteins | Retrieval of protein–protein interaction network | http://string-db.org/ |
| 33 | SwissTargetPrediction | Estimate the macromolecular targets of a small molecule | Identification of compound-related genes | http://www.swisstargetprediction.ch/ |
| 34 | Ucinet | Computer program for the analysis and visualization of networks | Network analysis | http://www.netminer.com/ |
| 35 | UniProtKB | Universal protein knowledge database | Analysis of protein | http://www.uniprot.org/uniprot/ |
| Sr. No. | Medicinal Plant/Traditional Formulations | Phytoconstituents | Biomolecular Targets/Genes | Therapeutic Role | Mechanism of Action | References |
|---|---|---|---|---|---|---|
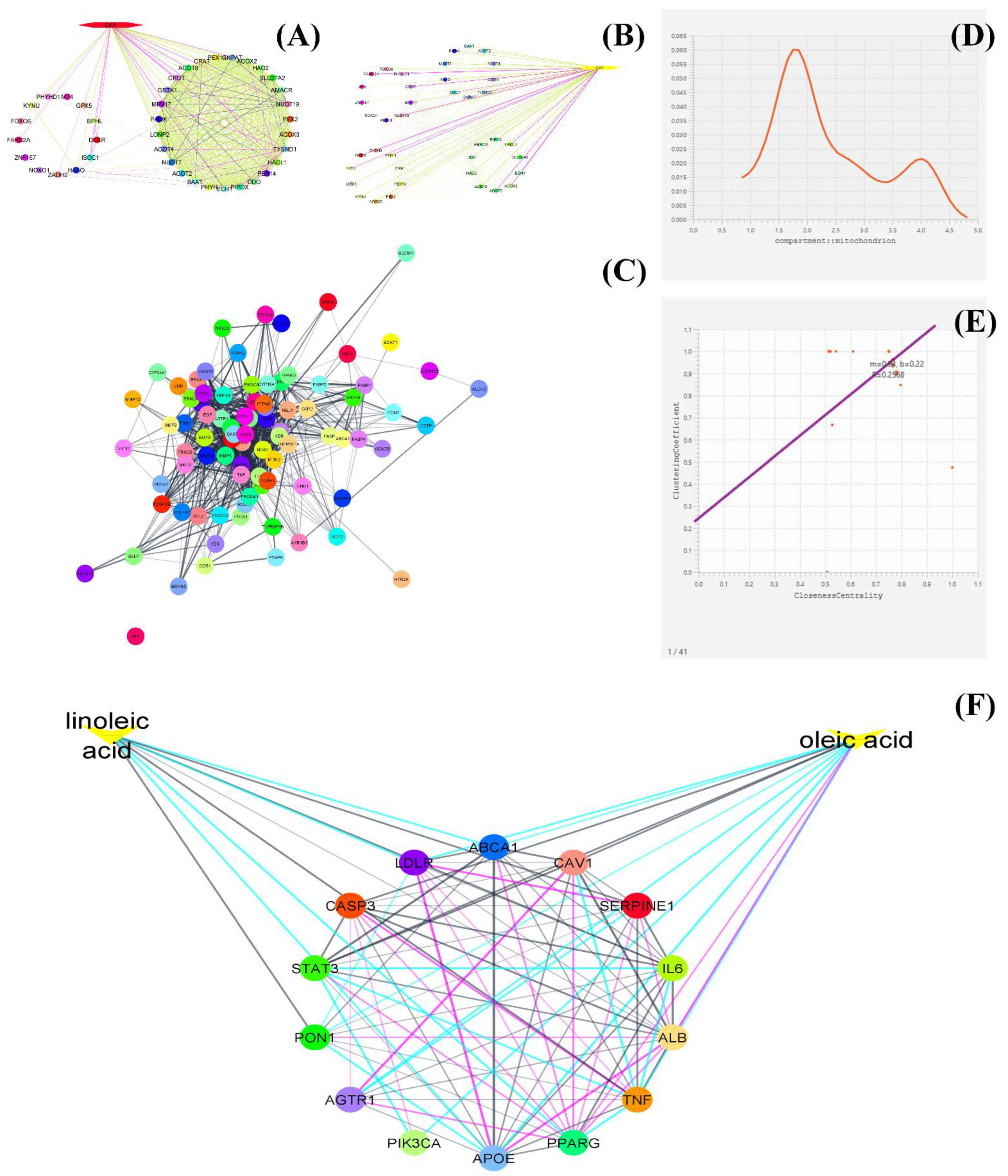
| 1 | Boerhaavia diffusa | Apigenin, ferulic acid and quercetin | ABCA1, AKT1, MMP9, LEPR, AKR1C3, KDR, PPARs, ESR1, ARNT, PON2, SESN2, IRS1, ILs, MAPKs, CAPSs, NOS, etc. | Nephroprotective | Reduction of oxidative and inflammatory stress, glomerulonephritis, reduction of vascular rigidity in hypertension | [33] |
| 2 | Tinospora cordifolia | Gallic acid and rutin | CASPs, MAPKs, NOS, PRNP, CTGF, SREBF1, and JUN | Nephroprotective | Reduction of oxidative and inflammatory stress, glomerulonephritis | [33] |
| 3 | Momordica charantia | Linalool, quercetin, gallic acid, apiole, ferulic acid, caffeic acid, limonene and catechin | NOS, ILs, CASPs, MAPKs, G6PD, MAPK1, MAPK3, MMPs, PTGS2, PON1, ILs, AKT1, JUN, MMPs, PPARG, TP53, etc. | Diabetic nephropathy | Amelioration in endothelial dysfunction, fatty liver disease, diabetes mellitus, acute kidney injury, fibrosis, hypertensive disease, obesity, etc. | [34] |
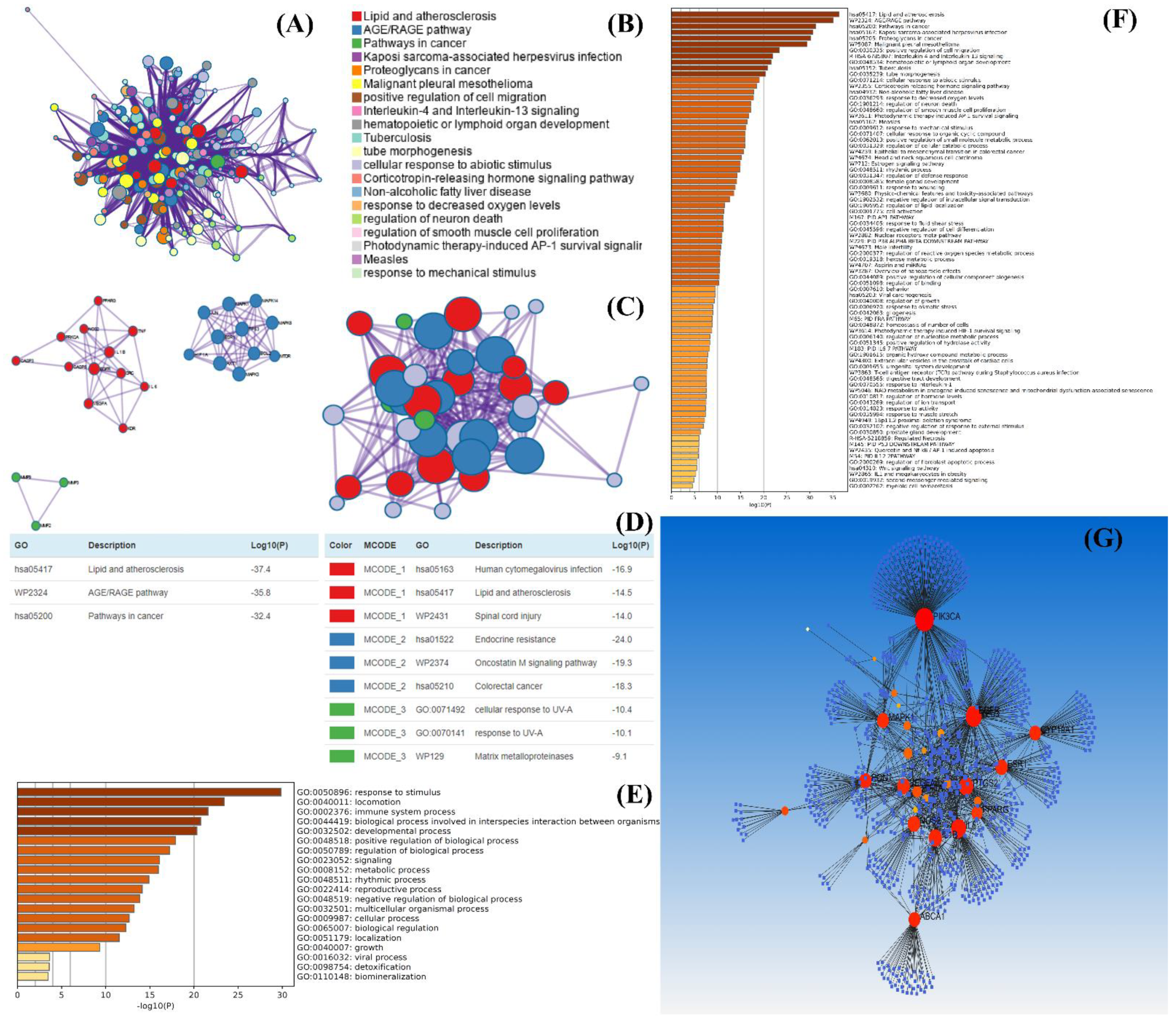
| 4 | - | Quercetin and kaempferol | TNF, JUN, IL6, STAT3, MAPK1, and MAPK3 | Acne Vulgaris | Anti-inflammatory effect and regulate the excessive lipogenesis in sebaceous glands via different signaling pathways | [104] |
| 5 | Gelsemium elegans | Gelegamine E, gelsesyringalidine, humantenine, gelsedine, 19-z-akuammidine, gelegamine B, tabersonine, koumine, gelsemine | MAPK3, HSP90AA1, JUN, EGFR, CDK1, TNF, CCND1, ESR1, PRKACA, CCNA2, CDC25C, CDK2, CCNB1, AR, CREBBP, AURKA, CDC25A, CHEK1, BCL2L1, and PIK3CD | Anti-cancer | Pathways in cancer, cell cycle, and colorectal cancer | [105] |
| 6 | Danning Tablets (A polyherbal formulation) | Quercetin, ß-sitosterol, luteolin, kaempferol, supraene, curcumenolactone C, and stigmasterol | IL6, MAPK8, VEGFA, CASP3, ALB, APP, MYC, PPARG, and RELA | Nonalcoholic Fatty Liver Disease | Lipid metabolism, inflammation, oxidation, insulin resistance (IR), atherosclerosis, and apoptosis | [106] |
| 7 | Panax notoginseng | Mandenol, beta-sitosterol, stigmasterol, ginsenoside rh2, ginsenoside f2, quercetin | VEGFA, MMP-9, MMP-2, FGF2, and COX-2 | Diabetic Retinopathy | Intervention of angiogenesis, inflammation, and apoptosis | [107] |
| 8 | Angelica sinensis | Isorhamnetin, jaranol, 3,9-di-O-methylnissolin, mairin, formononetin, isoflavanone, quercetin, kaempferol and (3R)-3-(2-hydroxy-3,4-dimethoxyphenyl)chroman-7-ol | VEGFA, TP53, IL-6, TNF, MARK1 | Diabetic nephropathy | Apoptosis, oxidative stress, inflammation, glucose, and lipid metabolism processes | [108] |
| 9 | Caesalpinia pulcherima | Gallic acid, catechin, ellagic acid, quercetina and cyanidin 3-glucoside | ESR-1, ESR-2, ESRRA, MET, VEGF, FGF, PI3K, PDK-1, MAPK, PLK-1, NEK-2, and GRK | Breast cancer | Breast cancer, endometrial cancer, and osteoporosis, estrogen signaling pathway, prolactin signaling pathway, thyroid hormone signaling pathway, and relaxin signaling pathway | [109] |
| 10 | Chinese herbal formula Shenyi (SY) | Moupinamide, quercetin, eriodictyol, hesperetin, formononetin, palmatine, norlobeline, taxifoline, sinoacutine, beta-sitosterol, kaempferol, stigmasterol, acacetin, linarin, and isovitexin | AKT1, TNF, IL6, TP53, VEGFA, EGFR, CASP3, JUN, IL1B, MYC, ESR1, HIF1A, HSP90AA1, EGF, PTGS2, MMP9, and CCND1 | Diabetic nephropathy | Glomerulosclerosis, nephrotoxicity, oxidative stress, inflammation, and hyperglycemia, inhibit RhoA/Rho kinase to ameliorate kidney injury and decrease fibrosis | [110] |
| 11 | Jasminum grandiflorum L. | Quercetin, kaempferol, myricitrin, laricitrin 3-o-glucoside, myricetin 3-xyloside, reynoutrin, kaempferitrin, oleuropein, multifloroside, isoquercitrin, oleuropein glucoside, verbascoside | MKK4, MKK7, I-CAM 1, IL-6, TNF, TRAF2, PI3K-Akt, MAPK, EGFR, | Nephroprotective | Reduction in tubular cell lining vacuolation and intratubular cast deposition in the renal tubules, oxidative stress, inflammation, and apoptosis | [111] |
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-Based Metabolomics. Mol. Biosyst. 2012, 8, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Gaurav; Zahiruddin, S.; Parveen, B.; Ibrahim, M.; Sharma, I.; Sharma, S.; Sharma, A.K.; Parveen, R.; Ahmad, S. TLC-MS Bioautography-Based Identification of Free-Radical Scavenging, α-Amylase, and α-Glucosidase Inhibitor Compounds of Antidiabetic Tablet BGR-34. ACS Omega 2020, 5, 29688–29697. [Google Scholar] [CrossRef] [PubMed]
- Gautam, G.; Parveen, B.; Umar Khan, M.; Sharma, I.; Kumar Sharma, A.; Parveen, R.; Ahmad, S. A Systematic Review on Nephron Protective AYUSH Drugs as Constituents of NEERI-KFT (A Traditional Indian Polyherbal Formulation) for the Management of Chronic Kidney Disease. Saudi J. Biol. Sci. 2021, 28, 6441–6453. [Google Scholar] [CrossRef] [PubMed]
- Karthikkeyan, G.; Pervaje, R.; Subbannayya, Y.; Patil, A.H.; Modi, P.K.; Prasad, T.S.K. Plant Omics: Metabolomics and Network Pharmacology of Liquorice, Indian Ayurvedic Medicine Yashtimadhu. Omi. A J. Integr. Biol. 2020, 24, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, L.; Xu, B.; Zhao, L.; Li, L.; Xu, K.; Tang, A.; Zhou, S.; Song, L.; Zhang, X.; et al. Based on Network Pharmacology Tools to Investigate the Molecular Mechanism of Cordyceps Sinensis on the Treatment of Diabetic Nephropathy. J. Diabetes Res. 2021, 2021, 8891093. [Google Scholar] [CrossRef]
- Jorge, T.F.; Mata, A.T.; António, C. Mass Spectrometry as a Quantitative Tool in Plant Metabolomics. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150370. [Google Scholar] [CrossRef]
- Sawada, Y.; Yokota Hirai, M. Integrated LC-MS/MS System for Plant Metabolomics. Comput. Struct. Biotechnol. J. 2013, 4, e201301011. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Yi, F.; Li, L.; Xu, L.J.; Meng, H.; Dong, Y.M.; Liu, H.B.; Xiao, P.G. In Silico Approach in Reveal Traditional Medicine Plants Pharmacological Material Basis. Chinese Med. 2018, 13, 33. [Google Scholar] [CrossRef]
- Yang, M.; Chen, J.; Xu, L.; Shi, X.; Zhou, X.; An, R.; Wang, X. A Network Pharmacology Approach to Uncover the Molecular Mechanisms of Herbal Formula Ban-Xia-Xie-Xin-Tang. Evid.-Based Complement. Altern. Med. 2018, 2018, 4050714. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Wang, X.; Hu, Y.; Su, S.; Li, W.; Li, S. Editorial: Network Pharmacology and Traditional Medicine. Front. Pharmacol. 2020, 11, 1194. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huang, X.; Wang, H.; Shao, J.; Luo, Y.; Zhao, K.; Liu, Y.; Wang, S. Integrated Metabolomics and Network Pharmacology Strategy for Ascertaining the Quality Marker of Flavonoids for Sophora Flavescens. J. Pharm. Biomed. Anal. 2020, 186, 113297. [Google Scholar] [CrossRef] [PubMed]
- Lotfi Shahreza, M.; Ghadiri, N.; Mousavi, S.R.; Varshosaz, J.; Green, J.R. A Review of Network-Based Approaches to Drug Repositioning. Brief. Bioinform. 2018, 19, 878–892. [Google Scholar] [CrossRef]
- Hao, H.; Zheng, X.; Wang, G. Insights into Drug Discovery from Natural Medicines Using Reverse Pharmacokinetics. Trends Pharmacol. Sci. 2014, 35, 168–177. [Google Scholar] [CrossRef]
- Li, M.; Chen, L.; Dai, Y.; Li, J.; Li, F.; Li, Q.; Yu, Z.; Chai, K.; Zhu, Y. Differential Metabolomics and Network Pharmacology Analysis of Silkworm Biotransformation between Mulberry Leaves and Silkworm Droppings. Evid.-Based Complement. Altern. Med. 2021, 2021, 8819538. [Google Scholar] [CrossRef]
- Emig, D.; Ivliev, A.; Pustovalova, O.; Lancashire, L.; Bureeva, S.; Nikolsky, Y.; Bessarabova, M. Drug Target Prediction and Repositioning Using an Integrated Network-Based Approach. PLoS ONE 2013, 8, e60618. [Google Scholar] [CrossRef]
- Chandran, U.; Mehendale, N.; Patil, S.; Chaguturu, R.; Patwardhan, B. Network Pharmacology. In Innovative Approaches in Drug Discovery: Ethnopharmacology, Systems Biology and Holistic Targeting; Academic Press: Cambridge, MA, USA, 2017; pp. 127–164. ISBN 9780128018224. [Google Scholar]
- Hao, D.C.; Xiao, P.G. Network Pharmacology: A Rosetta Stone for Traditional Chinese Medicine. Drug Dev. Res. 2014, 75, 299–312. [Google Scholar] [CrossRef]
- Yang, H.; Cheung, M.K.; Yue, G.G.L.; Leung, P.C.; Wong, C.K.; Lau, C.B.S. Integrated Network Pharmacology Analysis and in Vitro Validation Revealed the Potential Active Components and Underlying Mechanistic Pathways of Herba Patriniae in Colorectal Cancer. Molecules 2021, 26, 6032. [Google Scholar] [CrossRef]
- Orfali, R.; Perveen, S.; Siddiqui, N.A.; Alam, P.; Alhowiriny, T.A.; Al-Taweel, A.M.; Al-Yahya, S.; Ameen, F.; Majrashi, N.; Alluhayb, K.; et al. Pharmacological Evaluation of Secondary Metabolites and Their Simultaneous Determination in the Arabian Medicinal Plant Plicosepalus Curviflorus Using HPTLC Validated Method. J. Anal. Methods Chem. 2019, 2019, 7435909. [Google Scholar] [CrossRef]
- Buriani, A.; Garcia-Bermejo, M.L.; Bosisio, E.; Xu, Q.; Li, H.; Dong, X.; Simmonds, M.S.J.; Carrara, M.; Tejedor, N.; Lucio-Cazana, J.; et al. Omic Techniques in Systems Biology Approaches to Traditional Chinese Medicine Research: Present and Future. J. Ethnopharmacol. 2012, 140, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Buriani, A.; Fortinguerra, S.; Carrara, M.; Pelkonen, O. Systems Network Pharmaco-Toxicology in the Study of Herbal Medicines. In Toxicology of Herbal Products; Springer: Cham, Switzerland, 2017; pp. 129–164. ISBN 9783319438061. [Google Scholar]
- Majumder, S.; Ghosh, A.; Chakraborty, S.; Saha, S.; Bhattacharya, M. Metabolomics Affirms Traditional Alcoholic Beverage Raksi as a Remedy for High-Altitude Sickness. J. Ethn. Foods 2021, 8, 17. [Google Scholar] [CrossRef]
- Arnason, J.T.; Harris, C.S.; Guerrero-Analco, J.A. Phytochemistry in the Ethnopharmacology of North and Central America. Front. Pharmacol. 2022, 13, 815742. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Katiyar, C.K.; Ulrich-Merzenich, G.S.; Mukherjee, P.K. Editorial: Metabolomics and Ethnopharmacology in the Development of Herbal and Traditional Medicine. Front. Pharmacol. 2022, 13, 851023. [Google Scholar] [CrossRef]
- Alshibl, H.M.; Al-Abdullah, E.S.; Haiba, M.E.; Alkahtani, H.M.; Awad, G.E.A.; Mahmoud, A.H.; Ibrahim, B.M.M.; Bari, A.; Villinger, A. Synthesis and Evaluation of New Coumarin Derivatives as Antioxidant, Antimicrobial, and Anti-Inflammatory Agents. Molecules 2020, 25, 3251. [Google Scholar] [CrossRef]
- Alhozaimy, G.A.; Al-Sheddi, E.; Ibrahim, T.A. Biological Activity and Isolation of Compounds from Stem Bark of Plumeria Acutifolia. Pharmacogn. Mag. 2017, 13, S505–S511. [Google Scholar] [CrossRef]
- Gonulalan, E.M.; Nemutlu, E.; Demirezer, L.O. A New Perspective on Evaluation of Medicinal Plant Biological Activities: The Correlation between Phytomics and Matrix Metalloproteinases Activities of Some Medicinal Plants. Saudi Pharm. J. 2019, 27, 446–452. [Google Scholar] [CrossRef]
- Sagar, P.K.; Murugeswaran, R.; Meena, R.; Mageswari, S.; Sri, M.D.; Khair, S. Standardization and HPTLC, Fingerprinting Study of Poly Herbal Unani Formulation–Habb-e-Sara Khas. Medicine 2020, 5, 27. [Google Scholar]
- Ibrahim, M.; Parveen, B.; Zahiruddin, S.; Gautam, G.; Parveen, R.; Ahmed, M.; Arun, K.; Sayeed, G. Analysis of Polyphenols in Aegle Marmelos Leaf and Ameliorative Efficacy against Diabetic Mice through Restoration of Antioxidant and Anti-Inflammatory Status. J. Food Biochem. 2022, 46, e13852. [Google Scholar] [CrossRef]
- Koek, M.M.; Jellema, R.H.; van der Greef, J.; Tas, A.C.; Hankemeier, T. Quantitative Metabolomics Based on Gas Chromatography Mass Spectrometry: Status and Perspectives. Metabolomics 2011, 7, 307–328. [Google Scholar] [CrossRef]
- Gaurav; Khan, M.U.; Basist, P.; Zahiruddin, S.; Ibrahim, M.; Parveen, R.; Krishnan, A.; Ahmad, S. Nephroprotective Potential of Boerhaavia Diffusa and Tinospora Cordifolia Herbal Combination against Diclofenac Induced Nephrotoxicity. S. Afr. J. Bot. 2022. [Google Scholar] [CrossRef]
- Gautam, G. Network Pharmacology-Based Validation of Traditional Therapeutic Claim of Momordica Charantiain Alleviating Diabetic Nephropathy. J. CAM Res. Prog. 2022, 1, 102. [Google Scholar]
- Kibble, M.; Saarinen, N.; Tang, J.; Wennerberg, K.; Mäkelä, S.; Aittokallio, T. Network Pharmacology Applications to Map the Unexplored Target Space and Therapeutic Potential of Natural Products. Nat. Prod. Rep. 2015, 32, 1249–1266. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.R.; Habela, C.W.; Stafstrom, C.E. Pediatric Epilepsy Mechanisms: Expanding the Paradigm of Excitation/Inhibition Imbalance. Children 2019, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Goel, R.; Pathak, S.; Srivastava, A.; Singh, S.P.; Sangwan, R.S.; Asif, M.H.; Trivedi, P.K. De Novo Assembly, Functional Annotation and Comparative Analysis of Withania Somnifera Leaf and Root Transcriptomes to Identify Putative Genes Involved in the Withanolides Biosynthesis. PLoS ONE 2013, 8, e62714. [Google Scholar] [CrossRef] [PubMed]
- Vadivu, G.; Waheeta Hopper, S. Ontology Mapping of Indian Medicinal Plants with Standardized Medical Terms. J. Comput. Sci. 2012, 8, 1576–1584. [Google Scholar] [CrossRef]
- Lagunin, A.A.; Ivanov, S.M.; Gloriozova, T.A.; Pogodin, P.V.; Filimonov, D.A.; Kumar, S.; Goel, R.K. Combined Network Pharmacology and Virtual Reverse Pharmacology Approaches for Identification of Potential Targets to Treat Vascular Dementia. Sci. Rep. 2020, 10, 257. [Google Scholar] [CrossRef]
- Khanal, P.; Patil, B.M. Gene Ontology Enrichment Analysis of α-Amylase Inhibitors from Duranta Repens in Diabetes Mellitus. J. Diabetes Metab. Disord. 2020, 9, 735–747. [Google Scholar] [CrossRef]
- Wu, C.; Gudivada, R.C.; Aronow, B.J.; Jegga, A.G. Computational Drug Repositioning through Heterogeneous Network Clustering. BMC Syst. Biol. 2013, 7, S6. [Google Scholar] [CrossRef]
- Jung, Y.S.; Kim, Y.; Cho, Y.R. Comparative Analysis of Network-Based Approaches and Machine Learning Algorithms for Predicting Drug-Target Interactions. Methods 2022, 198, 19–31. [Google Scholar] [CrossRef]
- Cao, D.S.; Zhang, L.X.; Tan, G.S.; Xiang, Z.; Bin Zeng, W.; Xu, Q.S.; Chen, A.F. Computational Prediction of Drug-Target Interactions Using Chemical, Biological, and Network Features. Mol. Inform. 2014, 33, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Nezhadali, A.; Parsa, M. Study of the Volatile Compounds in Artemisia Sagebrush from Iran Using HS/SPME/GC/MS. Int. J. Environ. Sci. Dev. 2010, 1, 287. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, H.; Singh, R.; Srikanth, N. High-Performance Thin-Layer Chromatography Estimation of Boeravinone-B in Boerhavia diffusa L. and Its Polyherbal Dosage Form (Capsule). Pharmacogn. Res. 2019, 11, 267. [Google Scholar] [CrossRef]
- Mallavadhani, U.V.; Aparna, Y.; Mohapatra, S.; Mane, D.V. Quantitative Evaluation of Boerhavia Diffusa and Its Commercial Formulations with Respect to Its Major Bioactive Marker, Eupalitin Galactoside, Using High-Performance Thin-Layer Chromatography. J. Planar Chromatogr. Mod. TLC 2017, 30, 521–526. [Google Scholar] [CrossRef]
- Nipun, T.S.; Khatib, A.; Ibrahim, Z.; Ahmed, Q.U.; Redzwan, I.E.; Primaharinastiti, R.; Saiman, M.Z.; Fairuza, R.; Widyaningsih, T.D.; Alajmi, M.F.; et al. Gc-Ms-and Nmr-Based Metabolomics and Molecular Docking Reveal the Potential Alpha-Glucosidase Inhibitors from Psychotria Malayana Jack Leaves. Pharmaceuticals 2021, 14, 978. [Google Scholar] [CrossRef]
- Chester, K.; Zahiruddin, S.; Ahmad, A.; Khan, W.; Paliwal, S.; Ahmad, S. Bioautography-Based Identification of Antioxidant Metabolites of Solanum nigrum L. and Exploration Its Hepatoprotective Potential against D-Galactosamine-Induced Hepatic Fibrosis in Rats. Pharmacogn. Mag. 2017, 15, 104–110. [Google Scholar] [CrossRef]
- Abdullahi1, A.; Muhammad, M.T.; Suleiman, J.; Sokoto1, R.M. Isolation and Identification of Bacteria Associated with Aerial Part of Rice Plant from Kware Lake. Asian J. Res. Bot. 2018, 1, 1–8. [Google Scholar]
- Ehsani, A.; Alizadeh, O.; Hashemi, M.; Afshari, A.; Aminzare, M. Phytochemical, Antioxidant and Antibacterial Properties of Melissa Officinalis and Dracocephalum Moldavica Essential Oils. Vet. Res. Forum Int. Q. J. 2017, 8, 223–229. [Google Scholar]
- Peraza-Luna, F.; Rodríguez-Mendiola, M.; Arias-Castro, C.; Bessiere, J.M.; Calva-Calva, G. Sotolone Production by Hairy Root Cultures of Trigonella Foenum-Graecum in Airlift with Mesh Bioreactors. J. Agric. Food Chem. 2001, 49, 6012–6019. [Google Scholar] [CrossRef]
- Gul, I.; Nasrullah, N.; Nissar, U.; Saifi, M.; Abdin, M.Z. Development of DNA and GC-MS Fingerprints for Authentication and Quality Control of Piper nigrum L. and Its Adulterant Carica papaya L. Food Anal. Methods 2018, 11, 1209–1222. [Google Scholar] [CrossRef]
- Pieczykolan, A.; Pietrzak, W.; Szewczyk, K.D.S.; Gawlik-Dziki, U.; Nowak, R. LC-ESI-MS/MS Polyphenolic Profile and In Vitro Study of Cosmetic Potential of Aerva lanata (L.) Juss. Herb Extracts. Molecules 2022, 27, 1259. [Google Scholar] [CrossRef] [PubMed]
- Vidhya, R.; Udayakumar, R. Gas Chromatography-Mass Spectrometry (GC-MS) Analysis of Ethanolic Extracts of Aerva lanata (L.). Int. J. Biochem. Res. Rev. 2015, 7, 192–203. [Google Scholar] [CrossRef]
- Fahim, M.; Ibrahim, M.; Zahiruddin, S.; Parveen, R.; Khan, W.; Ahmad, S.; Shrivastava, B.; Shrivastava, A.K. TLC-Bioautography Identification and GC-MS Analysis of Antimicrobial and Antioxidant Active Compounds in Musa × Paradisiaca L. Fruit Pulp Essential Oil. Phytochem. Anal. 2019, 30, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Razack, S.; Kumar, K.H.; Nallamuthu, I.; Naika, M.; Khanum, F. Antioxidant, Biomolecule Oxidation Protective Activities of Nardostachys Jatamansi DC and Its Phytochemical Analysis by RP-HPLC and GC-MS. Antioxidants 2015, 4, 185–203. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Dowlath, M.J.H.; Karuppannan, S.K.; Saravanan, M.; Arunachalam, K.D. Effect of Solvent on the Phytochemical Extraction and Gc-Ms Analysis of Gymnema Sylvestre. Pharmacogn. J. 2020, 12, 749–761. [Google Scholar] [CrossRef]
- Singh, R.; Chahal, K.K. Cichorium intybus from India: GC-MS Profiling, Phenolic Content and In Vitro Antioxidant Capacity of Sequential Soxhlet Extracted Roasted Roots. Braz. Arch. Biol. Technol. 2019, 62, e19180370. [Google Scholar] [CrossRef]
- Chester, K.; Paliwal, S.; Khan, W.; Ahmad, S. UPLC-ESI-MS/MS and HPTLC Method for Quantitative Estimation of Cytotoxic Glycosides and Aglycone in Bioactivity Guided Fractions of Solanum nigrum L. Front. Pharmacol. 2017, 8, 434. [Google Scholar] [CrossRef]
- Aburjai, T.A.; Oun, I.M.; Auzi, A.A.; Hudaib, M.M. Volatile Oil Constituents of Fruits and Leaves of Solanum nigrum L. Growing in Libya. J. Essent. Oil-Bearing Plants 2014, 17, 397–404. [Google Scholar] [CrossRef]
- Keskes, H.; Belhadj, S.; Jlail, L.; El Feki, A.; Sayadi, S.; Allouche, N. LC–MS–MS and GC–MS Analyses of Biologically Active Extracts of Tunisian Fenugreek (Trigonella foenum-graecum L.) Seeds. J. Food Meas. Charact. 2018, 12, 209–220. [Google Scholar] [CrossRef]
- Prabhadevi, V.; Sahaya, S.S.; Johnson, M.; Venkatramani, B.; Janakiraman, N. Phytochemical Studies on Allamanda cathartica L. Using GC-MS. Asian Pac. J. Trop. Biomed. 2012, 2, S550–S554. [Google Scholar] [CrossRef]
- Gowda, P.J.; Ramakrishnaiah, H.; Krishna, V.; Narra, S.; Jagannath, N. Caryophyllene-Rich Essential Oil of Didymocarpus Tomentosa: Chemical Composition and Cytotoxic Activity. Nat. Prod. Commun. 2012, 7, 1535–1538. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.K.; Arumugam, G.; Kaur, R.; Ghasemzadeh, A.; Yusoff, M.M.; Sinniah, U.R. GC-MS Based Metabolite Profiling, Antioxidant and Antimicrobial Properties of Different Solvent Extracts of Malaysian Plectranthus Amboinicus Leaves. Evid.-Based Complement. Altern. Med. 2017, 2017, 1517683. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Tandon, S.; Nandi, S.P.; Kaur, T.; Tandon, C. Downregulation of Inflammatory Mediators by Ethanolic Extract of Bergenia ligulata (Wall.) in Oxalate Injured Renal Epithelial Cells. J. Ethnopharmacol. 2021, 275, 114104. [Google Scholar] [CrossRef]
- Juneja, K.; Mishra, R.; Chauhan, S.; Gupta, S.; Roy, P.; Sircar, D. Metabolite Profiling and Wound-Healing Activity of Boerhavia Diffusa Leaf Extracts Using In Vitro and In Vivo Models. J. Tradit. Complement. Med. 2020, 10, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Matulyte, I.; Marksa, M.; Ivanauskas, L.; Kalveniene, Z.; Lazauskas, R.; Bernatoniene, J. GC-MS Analysis of the Composition of the Extracts and Essential Oil from Myristica Fragrans Seeds Using Magnesium Aluminometasilicate as Excipient. Molecules 2019, 24, 1062. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Liang, Y.Z.; Cui, H.; Chau, F.T.; Chan, B.T.P. Determination of Volatile Components in Peptic Powder by Gas Chromatography-Mass Spectrometry and Chemometric Resolution. J. Chromatogr. A 2001, 909, 237–247. [Google Scholar] [CrossRef]
- Ahmad, A.; Ibrahim, M.; Chester, K.; Khan, W.; Ahmad, S.; Ansari, S. Antithrombocytopenic Potential of Bioactivity Guided Fractions of Traditionally Used Psidium Guajava Linn. Leaves in Busulfan Induced-Thrombocytopenic Rats. Pharmacogn. Mag. 2019, 15, 440. [Google Scholar] [CrossRef]
- Fasola, T.R.; Oloyede, G.K.; Aponjolosun, B.S. Chemical Composition, Toxicity and Antioxidant Activities of Essential Oils of Stem Bark of Nigerian Species of Guava (Psidium guajava Linn.). EXCLI J. 2011, 10, 34–43. [Google Scholar]
- Deore, S.L.; Khadabadi, S.S. Isolation and Characterization of Phytoconstituents from Chlorophytum Borivilianum. Pharmacognosy Res. 2010, 2, 343–349. [Google Scholar] [CrossRef]
- He, Y.; Li, Z.; Wang, W.; Sooranna, S.R.; Shi, Y.; Chen, Y.; Wu, C.; Zeng, J.; Tang, Q.; Xie, H. Chemical Profiles and Simultaneous Quantification of Aurantii Fructus by Use of Hplc-q-Tof-Ms Combined with Gc-Ms and Hplc Methods. Molecules 2018, 23, 2189. [Google Scholar] [CrossRef]
- Ulrich, D.; Olbricht, K. Diversity of Metabolite Patterns and Sensory Characters in Wild and Cultivated Strawberries. J. Berry Res. 2014, 4, 11–17. [Google Scholar] [CrossRef]
- Liu, H.; Kong, W.; Gong, B.; Miao, Q.; Qi, Y.; Yang, M. Rapid Analysis of Multi-Pesticides in Morinda Officinalis by GC-ECD with Accelerated Solvent Extraction Assisted Matrix Solid Phase Dispersion and Positive Confirmation by GC-MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015, 974, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Berkov, S.; Bastida, J.; Sidjimova, B.; Viladomat, F.; Codina, C. Phytochemical Differentiation of Galanthus Nivalis and Galanthus Elwesii (Amaryllidaceae): A Case Study. Biochem. Syst. Ecol. 2008, 36, 638–645. [Google Scholar] [CrossRef]
- Chludil, H.D.; Vilariño, M.D.P.; Franco, M.L.; Leicach, S.R. Changes in Lupinus Albus and Luplnus Angustlfollus Alkaloid Profiles in Response to Mechanical Damage. J. Agric. Food Chem. 2009, 57, 6107–6113. [Google Scholar] [CrossRef]
- Rathi, D.; Balasubramanian, P.L. Phytochemical Compound Analysis of Tinospora Cordifolia By GC-MS Method. Int. J. Recent Res. Asp. 2018, 5, 1–5. [Google Scholar]
- Wang, S.Q. Chemical Composition and Allelopathic Potential of Essential Oils from Eupatorium maculatum on Lolium perenne L. and Echinochloa crusgalli L. Allelopath. J. 2019, 49, 51–62. [Google Scholar] [CrossRef]
- Xinyu, Z.; Yi, W.; Xue, H.; Zixi, Y.; Weiqiong, Y.; Zhaolin, L. Characterization of Volatile Compounds in Five Blueberry Varieties Using Purge and Trap Coupled to Gas Chromatography-Mass Spectrometry. Ital. J. Food Sci. 2020, 32, 2020. [Google Scholar] [CrossRef]
- Ali, S.E.; El Gedaily, R.A.; Mocan, A.; Farag, M.A.; El-Seedi, H.R. Profiling Metabolites and Biological Activities of Sugarcane (Saccharum officinarum Linn.) Juice and Its Product Molasses via a Multiplex Metabolomics Approach. Molecules 2019, 24, 934. [Google Scholar] [CrossRef]
- Zainal, B.; Abdah, M.A.; Taufiq-Yap, Y.H.; Roslida, A.H.; Rosmin, K. Anticancer Agents from Non-Edible Parts of Theobroma Cacao. Nat. Prod. Chem. Res. 2014, 2, 4. [Google Scholar] [CrossRef]
- Sun, Q.L.; Hua, S.; Ye, J.H.; Zheng, X.Q.; Liang, Y.R. Flavonoids and Volatiles in Chrysanthemum Morifolium Ramat Flower from Tongxiang County in China. Afr. J. Biotechnol. 2010, 9, 1684–5315. [Google Scholar]
- Mari, A.; Lyon, D.; Fragner, L.; Montoro, P.; Piacente, S.; Wienkoop, S.; Egelhofer, V.; Weckwerth, W. Phytochemical Composition of Potentilla anserina L. Analyzed by an Integrative GC-MS and LC-MS Metabolomics Platform. Metabolomics 2013, 9, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Malar, J.; Vanmathi, J.; Chairman, K. Phytochemical Analysis of Lepidium sativum using UV-VIS and GC-MS. Int. J. Adv. Res. 2018, 6, 813–825. [Google Scholar] [CrossRef]
- González, J.; Gómez, E.; Pérez, J.C.; Monan, M.; François-Haugrin, F. Chemical Profile of the Leaves from Morus alba L. Using GC-MS. Acta Sci. Nutr. Health 2022, 6, 141–143. [Google Scholar] [CrossRef]
- Khan, H.J.; Ahmad, M.K.; Khan, A.R.; Rastogi, N.; Ansari, J.A.; Fatima, N.; Satyanarayan, G.N.V.; Mahdi, A.A. GC-MS/MS Based Identification of Bioactive Principles of Chloroform Fraction of Swertia Chirayatia (Chirata). Orient. J. Chem. 2016, 32, 875–883. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, T.; Wang, D.; Zhang, L.; Chen, G. Study on the Volatile Profile Characteristics of Oyster Crassostrea Gigas during Storage by a Combination Sampling Method Coupled with GC/MS. Food Chem. 2009, 115, 1150–1157. [Google Scholar] [CrossRef]
- Prakash, N.K.U.; Sripriya, N.S.; Raj, D.D.; Deepa, S.; Bhuvaneswari, S. Antioxidant Potency and GC-MS Composition of Origanum majorana Linn. Pak. J. Pharm. Sci. 2019, 32, 2117–2122. [Google Scholar]
- Shahmohamadi, R.; Sariri, R.; Rasa, M.; Ghafoori, H.; Aghamali, M.; Nasuti, S.; Tahery, M. Chemical Composition and Antimicrobial Activity of Flowering Aerial Parts Mentha Pulegium from Gilan. Pharmacologyonline 2011, 3, 651–659. [Google Scholar]
- Samejo, M.Q.; Memon, S.; Bhanger, M.I.; Khan, K.M. Comparison of Chemical Composition of Aerva Javanica Seed Essential Oils Obtained by Different Extraction Methods. Pak. J. Pharm. Sci. 2013, 26, 757–760. [Google Scholar]
- Pino, J.A.; Mesa, J.; Muñoz, Y.; Martí, M.P.; Marbot, R. Volatile Components from Mango (Mangifera indica L.) Cultivars. J. Agric. Food Chem. 2005, 53, 2213–2223. [Google Scholar] [CrossRef]
- Qadir, A.; Aqil, M.; Ali, A.; Ahmad, F.J.; Ahmad, S.; Arif, M.; Khan, N. GC-MS Analysis of the Methanolic Extracts of Smilax China and Salix Alba and Their Antioxidant Activity. Turkish J. Chem. 2020, 44, 352–363. [Google Scholar] [CrossRef]
- Bhalla, N.; Ingle, N.; Patri, S.V.; Haranath, D. Phytochemical Analysis of Moringa Oleifera Leaves Extracts by GC-MS and Free Radical Scavenging Potency for Industrial Applications. Saudi J. Biol. Sci. 2021, 28, 6915–6928. [Google Scholar] [CrossRef] [PubMed]
- Yasheshwar; Umar, S.; Sharma, M.P.; Khan, W.; Ahmad, S. Variation in Ornamental Traits, Physiological Responses of Tagetes erecta L. and T. patula L. in Relation to Antioxidant and Metabolic Profile under Deficit Irrigation Strategies. Sci. Hortic. 2017, 214, 200–208. [Google Scholar] [CrossRef]
- Renda, G.; Tosun, G.; Yaylı, N. SPME GC/MS Analysis of Three ornithogalum L. Species from Turkey. Rec. Nat. Prod. 2016, 10, 497. [Google Scholar]
- Mishra, D.; Patnaik, S. GC-MS Analysed Phyto-Chemicals and Antibacterial Activity of Withania somnifera (L.) Dunal Extract in the Context of Treatment to Liver Cirrhosis. Biomed. Pharmacol. J. 2020, 13, 71–78. [Google Scholar] [CrossRef]
- Lee, K.M.; Jeon, J.Y.; Lee, B.J.; Lee, H.; Choi, H.K. Application of Metabolomics to Quality Control of Natural Product Derived Medicines. Biomol. Ther. 2017, 25, 559–568. [Google Scholar] [CrossRef]
- Uduman, M.S.T.S.; Rathinam, P.; Karuru, Y.; Obili, G.; Chakka, G.; Janakiraman, A.K. GC-MS Analysis of Ethyl Acetate Extract of Whole Plant of Rostellularia Diffusa. Pharmacogn. J. 2017, 9, 70–72. [Google Scholar] [CrossRef]
- Liang, Z.; Li, K.; Wang, X.; Ke, Y.; Jin, Y.; Liang, X. Combination of Off-Line Two-Dimensional Hydrophilic Interaction Liquid Chromatography for Polar Fraction and Two-Dimensional Hydrophilic Interaction Liquid Chromatography×reversed-Phase Liquid Chromatography for Medium-Polar Fraction in a Traditional Chin. J. Chromatogr. A 2012, 1224, 61–69. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, X.; Xu, G. Development of a Comprehensive Two-Dimensional Hydrophilic Interaction Chromatography/Quadrupole Time-of-Flight Mass Spectrometry System and Its Application in Separation and Identification of Saponins from Quillaja Saponaria. J. Chromatogr. A 2008, 1181, 51–59. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, Z.; Feng, J.; Xue, X.; Zhang, F.; Xu, Q.; Liang, X. Development of Orthogonal Two-Dimensional Hydrophilic Interaction Chromatography Systems with the Introduction of Novel Stationary Phases. J. Sep. Sci. 2009, 32, 2871–2876. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, X.; Xu, G. Simultaneous Separation of Hydrophilic and Hydrophobic Compounds by Using an Online HILIC-RPLC System with Two Detectors. J. Sep. Sci. 2008, 31, 1564–1572. [Google Scholar] [CrossRef]
- Sentkowska, A.; Biesaga, M.; Pyrzynska, K. Application of Hydrophilic Interaction Liquid Chromatography for the Quantification of Flavonoids in Genista Tinctoria Extract. J. Anal. Methods Chem. 2016, 2016, 3789348. [Google Scholar] [CrossRef] [PubMed]
- Binsuwaidan, R.; Elekhnawy, E.; Elseady, W.S.; Keshk, W.A.; Shoeib, N.A.; Attallah, N.G.M.; Mokhtar, F.A.; Abd El Hadi, S.R.; Ahmed, E.; Magdeldin, S.; et al. Antibacterial Activity and Wound Healing Potential of Cycas Thouarsii R.Br n-Butanol Fraction in Diabetic Rats Supported with Phytochemical Profiling. Biomed. Pharmacother. 2022, 155, 113763. [Google Scholar] [CrossRef] [PubMed]
- Gholais, N.S.; Shi, C.; Zhang, J.; Liao, B.; Albarmaqi, R.A.; Tang, X.; Mi, L. Network Pharmacology-Based Investigation on the Mechanism of the JinGuanLan Formula in Treating Acne Vulgaris. Evid.-Based Complement. Altern. Med. 2022, 2022, 6944792. [Google Scholar] [CrossRef] [PubMed]
- Que, W.; Chen, M.; Yang, L.; Zhang, B.; Zhao, Z.; Liu, M.; Cheng, Y.; Qiu, H. A Network Pharmacology-Based Investigation on the Bioactive Ingredients and Molecular Mechanisms of Gelsemium Elegans Benth against Colorectal Cancer. BMC Complement. Med. Ther. 2021, 21, 99. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Li, L.; Liang, C.; Peng, L. Network Pharmacology-Based Investigation of the Therapeutic Mechanisms of Action of Danning Tablets in Nonalcoholic Fatty Liver Disease. Evid.-Based Complement. Altern. Med. 2021, 2021, 3495360. [Google Scholar] [CrossRef]
- Piao, C.; Sun, Z.; Jin, D.; Wang, H.; Wu, X.; Zhang, N.; Lian, F.; Tong, X. Network Pharmacology-Based Investigation of the Underlying Mechanism of Panax Notoginseng Treatment of Diabetic Retinopathy. Comb. Chem. High Throughput Screen. 2020, 23, 334–344. [Google Scholar] [CrossRef]
- Dong, Y.; Zhao, Q.; Wang, Y. Network Pharmacology-Based Investigation of Potential Targets of Astragalus Membranaceous-Angelica Sinensis Compound Acting on Diabetic Nephropathy. Sci. Rep. 2021, 11, 19496. [Google Scholar] [CrossRef]
- Sakle, N.S.; More, S.A.; Mokale, S.N. A Network Pharmacology-Based Approach to Explore Potential Targets of Caesalpinia Pulcherima: An Updated Prototype in Drug Discovery. Sci. Rep. 2020, 10, 17217. [Google Scholar] [CrossRef]
- Chen, K.; Deng, Y.; Shang, S.; Li, P.; Liu, L.; Chen, X. Network Pharmacology-Based Investigation of the Molecular Mechanisms of the Chinese Herbal Formula Shenyi in the Treatment of Diabetic Nephropathy. Front. Med. 2022, 9, 898624. [Google Scholar] [CrossRef]
- Alqahtani, M.J.; Mostafa, S.A.; Hussein, I.A.; Elhawary, S.; Mokhtar, F.A.; Albogami, S.; Tomczyk, M.; Batiha, G.E.S.; Negm, W.A. Metabolic Profiling of Jasminum Grandiflorum L. Flowers and Protective Role against Cisplatin-Induced Nephrotoxicity: Network Pharmacology and In Vivo Validation. Metabolites 2022, 12, 792. [Google Scholar] [CrossRef]
- Doke, S.K.; Dhawale, S.C. Alternatives to Animal Testing: A Review. Saudi Pharm. J. 2015, 23, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Schaduangrat, N.; Lampa, S.; Simeon, S.; Gleeson, M.P.; Spjuth, O.; Nantasenamat, C. Towards Reproducible Computational Drug Discovery. J. Cheminform. 2020, 12, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Moudgil, M.; Mandal, S.K. Rational Drug Design. Eur. J. Pharmacol. 2009, 625, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Pei, J.; Lai, L. Computational Multitarget Drug Design. J. Chem. Inf. Model. 2017, 57, 403–412. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, B.; Yadav, D.K. Metabolomics and Network Pharmacology in the Exploration of the Multi-Targeted Therapeutic Approach of Traditional Medicinal Plants. Plants 2022, 11, 3243. https://doi.org/10.3390/plants11233243
Sharma B, Yadav DK. Metabolomics and Network Pharmacology in the Exploration of the Multi-Targeted Therapeutic Approach of Traditional Medicinal Plants. Plants. 2022; 11(23):3243. https://doi.org/10.3390/plants11233243
Chicago/Turabian StyleSharma, Bharti, and Dinesh Kumar Yadav. 2022. "Metabolomics and Network Pharmacology in the Exploration of the Multi-Targeted Therapeutic Approach of Traditional Medicinal Plants" Plants 11, no. 23: 3243. https://doi.org/10.3390/plants11233243
APA StyleSharma, B., & Yadav, D. K. (2022). Metabolomics and Network Pharmacology in the Exploration of the Multi-Targeted Therapeutic Approach of Traditional Medicinal Plants. Plants, 11(23), 3243. https://doi.org/10.3390/plants11233243





